1. Introduction
Celiac disease is a gluten-sensitive immune-mediated enteropathy that occurs in genetically predisposed individuals [
1]. Diagnosis of celiac disease is made by combining clinical data, serological tests, and histopathological features [
1,
2]. Although celiac disease is a disease of infants, its onset usually occurs in patients aged between 10 and 40 years, when the typical signs of malabsorption are often replaced by an atypical presentation [
3,
4,
5,
6].
The clinical presentation is variable and exhibits a continuum spectra [
3,
4,
5,
6], with several degrees of severity correlated with histological severity and levels of tissue transglutaminase [
7,
8]. The “classical” gastrointestinal symptoms include persistent diarrhea, abdominal distension, weight loss, abdominal pain, constipation, and vomiting [
9]. Celiac disease is also associated with several non-gastrointestinal manifestations, such as growth and development alterations, neurologic and behavioral symptoms, liver disease, iron deficiency, skin alterations (dermatitis herpetiformis), dental and metabolic bone diseases, arthritis, and cardiomyopathy [
9,
10]
Histological characteristics of the small intestine (usually evaluated using duodenum biopsy) include mucosal inflammation, villous atrophy, and crypt hyperplasia that occur after exposure to dietary gluten; signs that improve after removing gluten from the diet [
11]. These histological features are variable and range from mild alteration with only increased numbers of intraepithelial lymphocytes, to severe atrophy and epithelial apoptosis [
12,
13,
14,
15,
16]. These alterations are assessed in several classifications, including the Marsh [
17], Marsh-Oberhuber [
18], Corazza-Villanacci [
19], Q-Marsh scale [
20], and Q-histology [
2].
The pathogenesis of celiac disease includes genetic factors (HLA DR3-DQ2, DR4-DQ8, several non-HLA loci, and autoimmune disorders), adaptive immune response (gliadin reactive T lymphocytes), autoantibodies and intraepithelial lymphocytes (IELs), and innate immune response. In patients with celiac disease, the immune response to fractions of gliadin results in an abnormal inflammatory reaction characterized by infiltration of the lamina propria and epithelium by chronic inflammatory cells and villous atrophy [
4]. A comprehensive review of the pathogenesis was conducted in our recent publication [
21].
Primary treatment for celiac disease is a gluten-free diet. Persistent or recurring symptoms may be due to a lack of adherence to dietary protocol, an incorrect initial diagnosis, or complications of refractory celiac disease and lymphoma [
1]. Among the different primary intestinal T-cell lymphomas, enteropathy-associated T-cell lymphoma (EATL) [
22,
23,
24,
25]. EATL may be preceded by refractory celiac disease [
26].
The diagnosis of celiac disease is based on the combination of clinical data (enterologist), serology (clinical pathologist), and duodenal biopsy with histological evaluation performed by a certified anatomical pathologist [
1]. Artificial intelligence technology allows computers to imitate human intellectual capacity and solve problems [
27]. Modern computer vision systems exhibit extraordinary image recognition and analysis accuracy. However, these systems do not understand what they observe. Several machine learning and deep learning algorithms have been developed to construct models that make predictions on images. Convolutional neural networks are supervised algorithms that are mostly used for image recognition workloads [
28]. This study used a convolutional neural network to classify images of celiac disease, small intestine control, duodenal inflammation and duodenal adenocarcinoma.
2. Materials and Methods
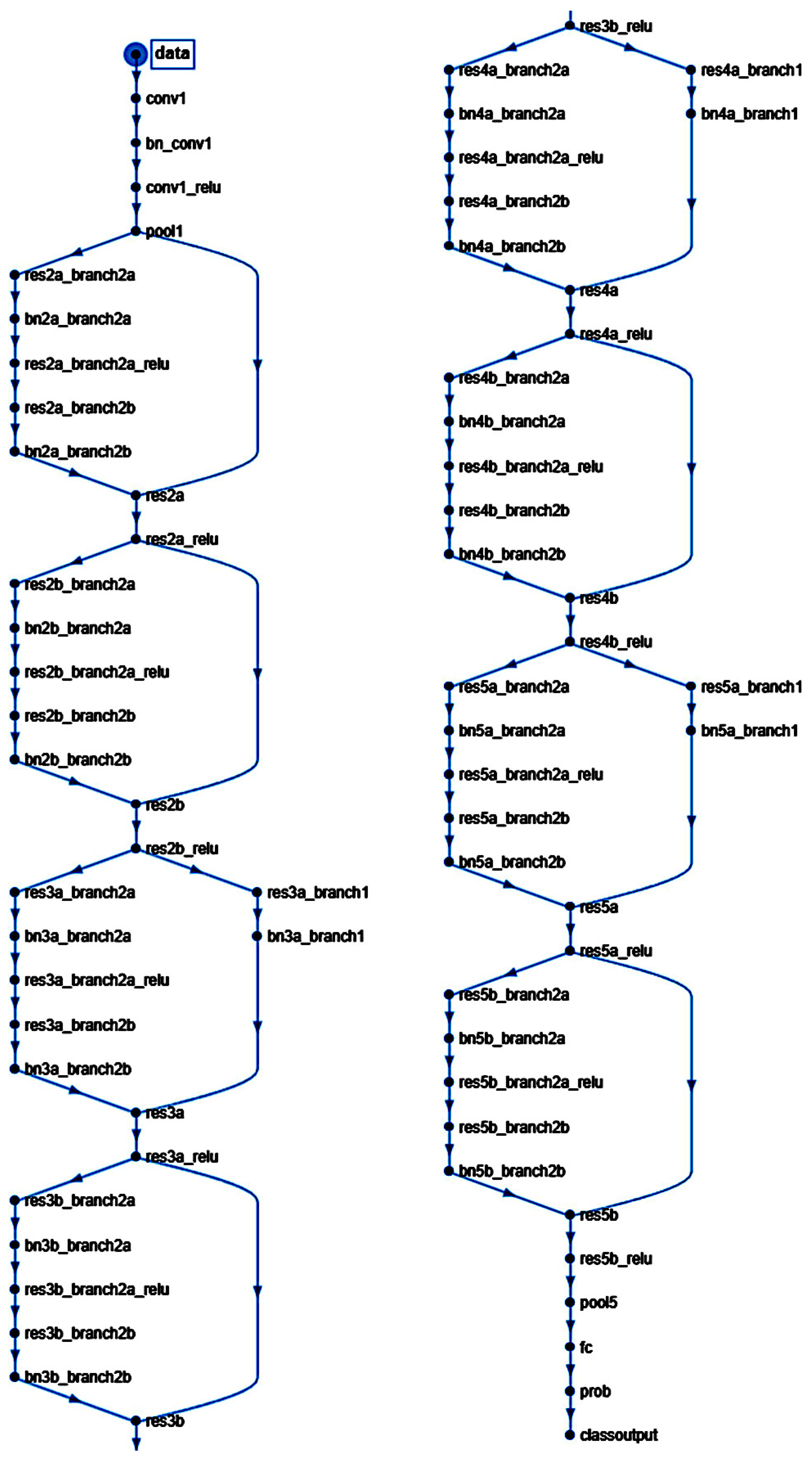
A script was written to create and train a deep learning network with 71 layers and 78 connections (
Figure 1 and
Figure 2). The script was run to create network layers (Appendix Table B), import training and validation data, and train the network. The code was created in MATLAB (R2023b Update 8 (23.2.0.25999560) 64-bit (win64) April 29, 2024) (MathWorks, Tokyo, Japan) and was based on transfer learning from the ResNet-18 (version 23.2.0) [
29] (
Figure 1 and
Figure 2). All analyses were performed using a desktop computer equipped with an AMD Ryzen 9 7950X CPU [
30], 32 Gb of RAM, and an Nvidia GeForce RTX 4080 super-graphics card [
31].
ResNet-18 is a pretrained model that was previously trained in a subset of images in the ImageNet database [
32]. This database includes 1000 types of objects and contains more than 1,000,000 images. ResNet-18 is a convolutional neural network with 18 layers. The input size is 224-by-224 (224×224×3). Size: 44.0 MB. Parameters: 11.7 M.
The analysis of the convolutional neural network (CNN) included the following steps: loading the pre-trained network, replacement of final layers, training of the network, prediction and assessment of network accuracy, and deployment of results.
