Submitted:
28 June 2024
Posted:
02 July 2024
You are already at the latest version
Abstract

Keywords:
1. Introduction
2. Materials and Methods
2.1. Animal Studies
2.2. Whole Transcriptomic mRNA Sequencing:
2.3. Single Cell Isolation
2.4. Single Cells Sequencing and Library Prep Protocol
2.5. Analysis of scRNAseq data
2.6. SoupX Analysis
2.7. Seurat Analysis
2.8. QC and Selecting Cells for Further Analysis
2.9. Unsupervised Cell Cluster Segregation and Cluster Identification
2.10. GTEX Data Analysis
2.11. Gene Set Enrichment Analysis
3. Results
3.1. Transcriptomic Changes in Pparg-/-epi Mice Shows a Strong Correlation to Cancer and Inflammatory Cell Recruitment and Mobilization
| Table 1A: Top 10 Disease or Biofunction terms that match with the DEG dataset from Pparg-/-epi mouse skin relative to WT skin (sorted by p-value) | ||
|---|---|---|
| Diseases or Functions Annotation | p-value | z-score |
| Non-hematological solid tumor | 1.87E-66 | 0.316 |
| Epithelial neoplasm | 1.71E-65 | 0.042 |
| Non-melanoma solid tumor | 1.93E-65 | -0.047 |
| Tumorigenesis of tissue | 2.76E-65 | -0.252 |
| Nonhematologic malignant neoplasm | 1.27E-64 | 0.295 |
| Carcinoma | 3.19E-63 | 0.192 |
| Solid tumor | 1.03E-58 | 0.869 |
| Malignant solid tumor | 2.09E-58 | 0.252 |
| Cancer | 1.06E-57 | 1.516 |
| Head and neck tumor | 4.16E-57 | -0.124 |
| Table 1B: Top 20 Disease or Biofunction terms that match with the DEG dataset from Pparg-/-epi mouse skin relative to WT skin (sorted by z-score) | ||
| Diseases or Functions Annotation | p-value | Activation z-score |
| Chemotaxis of leukocytes | 1.61E-12 | 5.052 |
| Homing of leukocytes | 8.69E-14 | 4.724 |
| Cell movement of tumor cell lines | 3.36E-11 | 4.686 |
| Chemotaxis | 2.68E-18 | 4.644 |
| Migration of cells | 1.13E-30 | 4.623 |
| Homing of blood cells | 4.72E-14 | 4.574 |
| Cell survival | 2.9E-15 | 4.568 |
| Leukocyte migration | 2.98E-29 | 4.468 |
| Cell movement | 2.97E-37 | 4.464 |
| Cell movement of blood cells | 1.82E-29 | 4.36 |
| Invasion of cells | 9.29E-15 | 4.359 |
| Homing of cells | 3.91E-21 | 4.249 |
| Inflammatory response | 9.14E-20 | 4.174 |
| Cell movement of myeloid cells | 2.13E-18 | 4.171 |
| Recruitment of myeloid cells | 1.19E-13 | 4.121 |
| Cell viability | 1.77E-14 | 4.086 |
| Recruitment of blood cells | 3.44E-19 | 4.07 |
| Cell movement of phagocytes | 5.77E-19 | 4.029 |
| Growth of lesion | 5.78E-30 | 3.992 |
| Growth of tumor | 1.15E-29 | 3.942 |
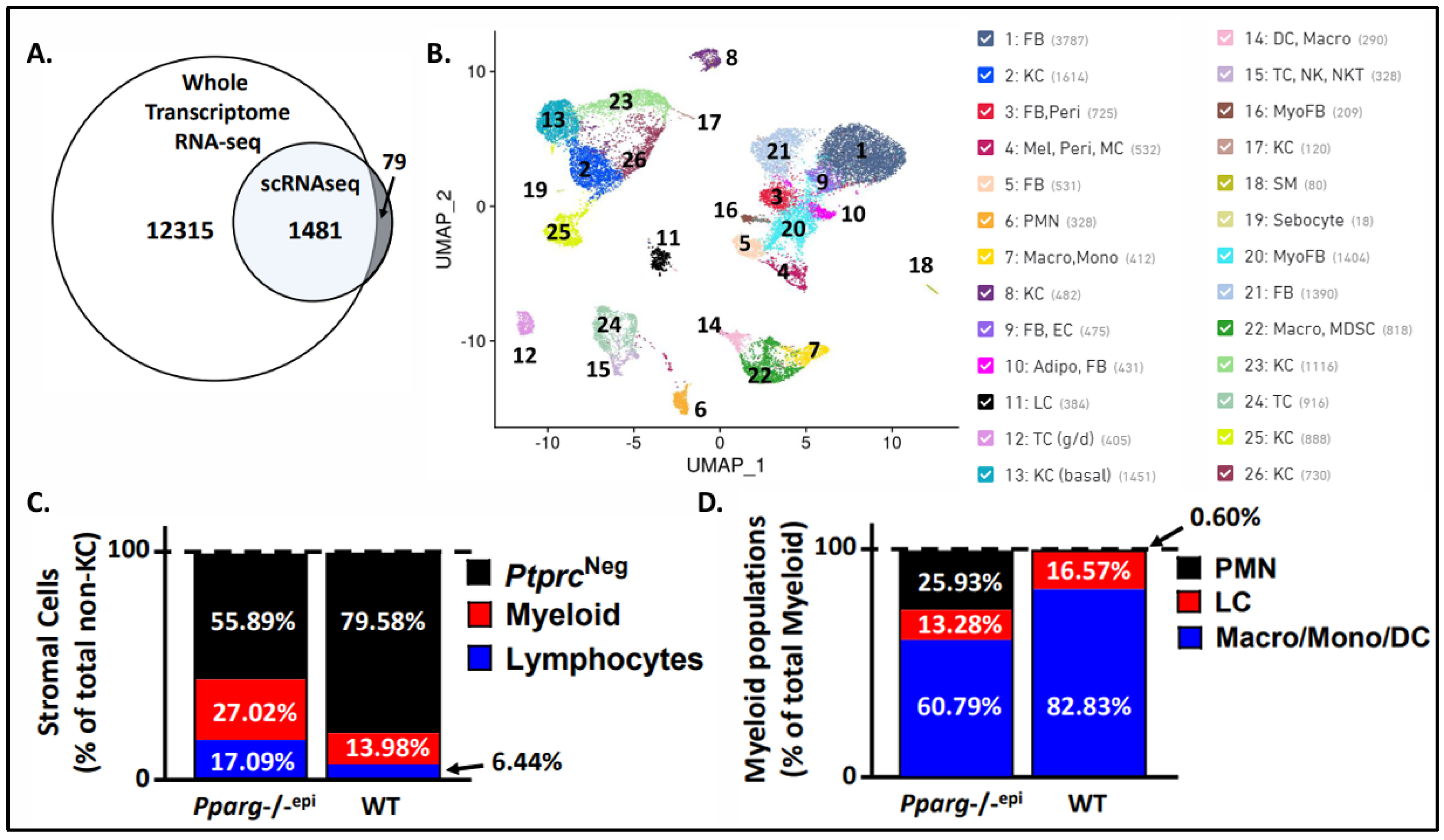
3.2. Single Cell Sequencing of Pparg-/-epi Mice Reveals an Increase in Immune Cells, Particularly Neutrophils
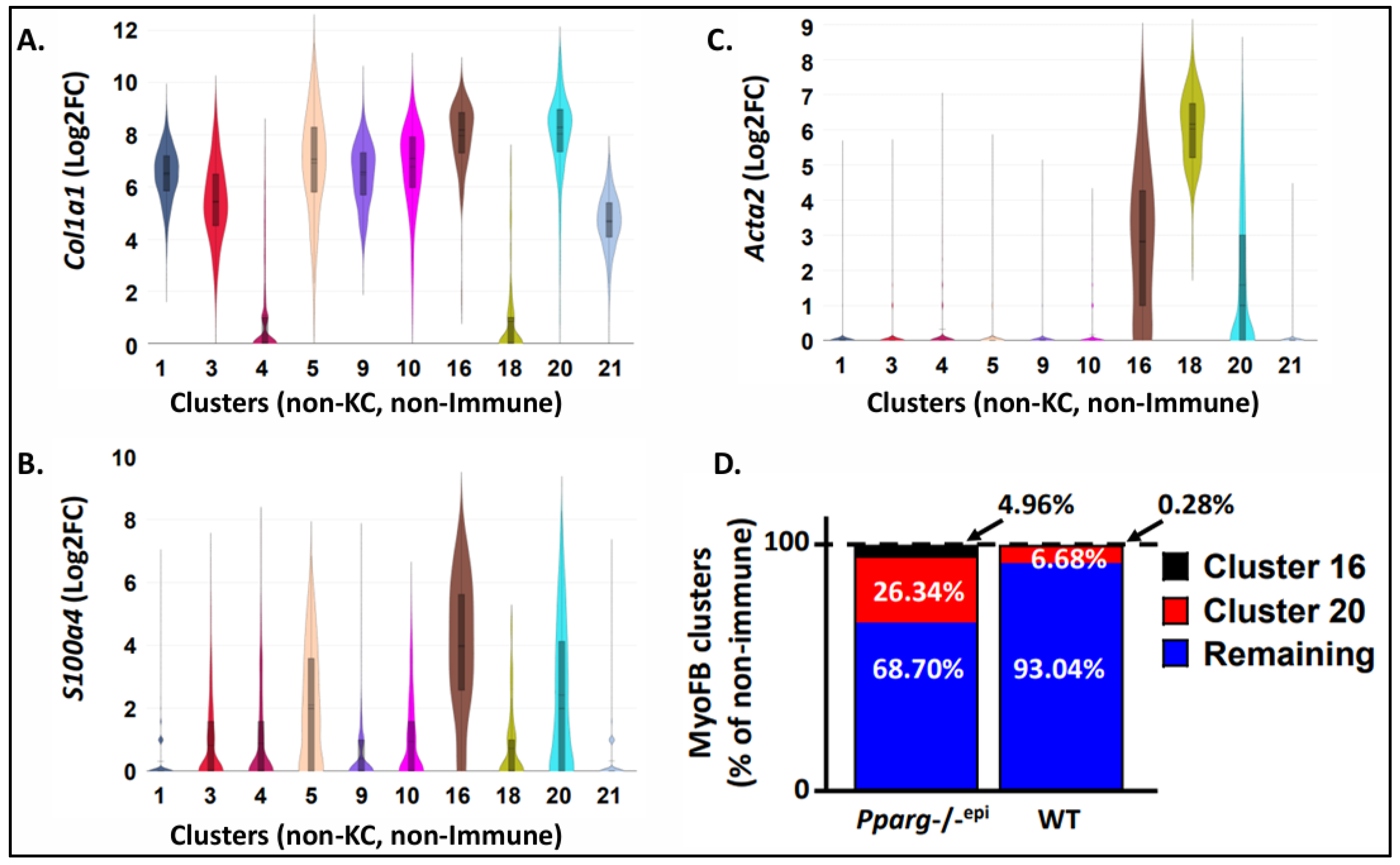
3.3. Fibroblasts Expressing Myofibroblast Markers are Increased in Pparg-/-epi Mouse Skin
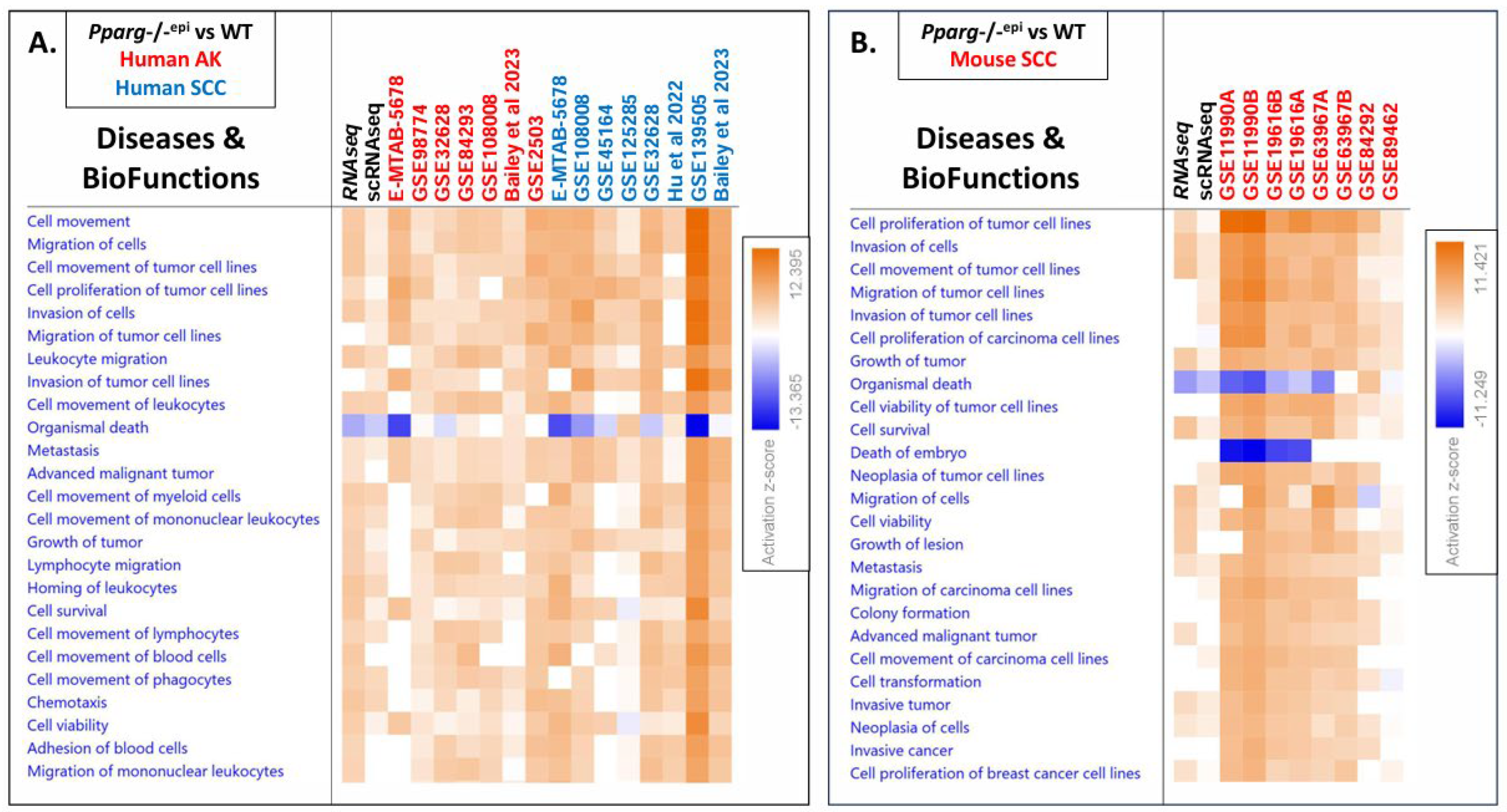
3.4. GSE Analysis Reveals Strong Similarity between the Pparg-/-epi Transcriptomic Data and that of Human Actinic Disease and Mouse and Human SCC
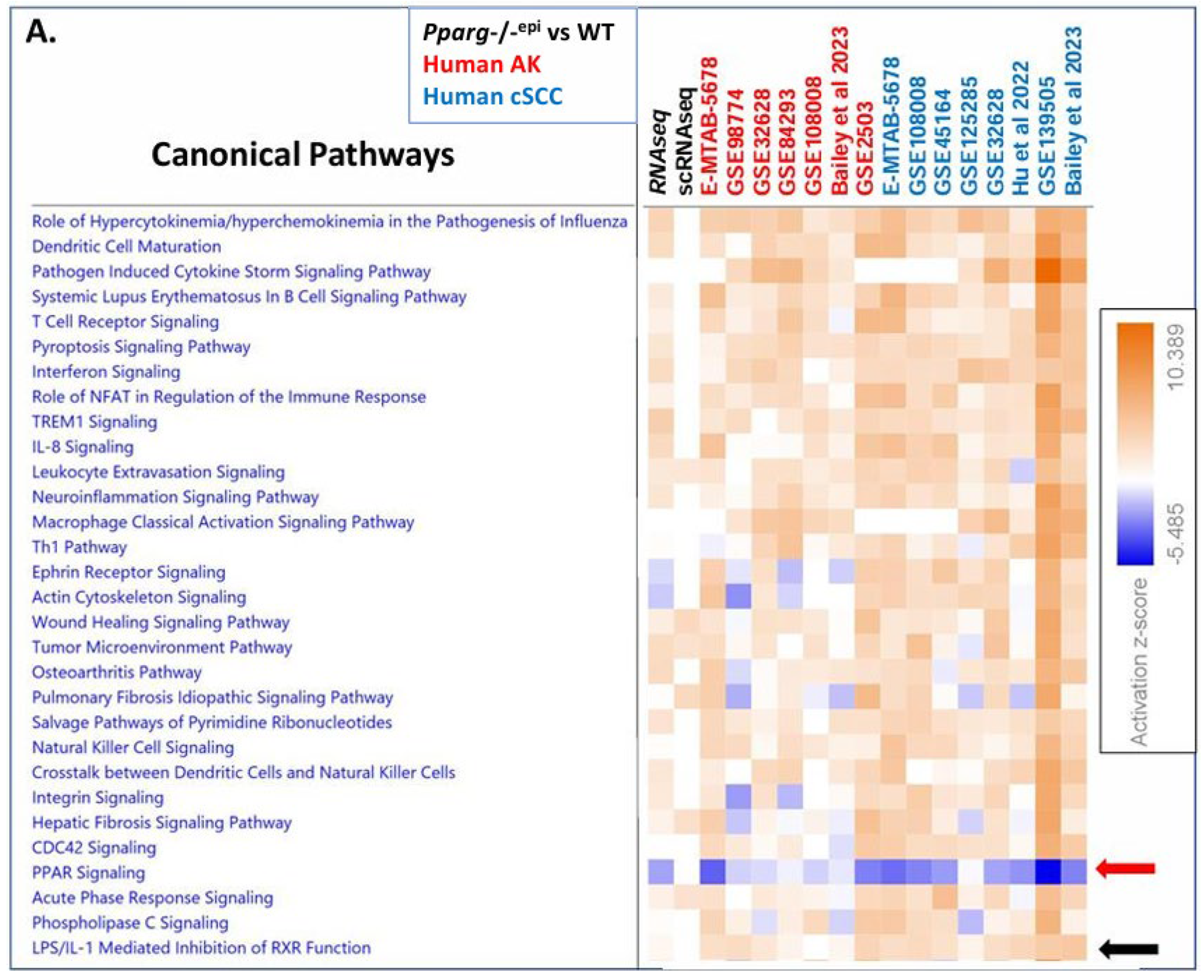
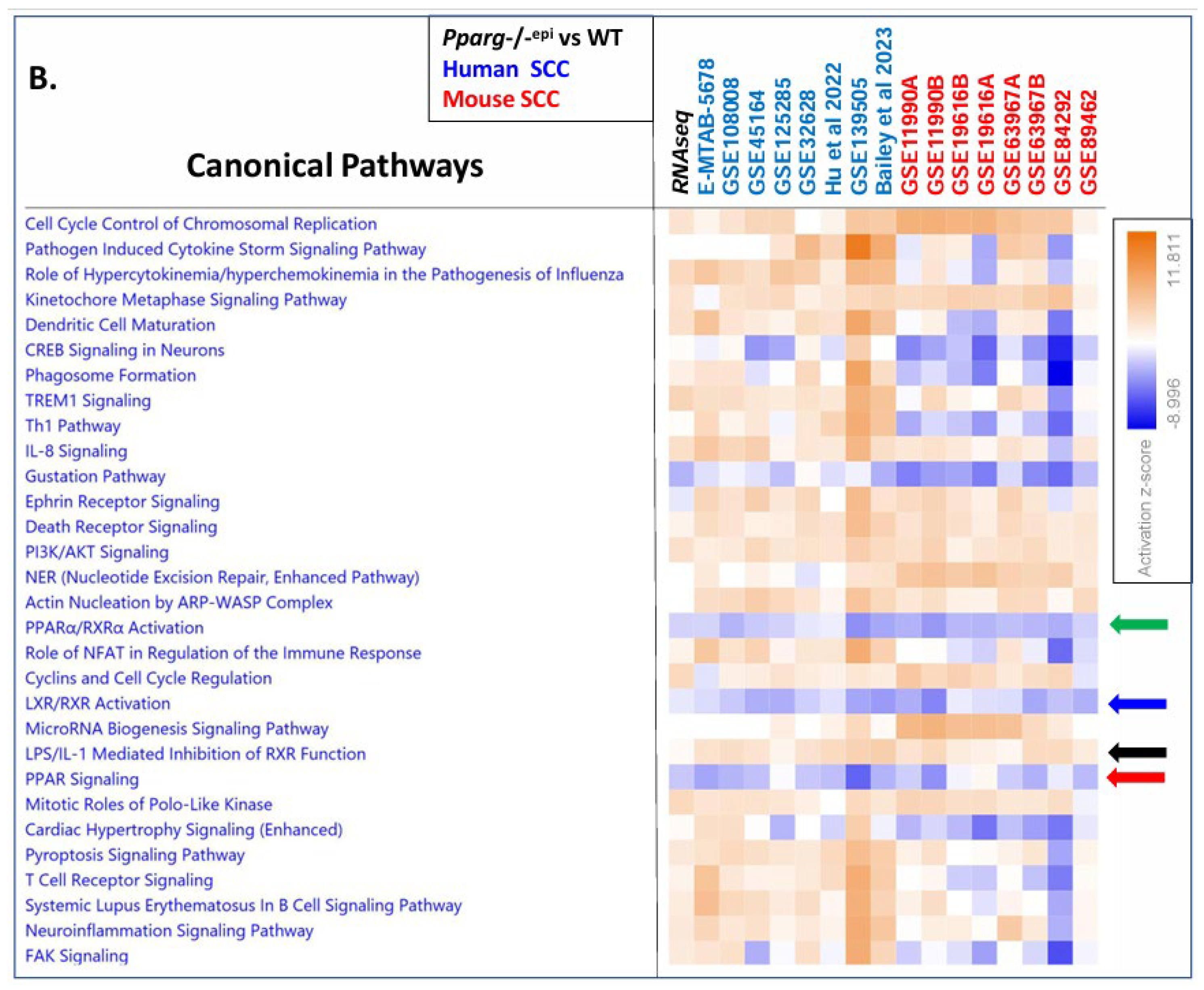
3.5. Canonical Pathway Analysis Shows Loss of PPAR Signaling is a Top Inhibited Canonical Pathway in Human AKs, Human SCCs and Mouse SCCs
