1. Introduction
The foundation of human civilization is the agricultural sector, which supplies the resources required for both survival and economic growth [
1]. Yet, there are many obstacles this vital sector must overcome, and among the most dangerous are plant diseases. Plant diseases reduce crop production while also compromising food quality and safety, resulting in major economic losses and affecting food security [
2,
3,
4,
5]. In order to effectively manage and mitigate these illnesses and maintain the stability and productivity of agricultural practices, early and precise detection of these diseases is necessary [
6,
7,
8].
Traditionally, plant disease diagnosis was mainly reliant on manual inspection by trained agronomists and farmers [
9]. Although this method worked well, it was labor-intensive, time-consuming, and frequently subjective, which results in inconsistent disease diagnosis. Furthermore, the swift dissemination of plant illnesses and the introduction of novel pathogens demand a more accurate and scalable diagnostic methodology. An encouraging answer to these problems is the development of deep learning and computer vision technologies, which make automated, precise, and effective plant disease identification through picture analysis possible [
10,
11]. One major example is the convolutional neural networks (CNNs).
With its exceptional performance in a variety of fields, such as autonomous driving [
12], facial recognition [
13], and medical imaging [
14], CNNs have completely changed the area of image classification. CNNs have demonstrated considerable potential in agriculture for recognizing diseases in plants from photos of its several parts, including leaves, stems, and fruits [
15,
16,
17,
18]. However, because there are intrinsic trade-offs between model complexity, depth, and feature extraction capabilities, no single CNN design can be said to be the best for all kinds of image classification applications. This in turn called for the idea of model hybridization.
Deep learning has witnessed a surge in interest in hybrid models in recent times. In order to take advantage of their complimentary qualities and improve overall performance, hybrid models blend several neural network architectures. The reasoning behind this strategy is that certain designs are better at identifying patterns and extracting features in different ways. Hybrid models, by combining these diverse qualities, can achieve higher accuracy and robustness than standalone models [
17,
19].
In a number of fields, including medical diagnosis, hybrid models have been effectively used. By fusing CNNs and Recurrent Neural Networks (RNNs) to investigate long-term survival of subjects in a lung cancer screening, illness prediction using sequential data has become more accurate [
20]. Hybrid models that combine CNNs and Transformers have improved sentiment analysis and text categorization in the field of natural language processing [
21].
In the context of agriculture, plant disease detection specifically, Poornim et al. (2023) [
22] designed a lightweight ‘VGG-ICNN’ hybrid model by training on PlantVillage dataset, Embrapa dataset and single crop datasets. They obtained a 99.61% accuracy on PlantVillage set, while establishing success for the other datasets utilized. Chin Poo Lee et al. (2023) [
19] developed a robust ensemble model called Plant-CNN-Vi by combining four pre-trained models; Vision Transformer, ResNet-50, DenseNet-201, and Xception, respectively. Other relevant literature were summarized and presented as
Table 1.