The diagnostic dataset included hematoxylin and eosin (H&E) staining of 16 celiac disease patients (57 biopsies), selected from the Department of Pathology, Hospital Clinic of Barcelona, Spain, as previously described [
21]. The clinicopathological characteristics such as age, sex, biopsy location, anatomical pathology diagnosis, and the Marsh-Oberhuber histological grade [
21,
33,
34] are shown in Appendix Table A.
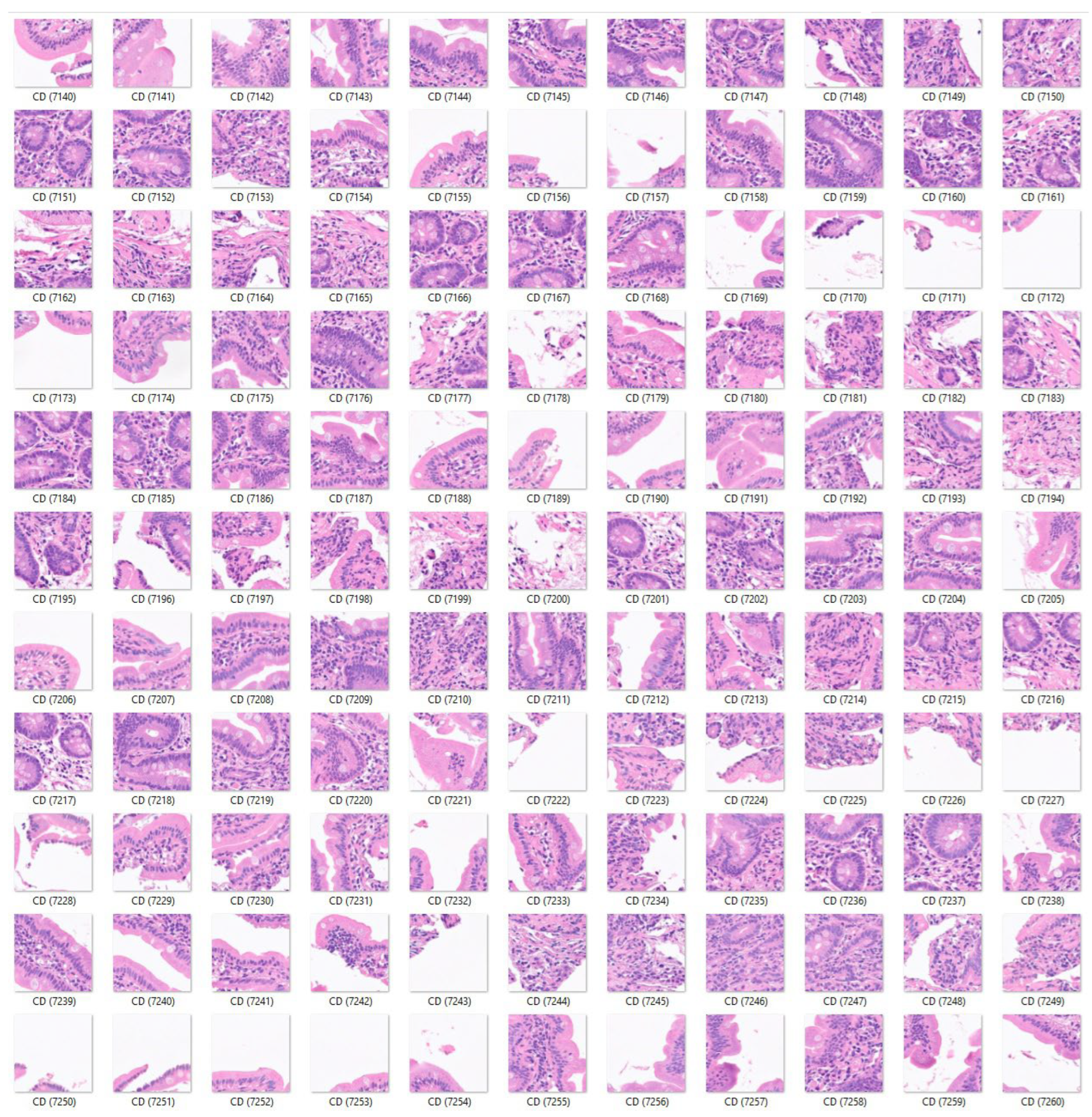
First, the input data for celiac disease included 7294 images, and the small intestine control included 11,642 images. The color images had three channels: red, green, and blue. An example is shown in
Figure 3,
Figure 4 and
Figure 5. The data (images) were split into three sets: a training set used for training the network (70%), a validation set used for testing its performance as it was trained (10%), and a test set used after training to assess how well the network performed on new data (20%). The order of the images was randomized to ensure that the network learned the classes at a more even rate. As transfer learning (adjustment of a pre-trained network) was performed on ResNet-18, the fully connected and classification layers were removed and replaced with new layers with an output size of 2. Augmentation was not performed during training. To avoid overfitting, the initial learning rate was set to 0.001. The number of maximum epochs was set to five.
Second, the analysis was repeated by adding a third histological subtype of nonspecific inflammation of the small intestine (duodenum). Therefore, in this analysis the input data included 7294 images of celiac disease, 11,642 images of small intestine control, and 5966 images of the small intestine (duodenum) with chronic and acute inflammation (
Figure 6).
Third, a fourth histological subtype of 3723 images of duodenal adenocarcinoma (
Figure 7) was added as test images of the previously trained convolutional neural network. The purpose of this analysis was to determine how the previously trained network, which was trained using celiac disease, small intestine control, and non-specific inflammation of the duodenum, could classify an unknown histological disease.
Finally, a convolutional neural network was trained, including as input all the histological subtypes of celiac disease, small intestine control (both duodenum and ileum), non-specific inflammation of the duodenum, and duodenal adenocarcinoma (
Figure 4,
Figure 5,
Figure 6 and
Figure 7).
2. Results
2.1. Celiac Disease vs. Small Intestine Control
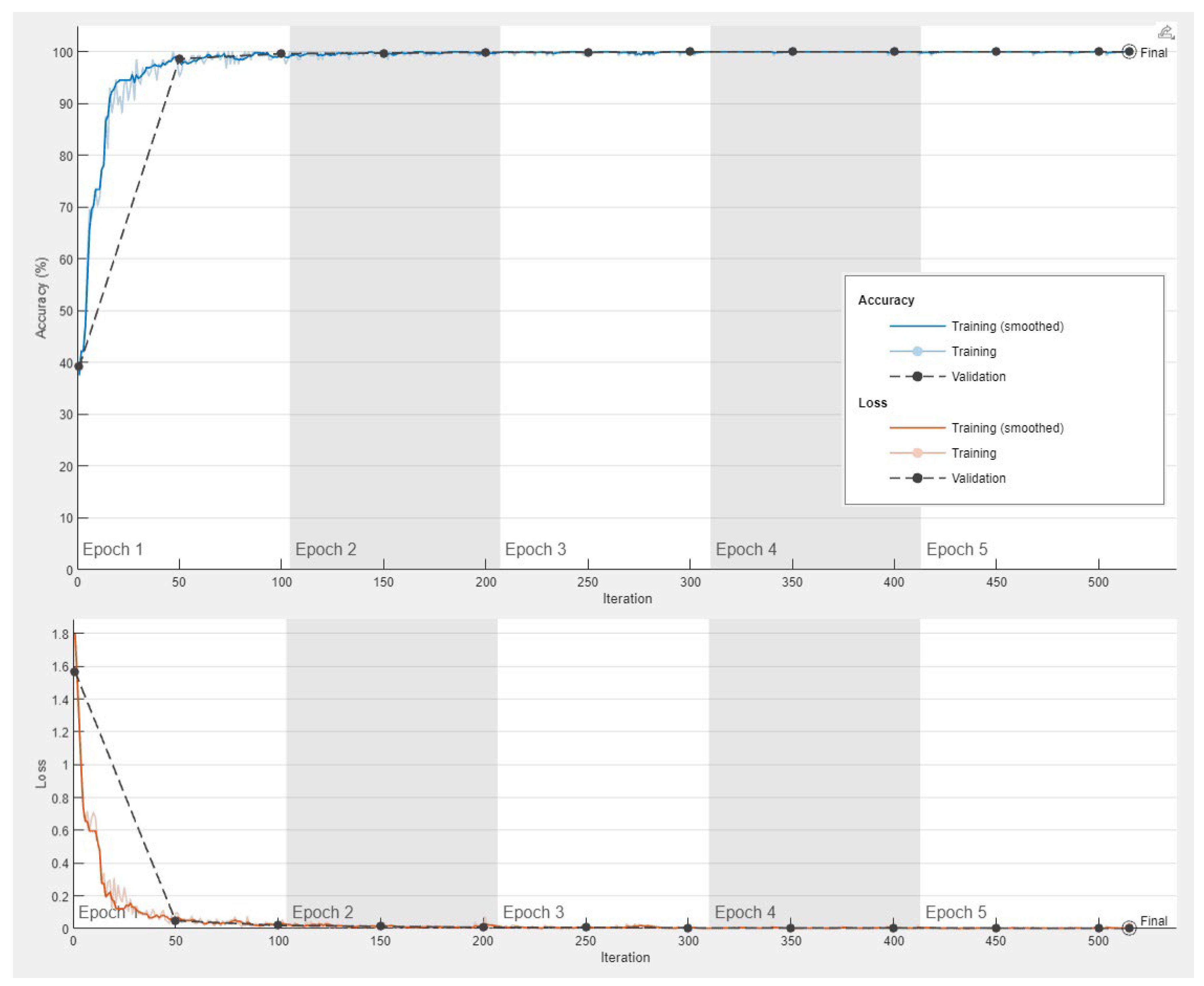
The progress of the convolutional neural network was satisfactory with a validation accuracy of 100%. The training cycle included 5 epochs, 515 iterations, and 103 iterations per epoch. The validation cycle included 50 iterations. Within the first 100 iterations, the accuracy percentage reached 100%, and the loss the 0 value (
Figure 8).