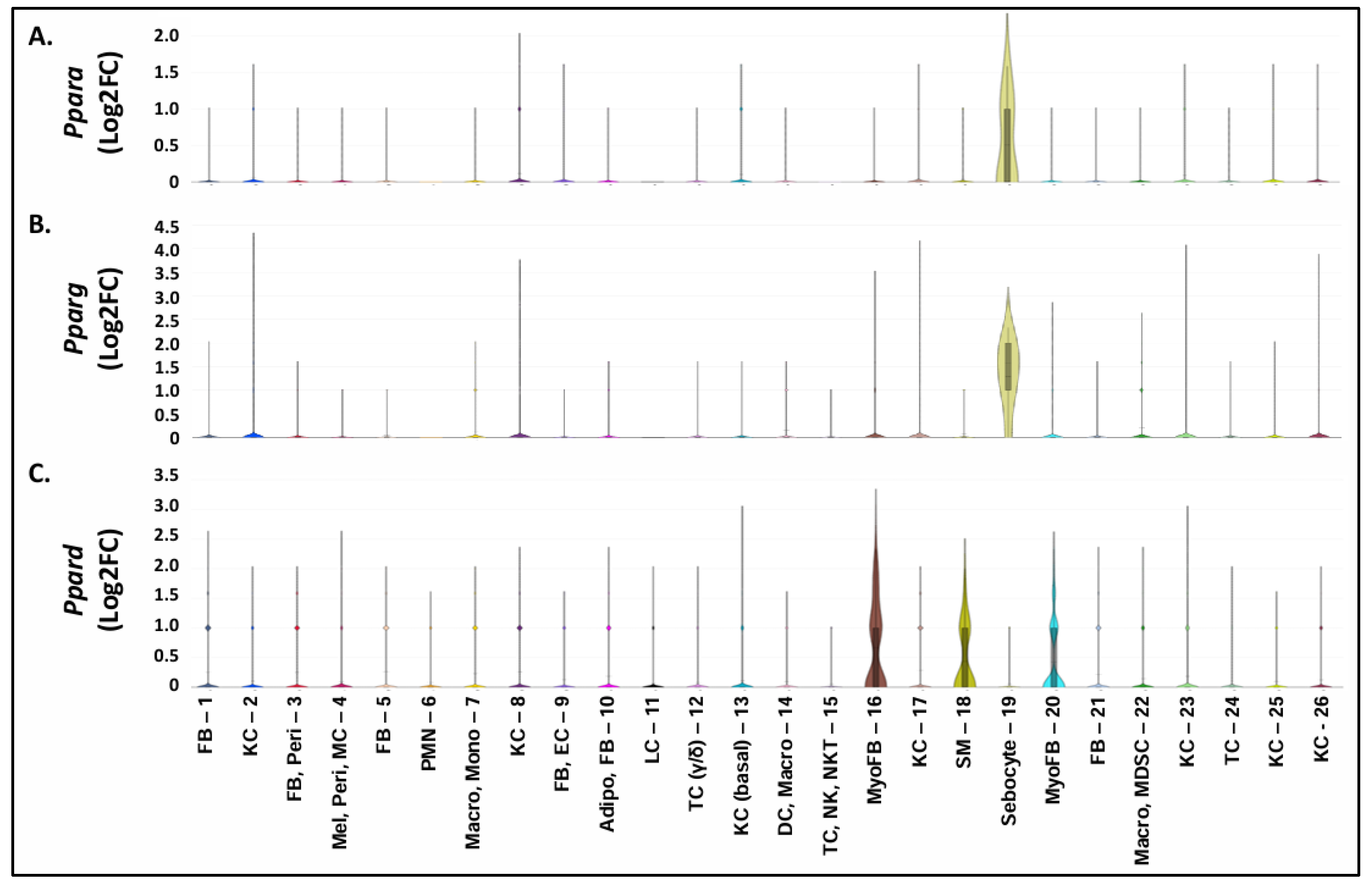
3.6. A Shift to Reduced PPAR and RXR Activity and Reduced PPARA/Ppara & PPARG/Pparg Expression but Increased PPARD/Ppard Expression Occurs during the Progression from Sun-Exposed skin to NMSC
3.7. Increased PPARD Expression Represents a Marker of Myofibroblast Populations Found in Pparg-/-epi Skin
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Germain, P.; Staels, B.; Dacquet, C.; Spedding, M.; Laudet, V. Overview of Nomenclature of Nuclear Receptors. Pharmacol Rev 2006, 58, 685–704. [Google Scholar] [CrossRef] [PubMed]
- Pirat, C.; Farce, A.; Lebègue, N.; Renault, N.; Furman, C.; Millet, R.; Yous, S.d.; Speca, S.; Berthelot, P.; Desreumaux, P. , et al. Targeting Peroxisome Proliferator-Activated Receptors (PPARs): Development of Modulators. J Med Chem 2012, 55, 4027–4061. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.D.; Wagner, N. Peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta) acts as regulator of metabolism linked to multiple cellular functions. Pharmacol Ther 2010, 125, 423–435. [Google Scholar] [CrossRef] [PubMed]
- Schmuth, M.; Moosbrugger-Martinz, V.; Blunder, S.; Dubrac, S. Role of PPAR, LXR, and PXR in epidermal homeostasis and inflammation. Biochim Biophys Acta - Mol Cell Biol Lipids 2014, 1841, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Pascual, G.; Fong, A.L.; Ogawa, S.; Gamliel, A.; Li, A.C.; Perissi, V.; Rose, D.W.; Willson, T.M.; Rosenfeld, M.G.; Glass, C.K. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-[gamma]. Nature 2005, 437, 759–763. [Google Scholar] [CrossRef] [PubMed]
- Konger, R.L.; Derr-Yellin, E.; Zimmers, T.A.; Katona, T.; Xuei, X.; Liu, Y.; Zhou, H.-M.; Simpson, E.R.; Turner, M.J. Epidermal PPARγ Is a Key Homeostatic Regulator of Cutaneous Inflammation and Barrier Function in Mouse Skin. Int J Mol Sci 2021, 22, 8634. [Google Scholar] [CrossRef] [PubMed]
- Mahgoub, D.; El Tawdy, A.M.; Metwally, D.; Manar, A.; Rashed, L. Estimation of peroxisomal proliferators-activated receptor g gene expression in inflammatory skin diseases: Atopic dermititis and psoriasis. Our Dermatol Online 2014, 5, 107–112. [Google Scholar] [CrossRef]
- Karnik, P.; Tekeste, Z.; McCormick, T.S.; Gilliam, A.C.; Price, V.H.; Cooper, K.D.; Mirmirani, P. Hair Follicle Stem Cell-Specific PPAR[gamma] Deletion Causes Scarring Alopecia. J Invest Dermatol 2008, 129, 1243–1257. [Google Scholar] [CrossRef] [PubMed]
- Sahu, R.P.; DaSilva, S.C.; Rashid, B.; Martel, K.C.; Jernigan, D.; Mehta, S.R.; Mohamed, D.R.; Rezania, S.; Bradish, J.R.; Armstrong, A.B. , et al. Mice lacking epidermal PPARγ exhibit a marked augmentation in photocarcinogenesis associated with increased UVB-induced apoptosis, inflammation and barrier dysfunction. Int J Cancer 2012, 131, E1055–E1066. [Google Scholar] [CrossRef]
- Konger, R.L.; Derr-Yellin, E.; Travers, J.B.; Ocana, J.A.; Sahu, R.P. Epidermal PPARg influences subcutaneous tumor growth and acts through TNF-a to regulate contact hypersensitivity and the acute photoresponse. Oncotarget 2017, 8, 98184–98199. [Google Scholar] [CrossRef]
- Indra, A.K.; Castaneda, E.; Antal, M.C.; Jiang, M.; Messaddeq, N.; Meng, X.; Loehr, C.V.; Gariglio, P.; Kato, S.; Wahli, W. , et al. Malignant transformation of DMBA/TPA-induced papillomas and nevi in the skin of mice selectively lacking retinoid-X-receptor alpha in epidermal keratinocytes. J Invest Dermatol 2007, 127, 1250–1260. [Google Scholar] [CrossRef] [PubMed]
- Nicol, C.J.; Yoon, M.; Ward, J.M.; Yamashita, M.; Fukamachi, K.; Peters, J.M.; Gonzalez, F.J. PPARgamma influences susceptibility to DMBA-induced mammary, ovarian and skin carcinogenesis. Carcinogenesis 2004, 25, 1747–1755. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Konger, R.L. Evidence that peroxisome proliferator-activated receptor γ suppresses squamous carcinogenesis through anti-inflammatory signaling and regulation of the immune response. Mol Carcinog 2019, 58, 1589–1601. [Google Scholar] [CrossRef] [PubMed]
- Konger, R.L.; Derr-Yellin, E.; Ermatov, N.; Ren, L.; Sahu, R.P. The PPARγ Agonist Rosiglitazone Suppresses Syngeneic Mouse SCC (Squamous Cell Carcinoma) Tumor Growth through an Immune-Mediated Mechanism. Molecules (Basel) 2019, 24, 2192. [Google Scholar] [CrossRef] [PubMed]
- Goyal, G.; Wong, K.; Nirschl, C.J.; Souders, N.; Neuberg, D.; Anandasabapathy, N.; Dranoff, G. PPARγ Contributes to Immunity Induced by Cancer Cell Vaccines That Secrete GM-CSF. Cancer Immunol Res 2018, 6, 723–732. [Google Scholar] [CrossRef] [PubMed]
- Ritter, B.; Greten, F.R. Modulating inflammation for cancer therapy. J Exp Med 2019, 216, 1234–1243. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.-H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A. , et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Behjati, S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience 2020, 9. [Google Scholar] [CrossRef]
- Satija, R.; Farrell, J.A.; Gennert, D.; Schier, A.F.; Regev, A. Spatial reconstruction of single-cell gene expression data. Nature Biotechnology 2015, 33, 495–502. [Google Scholar] [CrossRef]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Stuart, T.; Butler, A.; Hoffman, P.; Hafemeister, C.; Papalexi, E.; Mauck, W.M., III; Hao, Y.; Stoeckius, M.; Smibert, P.; Satija, R. Comprehensive Integration of Single-Cell Data. Cell 2019, 177, 1888–1902. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M., III; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M. , et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, D.J.; Campbell, K.R.; Lun, A.T.; Wills, Q.F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 2017, 33, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma'ayan, A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf 2013, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A. , et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016, 44, W90–97. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M. , et al. Gene Set Knowledge Discovery with Enrichr. Curr Protoc 2021, 1, e90. [Google Scholar] [CrossRef] [PubMed]
- Franzén, O.; Gan, L.-M.; Björkegren, J.L.M. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database 2019, 2019, baz046. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Andrade, J. DEApp: an interactive web interface for differential expression analysis of next generation sequence data. Source Code Biol Med 2017, 12, 2. [Google Scholar] [CrossRef] [PubMed]