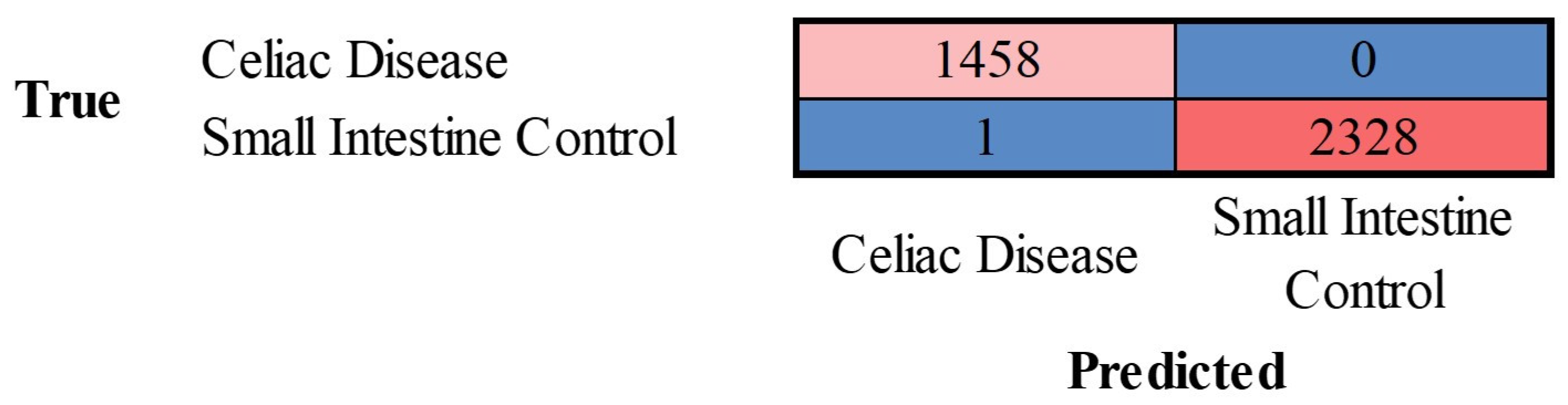
The images in the test set were classified by the trained network. The results are shown as a confusion matrix (
Figure 9). The performance parameters for celiac disease were as follows: accuracy, 99.97%; precision, 99.93%; recall, 100%; F1-Score, 99.97%; specificity, 100%, and false positive rate (FPR), 0.04% (
Table 1).
2.2. Celiac Disease vs. Small Intestine Control vs. Duodenal Inflammation
The progress of the training of the convolutional neural network is shown in
Appendix Figure C. The results are shown as a confusion matrix (
Figure 10).
The class-wise performance is summarized in
Table 2.
2.3. Test for Duodenal Adenocarcinoma on Previously Trained Network
Images of duodenal adenocarcinoma were tested directly on the previously trained network that had classified celiac disease, small intestine control, and duodenal inflammation. The analysis showed that the convolutional network classified duodenal adenocarcinoma as duodenal inflammation in 63.65% of images, small intestine control in 34.73%, and celiac disease in 1.61% of images. Therefore, a previously trained network can classify an unknown type of image but incorrectly diagnoses the image.
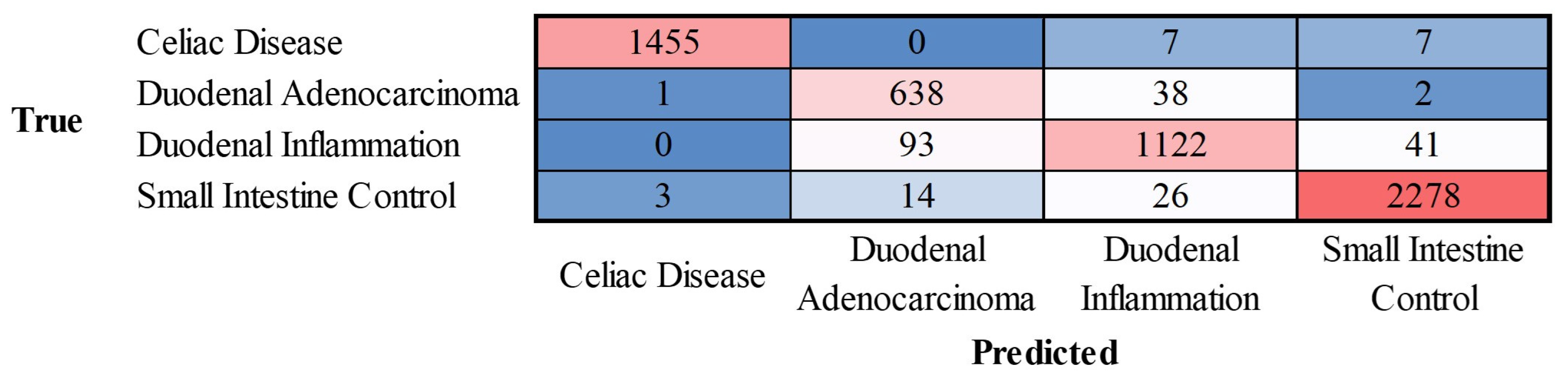
2.4. Celiac Disease vs. Small Intestine Control vs. Duodenal Inflammation vs. Duodenal Adenocarcinoma
The progress of training the convolutional neural network is shown in
Appendix Figure D. The results are shown as a confusion matrix (
Figure 11).
The class-wise performance is summarized in
Table 3.
3. Discussion
Within the specialty of computer science, computer vision is a technique that allows computers to recognize observable world. In the field of artificial intelligence, there are several machine learning and deep learning algorithms that build models that make predictions from images or videos [
35]. Convolution neural networks are a type of supervised deep learning algorithm that are used for image recognition. A simple convolutional network comprises several steps, including image channel, convolutions, pooling, convolutions, pooling, flattening, artificial neural network full connection, and prediction [
35].
The ResNet-18 network was used in this study. This convolutional neural network was a pretrained model trained on a subset of the ImageNet database. The network is trained is more than a million images and managed to classify them into 1000 different categories [
29]. In the medical field, this network has been used in several studies based on transfer learning, such as in the diagnosis of intracranial hemorrhage in CT scans [
36], heartbeat classification of electrocardiogram (ECG) signals [
37], dynamic gesture recognition [
38], selective transplanting of leafy vegetable seedlings [
39], automatic classification of malaria parasites on the blood smear [
40], prostate imaging [
41], classification of Alzheimer’s disease levels [
42], and diabetic retinopathy [
43], among others. Therefore, the ResNet-18 model is a useful network that can be applied to many types of studies, including our study of celiac disease.