- Krämer, A.; Green, J.; Pollard, J., Jr.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- Qie, C.; Jiang, J.; Liu, W.; Hu, X.; Chen, W.; Xie, X.; Liu, J. Single-cell RNA-Seq reveals the transcriptional landscape and heterogeneity of skin macrophages in Vsir(-/-) murine psoriasis. Theranostics 2020, 10, 10483–10497. [Google Scholar] [CrossRef]
- Apte, M.V.; Pirola, R.C.; Wilson, J.S. Pancreatic stellate cells: a starring role in normal and diseased pancreas. Front Physiol 2012, 3, 344. [Google Scholar] [CrossRef] [PubMed]
- McAndrews, K.M.; Miyake, T.; Ehsanipour, E.A.; Kelly, P.J.; Becker, L.M.; McGrail, D.J.; Sugimoto, H.; LeBleu, V.S.; Ge, Y.; Kalluri, R. Dermal aSMA+ myofibroblasts orchestrate skin wound repair via b1 integrin and independent of type I collagen production. EMBO J 2022, 41, e109470. [Google Scholar] [CrossRef] [PubMed]
- Cataldi, S.; Aprile, M.; Melillo, D.; Mucel, I.; Giorgetti-Peraldi, S.; Cormont, M.; Italiani, P.; Blüher, M.; Tanti, J.F.; Ciccodicola, A. , et al. TNFα Mediates Inflammation-Induced Effects on PPARG Splicing in Adipose Tissue and Mesenchymal Precursor Cells. Cells 2021, 11, 42. [Google Scholar] [CrossRef] [PubMed]
- Kita, R.; Fraser, H.B. Local Adaptation of Sun-Exposure-Dependent Gene Expression Regulation in Human Skin. PLOS Genet 2016, 12, e1006382. [Google Scholar] [CrossRef] [PubMed]
- Zou, D.-D.; Xu, D.; Deng, Y.-Y.; Wu, W.-J.; Zhang, J.; Huang, L.; He, L. Identification of key genes in cutaneous squamous cell carcinoma: a transcriptome sequencing and bioinformatics profiling study. Ann Transl Med 2021, 9, 1497. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, H.; Suárez-Fariñas, M.; Gulati, N.; Shah, K.R.; Cannizzaro, M.V.; Coats, I.; Felsen, D.; Krueger, J.G.; Carucci, J.A. Gene expression profiling of the leading edge of cutaneous squamous cell carcinoma: IL-24-driven MMP-7. J Invest Dermatol 2014, 134, 1418–1427. [Google Scholar] [CrossRef] [PubMed]
- Downie, M.M.; Sanders, D.A.; Maier, L.M.; Stock, D.M.; Kealey, T. Peroxisome proliferator-activated receptor and farnesoid X receptor ligands differentially regulate sebaceous differentiation in human sebaceous gland organ cultures in vitro. Br J Dermatol 2004, 151, 766–775. [Google Scholar] [CrossRef] [PubMed]
- Butti, R.; Nimma, R.; Kundu, G.; Bulbule, A.; Kumar, T.V.S.; Gunasekaran, V.P.; Tomar, D.; Kumar, D.; Mane, A.; Gill, S.S. , et al. Tumor-derived osteopontin drives the resident fibroblast to myofibroblast differentiation through Twist1 to promote breast cancer progression. Oncogene 2021, 40, 2002–2017. [Google Scholar] [CrossRef]
- Hansson, B.; Rippe, C.; Kotowska, D.; Wasserstrom, S.; Säll, J.; Göransson, O.; Swärd, K.; Stenkula, K. Rosiglitazone drives cavin-2/SDPR expression in adipocytes in a CEBPa-dependent manner. PLoS ONE 2017, 12, e0173412. [Google Scholar] [CrossRef]
- Cawthorn, W.P.; Scheller, E.L.; MacDougald, O.A. Adipose tissue stem cells meet preadipocyte commitment: going back to the future. J Lipid Res 2012, 53, 227–246. [Google Scholar] [CrossRef]
- Bochet, L.; Lehuédé, C.; Dauvillier, S.; Wang, Y.Y.; Dirat, B.; Laurent, V.; Dray, C.; Guiet, R.; Maridonneau-Parini, I.; Le Gonidec, S. , et al. Adipocyte-Derived Fibroblasts Promote Tumor Progression and Contribute to the Desmoplastic Reaction in Breast Cancer. Cancer Res 2013, 73, 5657–5668. [Google Scholar] [CrossRef] [PubMed]
- Costa-Guda, J.; Rosen, E.D.; Jensen, R.T.; Chung, D.C.; Arnold, A. Mutational analysis of PPARG as a candidate tumour suppressor gene in enteropancreatic endocrine tumours. Clin Endocrinol 2005, 62, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Ashton, K.J.; Weinstein, S.R.; Maguire, D.J.; Griffiths, L.R. Chromosomal Aberrations in Squamous Cell Carcinoma and Solar Keratoses Revealed by Comparative Genomic Hybridization. Arch Dermatol 2003, 139, 876–882. [Google Scholar] [CrossRef]
- Ashton, K.J.; Carless, M.A.; Griffiths, L.R. Cytogenetic alterations in nonmelanoma skin cancer: A review. Genes, Chromosomes and Cancer 2005, 43, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Popp, S.; Waltering, S.; Herbst, C.; Moll, I.; Boukamp, P. UV-B-type mutations and chromosomal imbalances indicate common pathways for the development of Merkel and skin squamous cell carcinomas. Int J Cancer 2002, 99, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Maestro, R.; Gasparotto, D.; Vukosavljevic, T.; Barzan, L.; Sulfaro, S.; Boiocchi, M. Three Discrete Regions of Deletion at 3p in Head and Neck Cancers. Cancer Res 1993, 53, 5775–5779. [Google Scholar] [PubMed]
- Yoo, W.J.; Cho, S.H.; Lee, Y.S.; Park, G.S.; Kim, M.S.; Kim, B.K.; Park, W.S.; Lee, J.Y.; Kang, C.S. Loss of heterozygosity on chromosomes 3p,8p,9p and 17p in the progression of squamous cell carcinoma of the larynx. J Korean Med Sci 2004, 19, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Rehman, I.; Takata, M.; Wu, Y.Y.; Rees, J.L. Genetic change in actinic keratoses. Oncogene 1996, 12, 2483–2490. [Google Scholar]
- Quinn, A.G.; Sikkink, S.; Rees, J.L. Basal Cell Carcinomas and Squamous Cell Carcinomas of Human Skin Show Distinct Patterns of Chromosome Loss1. Cancer Res 1994, 54, 4756–4759. [Google Scholar]
- Ikezoe, T.; Miller, C.W.; Kawano, S.; Heaney, A.; Williamson, E.A.; Hisatake, J.; Green, E.; Hofmann, W.; Taguchi, H.; Koeffler, H.P. Mutational analysis of the peroxisome proliferator-activated receptor gamma gene in human malignancies. Cancer Res 2001, 61, 5307–5310. [Google Scholar]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J. , et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017, 9, 34. [Google Scholar] [CrossRef]
- Aprile, M.; Ambrosio, M.R.; D'Esposito, V.; Beguinot, F.; Formisano, P.; Costa, V.; Ciccodicola, A. PPARG in Human Adipogenesis: Differential Contribution of Canonical Transcripts and Dominant Negative Isoforms. PPAR Res 2014, 2014, 537865. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Hwang, J.Y.; Kim, H.J.; Choi, W.S.; Lee, J.H.; Kim, H.J.; Chang, K.C.; Nishinaka, T.; Yabe-Nishimura, C.; Seo, H.G. Expression of a peroxisome proliferator-activated receptor gamma 1 splice variant that was identified in human lung cancers suppresses cell death induced by cisplatin and oxidative stress. Clin Cancer Res 2007, 13, 2577–2583. [Google Scholar] [CrossRef]