Convolutional neural networks and image recognition have also been applied to celiac disease research, including the analysis of whole slide images [
44,
45,
46,
47,
48], and endoscopic images [
49,
50]. Therefore, computer vision is a useful tool in the field of histopathology.
Our group has published several papers on the use of artificial intelligence, including machine learning and artificial neural networks, in the field of lymphoma research [
51,
52,
53,
54,
55,
56,
57]. In these publications, the focus was on data analysis of gene expression levels in the context of immuno-oncology in lymphoma and other hematological neoplasia [
51,
52,
53,
54,
55,
56,
57]. The most frequent lymphoma subtype that we analyzed was diffuse large b-cell lymphoma [
52,
53,
54,
55], which is one of the most frequent non-Hodgkin lymphomas [
26]. In addition, we have also published data analysis-based studies on celiac disease in which we highlighted the importance of the B and T lymphocyte associated (BTLA) gene [
21], and programed cell death 1 ligand 1 (CD274 antigen) in ulcerative colitis [
58]. The subject of this article represented a switch from data analytics to computer vision.
In this study, a confusion matrix was used to measure the performance of the trained network. The data (images) were split into three sets: a training set used for training (i.e., teaching) the network (70%), a validation set used for testing its performance as it was trained (10%), and a test set used after training to assess how well the network performed on new data (20%). The order of the images was randomized to ensure that the network learned the classes at a more even rate. In the results section, the confusion matrices of the test set were shown. Of note, if the data were imbalanced, the performance checking by accuracy could be deceptive. The confusion matrices of our study combined output data that was binary (
Figure 9) and multiclass (
Figure 10 and
Figure 11). All performance parameters were high, including accuracy (defined as the proportion of correct predictions), precision (used in information retrieval, pattern recognition), recall (what in medicine is called sensitivity), and F1-score (measure test of accuracy). The fundamentals of clinical data science and modeling methodology are well described in chapter 8 of the book written by Frank J.W.M. Dankers et al. [
59].
This study focused on the identification and classification of celiac disease images compared with normal small intestine images obtained from the duodenum and ileum. The accuracy of the network was very high. The model could handle and properly classify 3 classes with the addition of non-specific acute and chronic duodenal inflammation. Interestingly, when the 3 classes trained network was tested with duodenal adenocarcinoma, the network failed to realize that those samples were a different type of disease. Therefore, the use of automated computer vision analysis for the evaluation of histopathological slides is not recommended without the supervision of a pathology specialist. However, when the network was trained with 4 classes of histological subtypes, the network managed to differentiate celiac disease, duodenal inflammation, small intestine control, and duodenal adenocarcinoma with good performance, proving the usefulness of convolutional neural network for classifying histological images.
In conclusion, a convolutional neural network based on the transfer learning of ResNet-18 was able to classify celiac disease, other duodenal pathological diseases, and tissue control with good performance. However, all computer vision-based automated diagnoses should be supervised and validated by pathology medical specialist to identify other pathologies for which the network has not previously trained.
Author Contributions
Conceptualization, J.C.; methodology, formal analysis, investigation, writing—original draft preparation, writing—review and editing, J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ministry of Education, Culture, Sports, Science and Technology of Japan, KAKEN grants 23K06454, 18K15100, and 15K19061.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of TOKAI UNIVERSITY, SCHOOL OF MEDICINE (protocol code IRB14R-080 and IRB20-156).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All the data, including methodology, are available upon reasonable request to Dr. Joaquim Carreras (joaquim.carreras@tokai-u.jp).
Acknowledgments
We want to thank Josep A. Bombi from the Department of Pathology, Hospital Clinic of Barcelona (Spain) for the celiac disease cases.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Appendix Table A
Table A.
Clinicopathological characteristics of celiac disease cases.
Table A.
Clinicopathological characteristics of celiac disease cases.
| Age |
Sex |
Biopsy location |
Diagnosis |
Marsh-Oberhuber Classification |
| |
|
|
|
|
| 70 |
Male |
Duodenum |
Celiac Disease |
3a |
| 62 |
Male |
Pylorus/duodenum |
Celiac Disease/Chronic gastritis |
2 |
| 62 |
Male |
Duodenum |
Celiac Disease |
2 |
| 78 |
Female |
Duodenum |
Celiac Disease |
3b |
| 59 |
Male |
Duodenum |
Celiac Disease |
3a |
| 44 |
Female |
Duodenum |
Celiac Disease |
2 |
| 17 |
Female |
Duodenum |
Celiac Disease |
3b |
| 56 |
Female |
Duodenum |
Celiac Disease |
3a |
| 54 |
Female |
Duodenum |
Celiac Disease |
2 |
| 58 |
Female |
Duodenum |
Celiac Disease |
3b |
| 61 |
Female |
Duodenum |
Celiac Disease |
3c |
| 45 |
Male |
Duodenum |
Celiac Disease |
3a |
| 70 |
Female |
Duodenum |
Celiac Disease |
2 |
| 40 |
Female |
Duodenum |
Celiac Disease |
3a |
| 61 |
Female |
Duodenum |
Celiac Disease |
3c |
| 44 |
Female |
Duodenum |
Celiac Disease |
3a |
Appendix Figure C
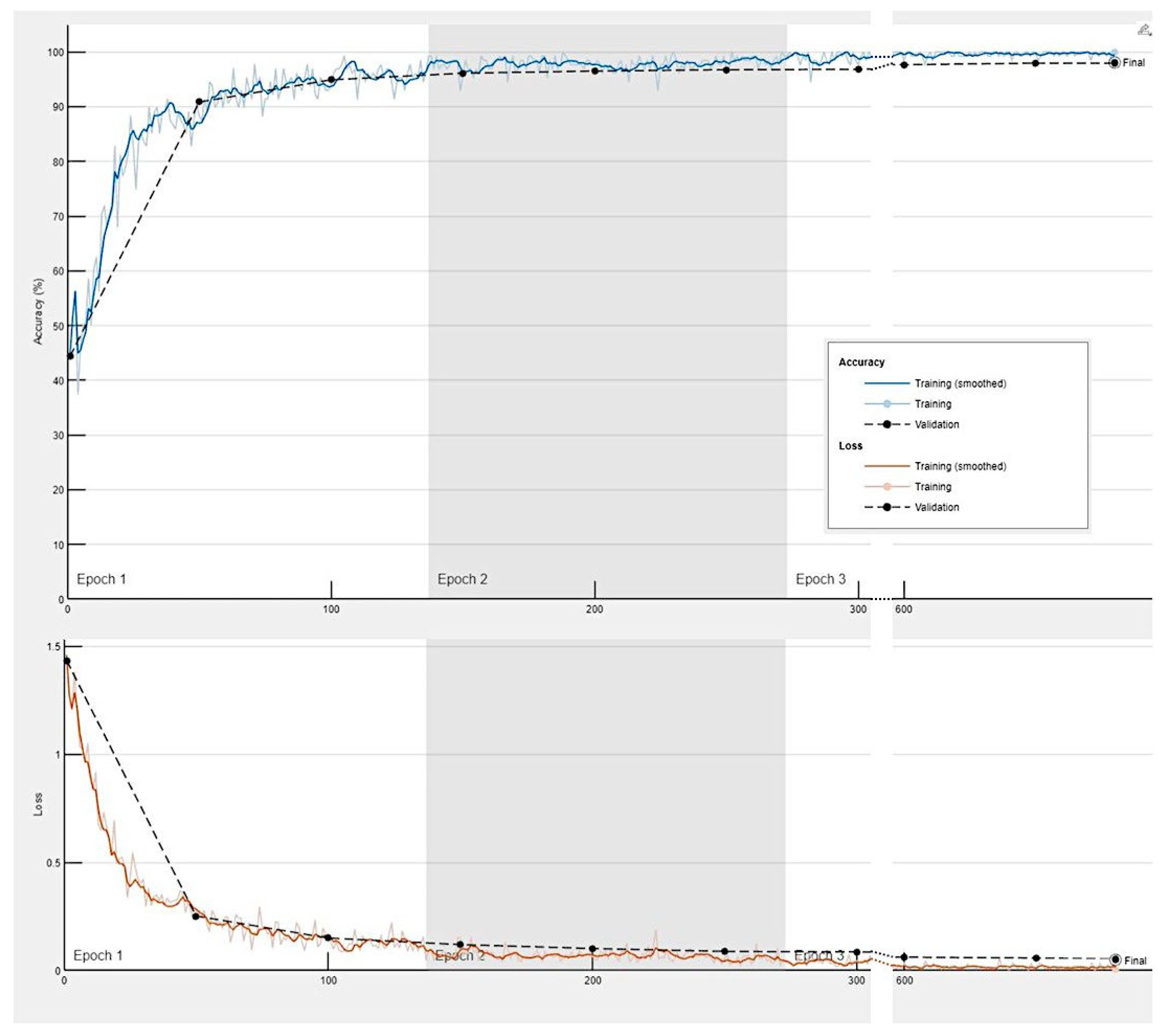
Training progress of the convolutional neural network for the classification of celiac disease, small intestine control and duodenal inflammation.