- Hernandez-Quiles, M.; Broekema, M.F.; Kalkhoven, E. PPARgamma in Metabolism, Immunity, and Cancer: Unified and Diverse Mechanisms of Action. Front Endocrinol (Lausanne) 2021, 12, 624112. [Google Scholar] [CrossRef] [PubMed]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Motawi, T.K.; Shaker, O.G.; Ismail, M.F.; Sayed, N.H. Peroxisome Proliferator-Activated Receptor Gamma in Obesity and Colorectal Cancer: the Role of Epigenetics. Sci Rep 2017, 7, 10714. [Google Scholar] [CrossRef]
- Sand, M.; Skrygan, M.; Georgas, D.; Sand, D.; Hahn, S.A.; Gambichler, T.; Altmeyer, P.; Bechara, F.G. Microarray analysis of microRNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci 2012, 68, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Kazeminasab, F.; Baharlooie, M.; Ghaedi, K. Noncoding RNAs Associated with PPARs in Etiology of MAFLD as a Novel Approach for Therapeutics Targets. PPAR Res 2022, 2022, 6161694. [Google Scholar] [CrossRef]
- Li, S.-S.; Zhou, L.; Gao, L.; Wang, Y.-H.; Ding, Z.-H. Role of long noncoding RNA MALAT1 promotes the occurrence and progression of cutaneous squamous cell carcinoma. Nan Fang Yi Ke Da Xue Xue Bao 2018, 38, 421–427. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, J.; Guo, L.; Peng, M. Long non-coding RNA MALAT1 regulates cell proliferation, invasion and apoptosis by modulating the Wnt signaling pathway in squamous cell carcinoma. Am J Transl Res 2021, 13, 9233–9240. [Google Scholar] [PubMed]
- Xu, Y.; Dong, Y.; Deng, Y.; Qi, Q.; Wu, M.; Liang, H.; She, Q.; Guo, Q. Identifying an lncRNA-Related ceRNA Network to Reveal Novel Targets for a Cutaneous Squamous Cell Carcinoma. Biology (Basel) 2021, 10. [Google Scholar] [CrossRef]
- Pancione, M.; Sabatino, L.; Fucci, A.; Carafa, V.; Nebbioso, A.; Forte, N.; Febbraro, A.; Parente, D.; Ambrosino, C.; Normanno, N. , et al. Epigenetic Silencing of Peroxisome Proliferator-Activated Receptor γ Is a Biomarker for Colorectal Cancer Progression and Adverse Patients' Outcome. PLoS ONE 2010, 5, e14229. [Google Scholar] [CrossRef] [PubMed]
- Gilmore, T.D. NF-κB and Human Cancer: What Have We Learned over the Past 35 Years? Biomedicines 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wang, G.; Qu, P.; Yan, C.; Du, H. Overexpression of Dominant Negative Peroxisome Proliferator-Activated Receptor-γ (PPARγ) in Alveolar Type II Epithelial Cells Causes Inflammation and T-Cell Suppression in the Lung. Am J Pathol 2011, 178, 2191–2204. [Google Scholar] [CrossRef]
- Grygiel-Górniak, B. Peroxisome proliferator-activated receptors and their ligands: nutritional and clinical implications--a review. Nutr J 2014, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.-D.; Du, S.; Martin, L.; Leccia, N.; Michiels, J.-F.; Wagner, N. Vascular PPARβ/δ Promotes Tumor Angiogenesis and Progression. Cells 2019, 8, 1623. [Google Scholar] [CrossRef] [PubMed]
- Ham, S.A.; Hwang, J.S.; Yoo, T.; Lee, W.J.; Paek, K.S.; Oh, J.-W.; Park, C.-K.; Kim, J.-H.; Do, J.T.; Kim, J.-H. , et al. Ligand-activated PPARδ upregulates α-smooth muscle actin expression in human dermal fibroblasts: A potential role for PPARδ in wound healing. J Dermatol Sci 2015, 80, 186–195. [Google Scholar] [CrossRef] [PubMed]
- Tai, Y.; Woods, E.L.; Dally, J.; Kong, D.; Steadman, R.; Moseley, R.; Midgley, A.C. Myofibroblasts: Function, Formation, and Scope of Molecular Therapies for Skin Fibrosis. Biomolecules 2021, 11. [Google Scholar] [CrossRef]
- Procopio, M.G.; Laszlo, C.; Al Labban, D.; Kim, D.E.; Bordignon, P.; Jo, S.H.; Goruppi, S.; Menietti, E.; Ostano, P.; Ala, U. , et al. Combined CSL and p53 downregulation promotes cancer-associated fibroblast activation. Nat Cell Biol 2015, 17, 1193–1204. [Google Scholar] [CrossRef]
- Lecarpentier, Y.; Schussler, O.; Claes, V.; Vallée, A. The Myofibroblast: TGFβ-1, A Conductor which Plays a Key Role in Fibrosis by Regulating the Balance between PPARγ and the Canonical WNT Pathway. Nucl Recept Res 2017, 4, Article. [Google Scholar] [CrossRef] [PubMed]
- Chitsazzadeh, V.; Coarfa, C.; Drummond, J.A.; Nguyen, T.; Joseph, A.; Chilukuri, S.; Charpiot, E.; Adelmann, C.H.; Ching, G.; Nguyen, T.N. , et al. Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat Commun 2016, 7, 12601. [Google Scholar] [CrossRef] [PubMed]
- McCreery, M.Q.; Halliwill, K.D.; Chin, D.; Delrosario, R.; Hirst, G.; Vuong, P.; Jen, K.Y.; Hewinson, J.; Adams, D.J.; Balmain, A. Evolution of metastasis revealed by mutational landscapes of chemically induced skin cancers. Nat Med 2015, 21, 1514–1520. [Google Scholar] [CrossRef] [PubMed]
- García-Escudero, R.; Martínez-Cruz, A.B.; Santos, M.; Lorz, C.; Segrelles, C.; Garaulet, G.; Saiz-Ladera, C.; Costa, C.; Buitrago-Pérez, A.; Dueñas, M. , et al. Gene expression profiling of mouse p53-deficient epidermal carcinoma defines molecular determinants of human cancer malignancy. Mol Cancer 2010, 9, 193. [Google Scholar] [CrossRef] [PubMed]
- Segura, S.; Gadea, A.; Nonell, L.; Andrades, E.; Sánchez, S.; Pujol, R.; Hernández-Muñoz, I.; Toll, A. Identification of differentially expressed genes in actinic keratosis samples treated with ingenol mebutate gel. PLoS ONE 2020, 15, e0232146. [Google Scholar] [CrossRef] [PubMed]
- Hoang, V.L.T.; Tom, L.N.; Quek, X.-C.; Tan, J.-M.; Payne, E.J.; Lin, L.L.; Sinnya, S.; Raphael, A.P.; Lambie, D.; Frazer, I.H. , et al. RNA-seq reveals more consistent reference genes for gene expression studies in human non-melanoma skin cancers. PeerJ 2017, 5, e3631. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.; Ridgway, R.A.; Cammareri, P.; Treanor-Taylor, M.; Bailey, U.-M.; Schoenherr, C.; Bone, M.; Schreyer, D.; Purdie, K.; Thomson, J. , et al. Driver gene combinations dictate cutaneous squamous cell carcinoma disease continuum progression. Nat Commun 2023, 14, 5211. [Google Scholar] [CrossRef] [PubMed]
- Nindl, I.; Dang, C.; Forschner, T.; Kuban, R.J.; Meyer, T.; Sterry, W.; Stockfleth, E. Identification of differentially expressed genes in cutaneous squamous cell carcinoma by microarray expression profiling. Mol Cancer 2006, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Hameetman, L.; Commandeur, S.; Bavinck, J.N.; Wisgerhof, H.C.; de Gruijl, F.R.; Willemze, R.; Mullenders, L.; Tensen, C.P.; Vrieling, H. Molecular profiling of cutaneous squamous cell carcinomas and actinic keratoses from organ transplant recipients. BMC Cancer 2013, 13, 58. [Google Scholar] [CrossRef]