Appendix Figure D
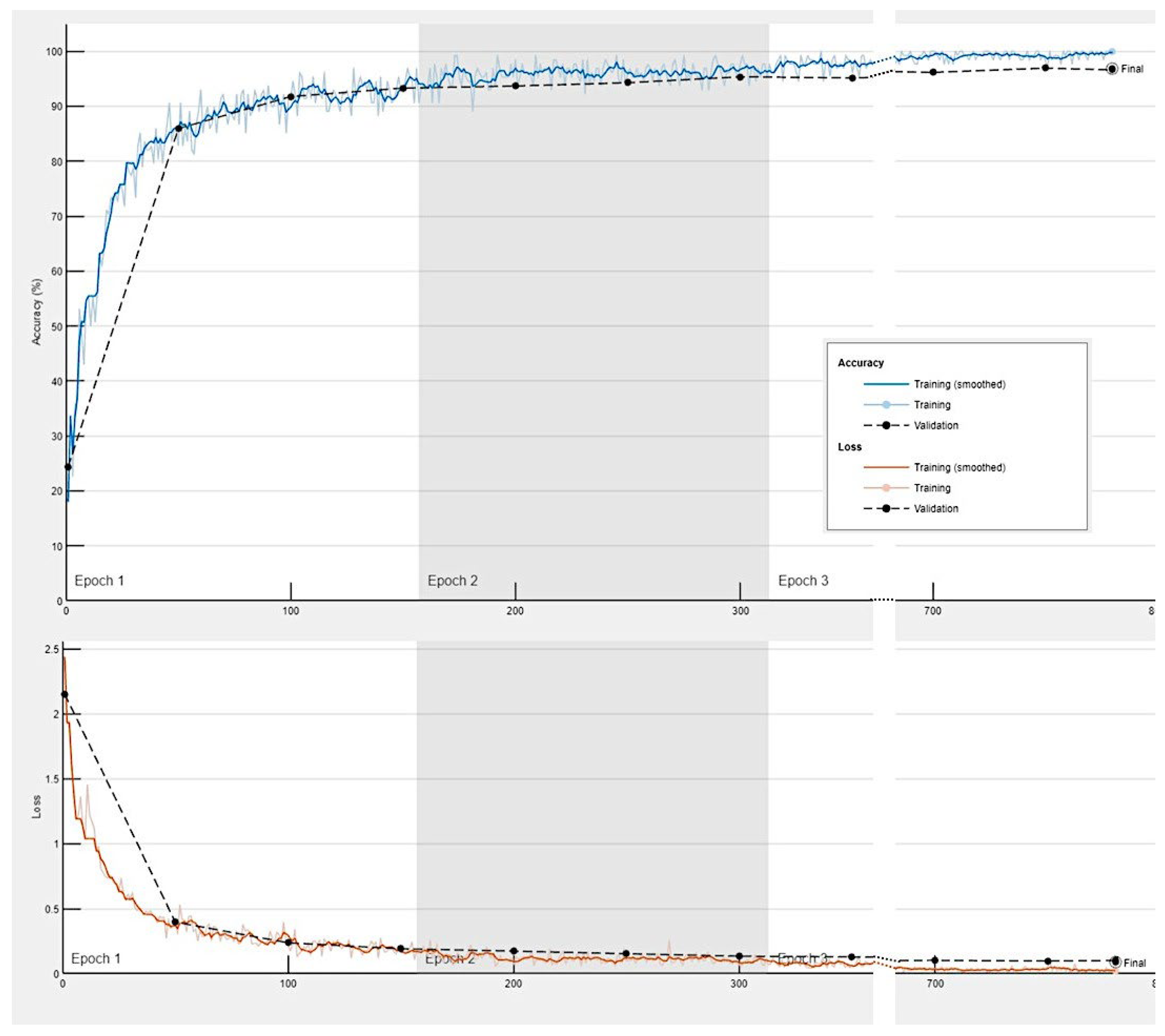
Training progress of the convolutional neural network for the classification of celiac disease, small intestine control, duodenal inflammation and duodenal adenocarcinoma.
References
- Al-Toma, A.; Volta, U.; Auricchio, R.; Castillejo, G.; Sanders, D.S.; Cellier, C.; Mulder, C.J.; Lundin, K.E.A. European Society for the Study of Coeliac Disease (ESsCD) guideline for coeliac disease and other gluten-related disorders. United European Gastroenterol J 2019, 7, 583–613. [Google Scholar] [CrossRef] [PubMed]
- Vargas, M.M.; Artigiani Neto, R.; Sdepanian, V.L. Quantitative histology as a diagnostic tool for celiac disease in children and adolescents. Ann Diagn Pathol 2022, 61, 152031. [Google Scholar] [CrossRef] [PubMed]
- Catassi, C.; Verdu, E.F.; Bai, J.C.; Lionetti, E. Coeliac disease. Lancet 2022, 399, 2413–2426. [Google Scholar] [CrossRef] [PubMed]
- Detlef Schuppan, W.D. Epidemiology, pathogenesis, and clinical manifestations of celiac disease in adults. In: UpToDate, Karen Law, J Thomas Lamont (Eds), Wolters Kluwer. (Accessed on June 20, 2024.), 2023. [Google Scholar]
- Oxentenko, A.S.; Rubio-Tapia, A. Celiac Disease. Mayo Clin Proc 2019, 94, 2556–2571. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Sanchez, M.I.; Silvester, J.A.; Lebwohl, B.; Leffler, D.A.; Anderson, R.P.; Therrien, A.; Kelly, C.P.; Verdu, E.F. Society for the Study of Celiac Disease position statement on gaps and opportunities in coeliac disease. Nat Rev Gastroenterol Hepatol 2021, 18, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Taavela, J.; Kurppa, K.; Collin, P.; Lahdeaho, M.L.; Salmi, T.; Saavalainen, P.; Haimila, K.; Huhtala, H.; Laurila, K.; Sievanen, H.; et al. Degree of damage to the small bowel and serum antibody titers correlate with clinical presentation of patients with celiac disease. Clin Gastroenterol Hepatol 2013, 11, 166–171. [Google Scholar] [CrossRef] [PubMed]
- West, J.; Logan, R.F.; Hill, P.G.; Khaw, K.T. The iceberg of celiac disease: what is below the waterline? Clin Gastroenterol Hepatol 2007, 5, 59–62. [Google Scholar] [CrossRef]
- Marisa Stahl, E.L. Diagnosis of celiac disease in children. In: UpToDate, B UK Li, Alison G Hoppin (Eds), Wolters Kluwer. (Accessed on June 20, 2024.), 2023. [Google Scholar]
- Sahin, Y. Celiac disease in children: A review of the literature. World J Clin Pediatr 2021, 10, 53–71. [Google Scholar] [CrossRef]
- Tye-Din, J.A.; Galipeau, H.J.; Agardh, D. Celiac Disease: A Review of Current Concepts in Pathogenesis, Prevention, and Novel Therapies. Front Pediatr 2018, 6, 350. [Google Scholar] [CrossRef]
- Ferguson, A.; Arranz, E.; O’Mahony, S. Clinical and pathological spectrum of coeliac disease--active, silent, latent, potential. Gut 1993, 34, 150–151. [Google Scholar] [CrossRef]
- Fry, L.; Seah, P.P.; McMinn, R.M.; Hoffbrand, A.V. Lymphocytic infiltration of epithelium in diagnosis of gluten-sensitive enteropathy. Br Med J 1972, 3, 371–374. [Google Scholar] [CrossRef] [PubMed]
- Hvas, C.L.; Jensen, M.D.; Reimer, M.C.; Riis, L.B.; Rumessen, J.J.; Skovbjerg, H.; Teisner, A.; Wildt, S. Celiac disease: diagnosis and treatment. Dan Med J 2015, 62, C5051. [Google Scholar] [PubMed]
- Marsh, M.N.; Crowe, P.T. Morphology of the mucosal lesion in gluten sensitivity. Baillieres Clin Gastroenterol 1995, 9, 273–293. [Google Scholar] [CrossRef] [PubMed]
- Troncone, R.; Greco, L.; Mayer, M.; Paparo, F.; Caputo, N.; Micillo, M.; Mugione, P.; Auricchio, S. Latent and potential coeliac disease. Acta Paediatr Suppl 1996, 412, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Marsh, M.N. Gluten, major histocompatibility complex, and the small intestine. A molecular and immunobiologic approach to the spectrum of gluten sensitivity (‘celiac sprue’). Gastroenterology 1992, 102, 330–354. [Google Scholar] [CrossRef] [PubMed]
- Oberhuber, G. Histopathology of celiac disease. Biomed Pharmacother 2000, 54, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Corazza, G.R.; Villanacci, V. Coeliac disease. J Clin Pathol 2005, 58, 573–574. [Google Scholar] [CrossRef] [PubMed]
- Adelman, D.C.; Murray, J.; Wu, T.T.; Maki, M.; Green, P.H.; Kelly, C.P. Measuring Change In Small Intestinal Histology In Patients With Celiac Disease. Am J Gastroenterol 2018, 113, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Carreras, J. Artificial Intelligence Analysis of Celiac Disease Using an Autoimmune Discovery Transcriptomic Panel Highlighted Pathogenic Genes including BTLA. Healthcare (Basel) 2022, 10. [Google Scholar] [CrossRef]
- Martin-Masot, R.; Herrador-Lopez, M.; Navas-Lopez, V.M.; Carmona, F.D.; Nestares, T.; Bossini-Castillo, L. Celiac Disease Is a Risk Factor for Mature T and NK Cell Lymphoma: A Mendelian Randomization Study. Int J Mol Sci 2023, 24. [Google Scholar] [CrossRef]
- Abdullah, S.A.A.; Goa, P.; Vandenberghe, E.; Flavin, R. Update on the Pathogenesis of Enteropathy-Associated T-Cell Lymphoma. Diagnostics (Basel) 2023, 13. [Google Scholar] [CrossRef] [PubMed]
- Al Somali, Z.; Hamadani, M.; Kharfan-Dabaja, M.; Sureda, A.; El Fakih, R.; Aljurf, M. Enteropathy-Associated T cell Lymphoma. Curr Hematol Malig Rep 2021, 16, 140–147. [Google Scholar] [CrossRef]
- Rishi, A.R.; Rubio-Tapia, A.; Murray, J.A. Refractory celiac disease. Expert Rev Gastroenterol Hepatol 2016, 10, 537–546. [Google Scholar] [CrossRef]
- Campo, E.; Jaffe, E.S.; Cook, J.R.; Quintanilla-Martinez, L.; Swerdlow, S.H.; Anderson, K.C.; Brousset, P.; Cerroni, L.; de Leval, L.; Dirnhofer, S.; et al. The International Consensus Classification of Mature Lymphoid Neoplasms: a report from the Clinical Advisory Committee. Blood 2022, 140, 1229–1253. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z. A gentle introduction to artificial neural networks. Ann Transl Med 2016, 4, 370. [Google Scholar] [CrossRef]
- Pang, S.; Du, A.; Orgun, M.A.; Yu, Z. A novel fused convolutional neural network for biomedical image classification. Med Biol Eng Comput 2019, 57, 107–121. [Google Scholar] [CrossRef]
- Kaiming He, X.Z. , Shaoqing Ren, Jian Sun. Deep Residual Learning for Image Recognition. Arxiv, Computer Science, Computer Vision and Pattern Recognition, 2015. [Google Scholar] [CrossRef]
- Advanced Micro Devices, I. AMD Ryzen™ 9 7950X Desktop Processor. Available online: https://www.amd.com/en/products/processors/desktops/ryzen/7000-series/amd-ryzen-9-7950x.html (accessed on 2024/06/27).
- Corporation, N. GeForce RTX 4080 Family. Available online: https://www.nvidia.com/en-us/geforce/graphics-cards/40-series/rtx-4080-family/ (accessed on 2024/06/27).
- Stanford Vision Lab, S.U.; , Princeton University. ImageNet. Available online: https://www.image-net.org/ (accessed on 2024/06/01).
- Kelly, C.P. Diagnosis of Celiac Disease in Adults. In: UpToDate, J Thomas Lamont, Karen Law (Eds), Wolters Kluwer. (Accessed on June 20, 2023.), 2023. [Google Scholar]
- Oberhuber, G.; Granditsch, G.; Vogelsang, H. The histopathology of coeliac disease: time for a standardized report scheme for pathologists. Eur J Gastroenterol Hepatol 1999, 11, 1185–1194. [Google Scholar] [CrossRef] [PubMed]
- Samaya Madhavan. Introduction to convolutional neural networks. 12 July 2021. IBM Corporation. IBM Developer. Available online: https://developer.ibm.com/articles/introduction-to-convolutional-neural-networks/ (accessed on 2024/06/26).
- Zhou, Q.; Zhu, W.; Li, F.; Yuan, M.; Zheng, L.; Liu, X. Transfer Learning of the ResNet-18 and DenseNet-121 Model Used to Diagnose Intracranial Hemorrhage in CT Scanning. Curr Pharm Des 2022, 28, 287–295. [Google Scholar] [CrossRef]
- Jing, E.; Zhang, H.; Li, Z.; Liu, Y.; Ji, Z.; Ganchev, I. ECG Heartbeat Classification Based on an Improved ResNet-18 Model. Comput Math Methods Med 2021, 2021, 6649970. [Google Scholar] [CrossRef]
- Zhang, Y.; Peng, L.; Ma, G.; Man, M.; Liu, S. Dynamic Gesture Recognition Model Based on Millimeter-Wave Radar With ResNet-18 and LSTM. Front Neurorobot 2022, 16, 903197. [Google Scholar] [CrossRef]
- Jin, X.; Tang, L.; Li, R.; Ji, J.; Liu, J. Selective transplantation method of leafy vegetable seedlings based on ResNet 18 network. Front Plant Sci 2022, 13, 893357. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, S.; Zhang, Y. ROENet: A ResNet-Based Output Ensemble for Malaria Parasite Classification. Electronics (Basel) 2022, 11, 2040. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Xiao, E.; Li, Z.; Wang, L. Deep Learning Based on ResNet-18 for Classification of Prostate Imaging-Reporting and Data System Category 3 Lesions. Acad Radiol 2024. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, Q.; Liu, X.; Hu, B. An Attention-Based CoT-ResNet With Channel Shuffle Mechanism for Classification of Alzheimer’s Disease Levels. Front Aging Neurosci 2022, 14, 930584. [Google Scholar] [CrossRef] [PubMed]
- Amin, J.; Shazadi, I.; Sharif, M.; Yasmin, M.; Almujally, N.A.; Nam, Y. Localization and grading of NPDR lesions using ResNet-18-YOLOv8 model and informative features selection for DR classification based on transfer learning. Heliyon 2024, 10, e30954. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.W.; Wei, J.W.; Jackson, C.R.; Ren, B.; Suriawinata, A.A.; Hassanpour, S. Automated Detection of Celiac Disease on Duodenal Biopsy Slides: A Deep Learning Approach. J Pathol Inform 2019, 10, 7. [Google Scholar] [CrossRef]