- García-Díez, I.; Hernández-Muñoz, I.; Hernández-Ruiz, E.; Nonell, L.; Puigdecanet, E.; Bódalo-Torruella, M.; Andrades, E.; Pujol, R.M.; Toll, A. Transcriptome and cytogenetic profiling analysis of matched in situ/invasive cutaneous squamous cell carcinomas from immunocompetent patients. Genes Chromosomes Cancer 2019, 58, 164–174. [Google Scholar] [CrossRef]
- Hu, Y.; Li, R.; Chen, H.; Chen, L.; Zhou, X.; Liu, L.; Ju, M.; Chen, K.; Huang, D. Comprehensive analysis of lncRNA-mRNAs co-expression network identifies potential lncRNA biomarkers in cutaneous squamous cell carcinoma. BMC Genom 2022, 23, 274. [Google Scholar] [CrossRef] [PubMed]
- Das Mahapatra, K.; Pasquali, L.; Søndergaard, J.N.; Lapins, J.; Nemeth, I.B.; Baltás, E.; Kemény, L.; Homey, B.; Moldovan, L.-I.; Kjems, J. , et al. A comprehensive analysis of coding and non-coding transcriptomic changes in cutaneous squamous cell carcinoma. Sci Rep 2020, 10, 3637. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Dai, H.; Zhang, X.; Liu, S.; Lin, Y.; Somani, A.K.; Xie, J.; Han, J. Distinct transcriptomic landscapes of cutaneous basal cell carcinomas and squamous cell carcinomas. Genes Dis 2021, 8, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Brooks, Y.S.; Ostano, P.; Jo, S.H.; Dai, J.; Getsios, S.; Dziunycz, P.; Hofbauer, G.F.; Cerveny, K.; Chiorino, G.; Lefort, K. , et al. Multifactorial ERβ and NOTCH1 control of squamous differentiation and cancer. J Clin Invest 2014, 124, 2260–2276. [Google Scholar] [CrossRef] [PubMed]
- Haider, A.S.; Peters, S.B.; Kaporis, H.; Cardinale, I.; Fei, J.; Ott, J.; Blumenberg, M.; Bowcock, A.M.; Krueger, J.G.; Carucci, J.A. Genomic Analysis Defines a Cancer-Specific Gene Expression Signature for Human Squamous Cell Carcinoma and Distinguishes Malignant Hyperproliferation from Benign Hyperplasia. J Invest Dermatol 2006, 126, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Riker, A.I.; Enkemann, S.A.; Fodstad, O.; Liu, S.; Ren, S.; Morris, C.; Xi, Y.; Howell, P.; Metge, B.; Samant, R.S. , et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics 2008, 1, 13. [Google Scholar] [CrossRef] [PubMed]
- Lo, B.K.; Yu, M.; Zloty, D.; Cowan, B.; Shapiro, J.; McElwee, K.J. CXCR3/ligands are significantly involved in the tumorigenesis of basal cell carcinomas. Am J Pathol 2010, 176, 2435–2446. [Google Scholar] [CrossRef] [PubMed]
- Jee, B.A.; Lim, H.; Kwon, S.M.; Jo, Y.; Park, M.C.; Lee, I.J.; Woo, H.G. Molecular classification of basal cell carcinoma of skin by gene expression profiling. Mol Carcinog 2015, 54, 1605–1612. [Google Scholar] [CrossRef]
- Atwood, S.X.; Sarin, K.Y.; Whitson, R.J.; Li, J.R.; Kim, G.; Rezaee, M.; Ally, M.S.; Kim, J.; Yao, C.; Chang, A.L. , et al. Smoothened variants explain the majority of drug resistance in basal cell carcinoma. Cancer Cell 2015, 27, 342–353. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