- Gruver, A.M.; Lu, H.; Zhao, X.; Fulford, A.D.; Soper, M.D.; Ballard, D.; Hanson, J.C.; Schade, A.E.; Hsi, E.D.; Gottlieb, K.; et al. Pathologist-trained machine learning classifiers developed to quantitate celiac disease features differentiate endoscopic biopsies according to modified marsh score and dietary intervention response. Diagn Pathol 2023, 18, 122. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, B.A.; Denholm, J.; Jaeckle, F.; Arends, M.J.; Branson, K.M.; Schonlieb, C.B.; Soilleux, E.J. Rapid artefact removal and H&E-stained tissue segmentation. Sci Rep 2024, 14, 309. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, B.A.; Denholm, J.; Gilbey, J.D.; Schonlieb, C.B.; Soilleux, E.J. Stain normalization gives greater generalizability than stain jittering in neural network training for the classification of coeliac disease in duodenal biopsy whole slide images. J Pathol Inform 2023, 14, 100324. [Google Scholar] [CrossRef]
- Denholm, J.; Schreiber, B.A.; Evans, S.C.; Crook, O.M.; Sharma, A.; Watson, J.L.; Bancroft, H.; Langman, G.; Gilbey, J.D.; Schonlieb, C.B.; et al. Multiple-instance-learning-based detection of coeliac disease in histological whole-slide images. J Pathol Inform 2022, 13, 100151. [Google Scholar] [CrossRef]
- Wimmer, G.; Vecsei, A.; Hafner, M.; Uhl, A. Fisher encoding of convolutional neural network features for endoscopic image classification. J Med Imaging (Bellingham) 2018, 5, 034504. [Google Scholar] [CrossRef]
- Zhou, T.; Han, G.; Li, B.N.; Lin, Z.; Ciaccio, E.J.; Green, P.H.; Qin, J. Quantitative analysis of patients with celiac disease by video capsule endoscopy: A deep learning method. Comput Biol Med 2017, 85, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Carreras, J.; Yukie Kikuti, Y.; Miyaoka, M.; Miyahara, S.; Roncador, G.; Hamoudi, R.; Nakamura, N. Artificial Intelligence Analysis and Reverse Engineering of Molecular Subtypes of Diffuse Large B-Cell Lymphoma Using Gene Expression Data. BioMedInformatics 2024, 4, 295–320. [Google Scholar] [CrossRef]
- Carreras, J.; Nakamura, N. Artificial Intelligence, Lymphoid Neoplasms, and Prediction of MYC, BCL2, and BCL6 Gene Expression Using a Pan-Cancer Panel in Diffuse Large B-Cell Lymphoma. Hemato 2024, 5, 119–143. [Google Scholar] [CrossRef]
- Carreras, J.; Hamoudi, R. Anomaly Detection and Artificial Intelligence Identified the Pathogenic Role of Apoptosis and RELB Proto-Oncogene, NF-kB Subunit in Diffuse Large B-Cell Lymphoma. BioMedInformatics 2024, 4, 1480–1505. [Google Scholar] [CrossRef]
- Carreras, J.; Hiraiwa, S.; Kikuti, Y.Y.; Miyaoka, M.; Tomita, S.; Ikoma, H.; Ito, A.; Kondo, Y.; Roncador, G.; Garcia, J.F.; et al. Artificial Neural Networks Predicted the Overall Survival and Molecular Subtypes of Diffuse Large B-Cell Lymphoma Using a Pancancer Immune-Oncology Panel. Cancers (Basel) 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Carreras, J.; Hamoudi, R.; Nakamura, N. Artificial Intelligence Analysis of Gene Expression Data Predicted the Prognosis of Patients with Diffuse Large B-Cell Lymphoma. Tokai J Exp Clin Med 2020, 45, 37–48. [Google Scholar] [PubMed]
- Carreras, J.; Nakamura, N.; Hamoudi, R. Artificial Intelligence Analysis of Gene Expression Predicted the Overall Survival of Mantle Cell Lymphoma and a Large Pan-Cancer Series. Healthcare (Basel) 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Carreras, J.; Roncador, G.; Hamoudi, R. Artificial Intelligence Predicted Overall Survival and Classified Mature B-Cell Neoplasms Based on Immuno-Oncology and Immune Checkpoint Panels. Cancers (Basel) 2022, 14. [Google Scholar] [CrossRef]
- Carreras, J. Artificial Intelligence Analysis of Ulcerative Colitis Using an Autoimmune Discovery Transcriptomic Panel. Healthcare (Basel) 2022, 10. [Google Scholar] [CrossRef]
- Dankers, F.; Traverso, A.; Wee, L.; van Kuijk, S.M.J. Prediction Modeling Methodology. In Fundamentals of Clinical Data Science, Kubben, P., Dumontier, M., Dekker, A., Eds.; Cham (CH), 2019; pp. 101-120.
Figure 1.
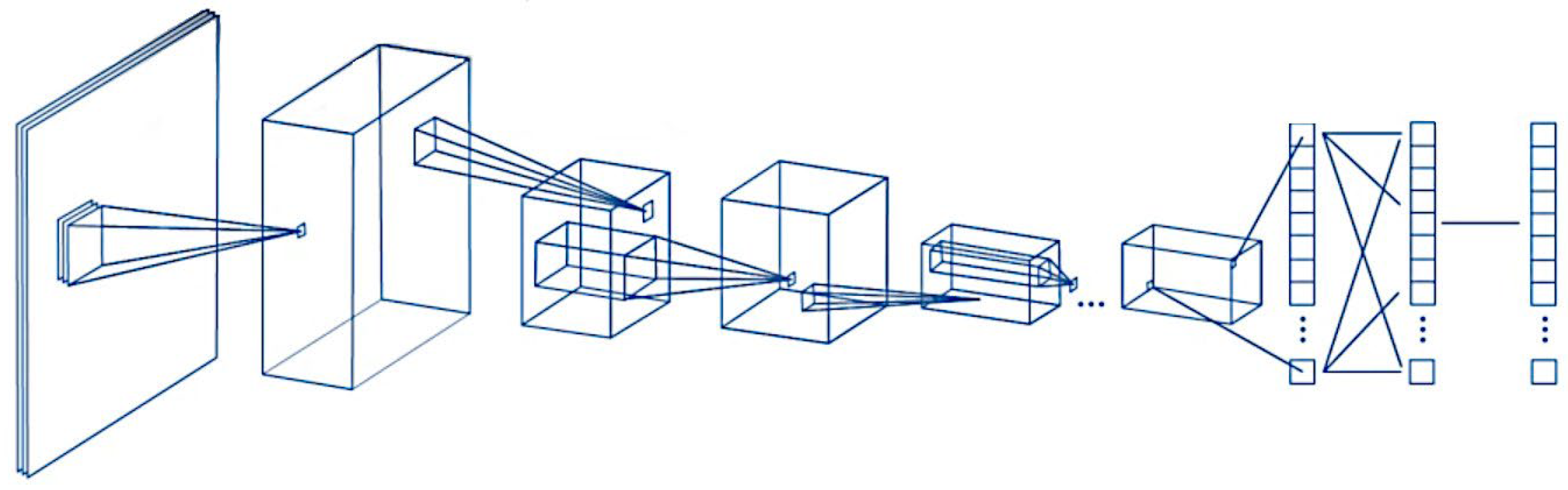
General design of the convolutional neural network. A convolutional neural network (CNN) is a deep learning algorithm that takes an input image, assigns weights/biases to different components of the image, and classifies all the image. There are three major components of the network: the convolutional layer, the pooling layer, and the fully connected layer.
Figure 1.
General design of the convolutional neural network. A convolutional neural network (CNN) is a deep learning algorithm that takes an input image, assigns weights/biases to different components of the image, and classifies all the image. There are three major components of the network: the convolutional layer, the pooling layer, and the fully connected layer.
Figure 2.
Structure of the convolutional neural network of this study (based on ResNet-18).
Figure 2.
Structure of the convolutional neural network of this study (based on ResNet-18).
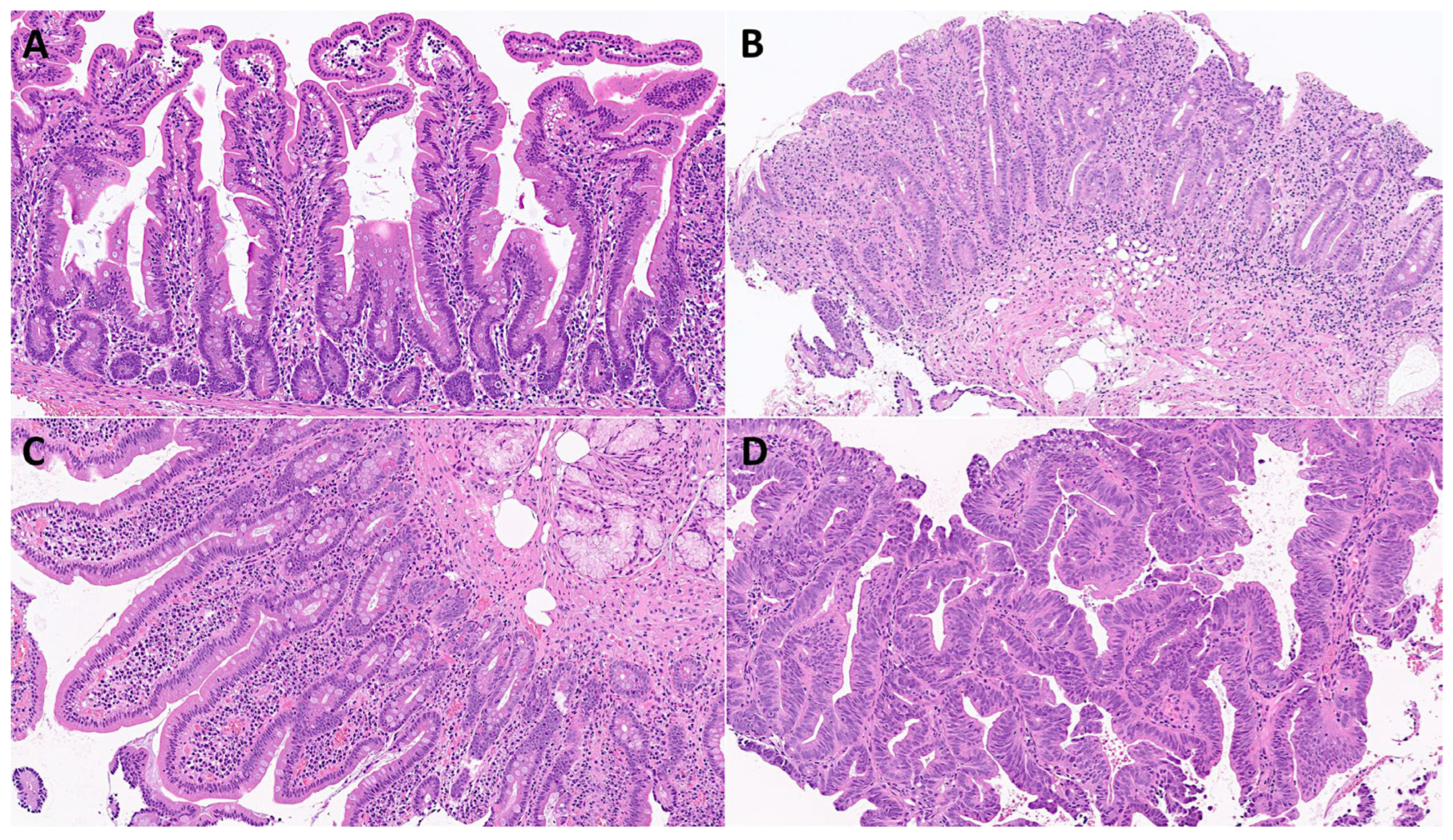
Figure 3.
Characteristic histological images of the small intestine (duodenum). Duodenal control (A); celiac disease (C); inflammatory duodenum (D); duodenal adenocarcinoma (E).
Figure 3.
Characteristic histological images of the small intestine (duodenum). Duodenal control (A); celiac disease (C); inflammatory duodenum (D); duodenal adenocarcinoma (E).
Figure 4.
Images of celiac disease. The input size is 224-by-224 (224×224×3).
Figure 4.
Images of celiac disease. The input size is 224-by-224 (224×224×3).
Figure 5.
Images of small intestine control. The input size is 224-by-224 (224×224×3). This figure shows images of the ileum. Additionally, images obtained from the duodenum were included in the dataset.
Figure 5.
Images of small intestine control. The input size is 224-by-224 (224×224×3). This figure shows images of the ileum. Additionally, images obtained from the duodenum were included in the dataset.
Figure 6.
Images of inflammatory small intestine. The input size is 224-by-224 (224×224×3).
Figure 6.
Images of inflammatory small intestine. The input size is 224-by-224 (224×224×3).
Figure 7.
Images of duodenal adenocarcinoma. The input size is 224-by-224 (224×224×3).
Figure 7.
Images of duodenal adenocarcinoma. The input size is 224-by-224 (224×224×3).
Figure 8.
Training progress of the convolutional neural network for the classification of celiac disease and small intestine control.
Figure 8.
Training progress of the convolutional neural network for the classification of celiac disease and small intestine control.
Figure 9.
Confusion matrix of celiac disease and small intestine control. This image shows the confusion matrix of the test set, which includes the analysis of images not previously used in the training and validation steps (i.e., the holdout data). The accuracy of predicting celiac disease was 99.97%.
Figure 9.
Confusion matrix of celiac disease and small intestine control. This image shows the confusion matrix of the test set, which includes the analysis of images not previously used in the training and validation steps (i.e., the holdout data). The accuracy of predicting celiac disease was 99.97%.
Figure 10.
Confusion matrix of celiac disease, duodenal inflammation, and small intestine control. This image shows the confusion matrix of the test set, which includes the analysis of images not previously used in the training and validation steps (i.e., the holdout data).
Figure 10.
Confusion matrix of celiac disease, duodenal inflammation, and small intestine control. This image shows the confusion matrix of the test set, which includes the analysis of images not previously used in the training and validation steps (i.e., the holdout data).
Figure 11.
Confusion matrix of celiac disease, duodenal adenocarcinoma, duodenal inflammation, and small intestine control. This image shows the confusion matrix of the test set, which includes the analysis of images not previously used in the training and validation steps (i.e., the holdout data).
Figure 11.
Confusion matrix of celiac disease, duodenal adenocarcinoma, duodenal inflammation, and small intestine control. This image shows the confusion matrix of the test set, which includes the analysis of images not previously used in the training and validation steps (i.e., the holdout data).
Table 1.
Performance parameters of the classification into 2 classes.
Table 1.
Performance parameters of the classification into 2 classes.
| Class |
Accuracy (%) |
Precision (%) |
Recall (%) |
F1-score (%) |
Specificity (%) |
False positive rate (%) |
| Celiac disease |
99.97 |
99.93 |
100 |
99.97 |
99.96 |
0.04 |
| Small intestine control |
99.97 |
100 |
99.96 |
99.96 |
100 |
0 |
Table 2.
Performance parameters of the classification into 3 classes.
Table 2.
Performance parameters of the classification into 3 classes.
| Class |
Accuracy (%) |
Precision (%) |
Recall (%) |
F1-score (%) |
Specificity (%) |
False positive rate (%) |
| Celiac disease |
99.68 |
99.59 |
99.32 |
99.45 |
99.83 |
0.17 |
| Duodenal inflammation |
98.37 |
96.48 |
96.72 |
96.60 |
98.89 |
1.11 |
| Small intestine control |
98.41 |
98.28 |
98.32 |
98.30 |
98.49 |
1.51 |
Table 3.
Performance parameters of the classification into 4 classes.
Table 3.
Performance parameters of the classification into 4 classes.
| Class |
Accuracy (%) |
Precision (%) |
Recall (%) |
F1-score (%) |
Specificity (%) |
False positive rate (%) |
| Celiac disease |
99.69 |
99.73 |
99.05 |
99.39 |
99.91 |
0.09 |
| Duodenal adenocarcinoma |
97.41 |
85.64 |
93.96 |
89.61 |
97.88 |
2.12 |
| Duodenal inflammation |
96.42 |
94.05 |
89.33 |
91.63 |
98.41 |
1.59 |
| Small intestine control |
98.38 |
97.85 |
98.15 |
98.00 |
98.53 |
1.47 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).