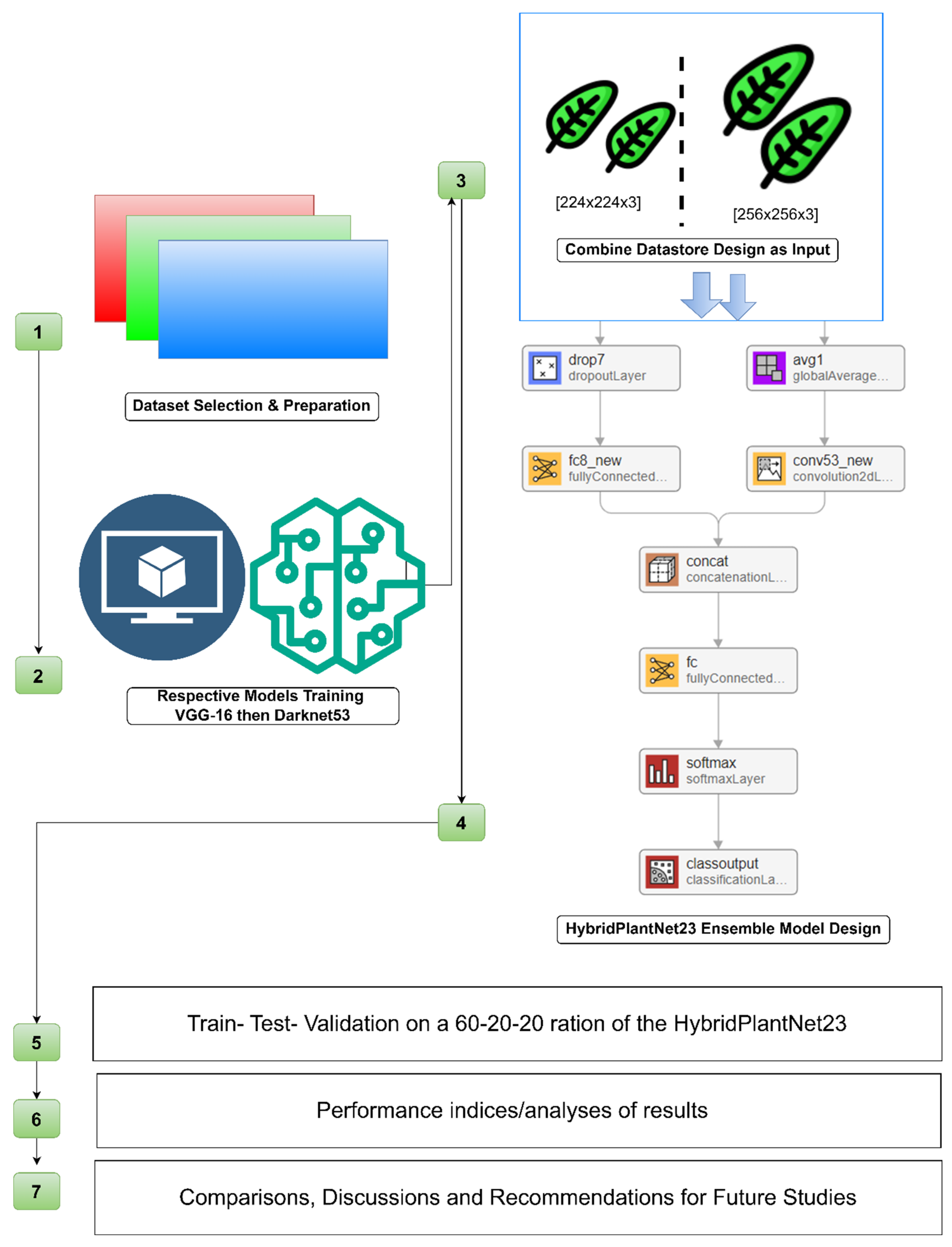
In this study, we investigated the effectiveness of integrating two robust CNN architectures, VGG16 and Darknet53, into HybridPlantNet23, to improve plant disease classification systems. VGG16, created by the Visual Geometry Group at Oxford, is known for its simplicity and depth, with 16 layers that excel in extracting hierarchical elements from images. Darknet53, the backbone of the YOLO (You Only Look Once) object identification framework, is a more complicated and deep network with 53 convolutional layers that is well-known for its resilient feature extraction and exceptional performance in a variety of computer vision tasks. Utilizing the complementing qualities of both architectures is the driving force behind this hybrid strategy. The design of VGG16 makes it easier to extract fine-grained, low-level characteristics, which are important for recognizing small variations in plant disease symptoms. Darknet53’s richer structure allows it to capture high-level, abstract information, which improves the model’s capacity to generalize across disease kinds and environmental situations. We hope to construct a more robust and accurate model capable of classifying a diverse spectrum of plant illnesses by merging these architectures. The study has practical ramifications for farmers, agronomists, and agricultural researchers, making it significant outside the confines of academia. A precise and dependable plant disease classification system can enable prompt and targeted interventions, lowering crop losses and increasing output quality. In addition, the implementation of these systems in farming operations can improve disease surveillance, facilitate precision farming methods, and foster the growth of sustainable agriculture.
Figure 1.
Values of Ensemble Modeling.
Figure 1.
Values of Ensemble Modeling.
2. Materials and Methods
2.1. Dataset
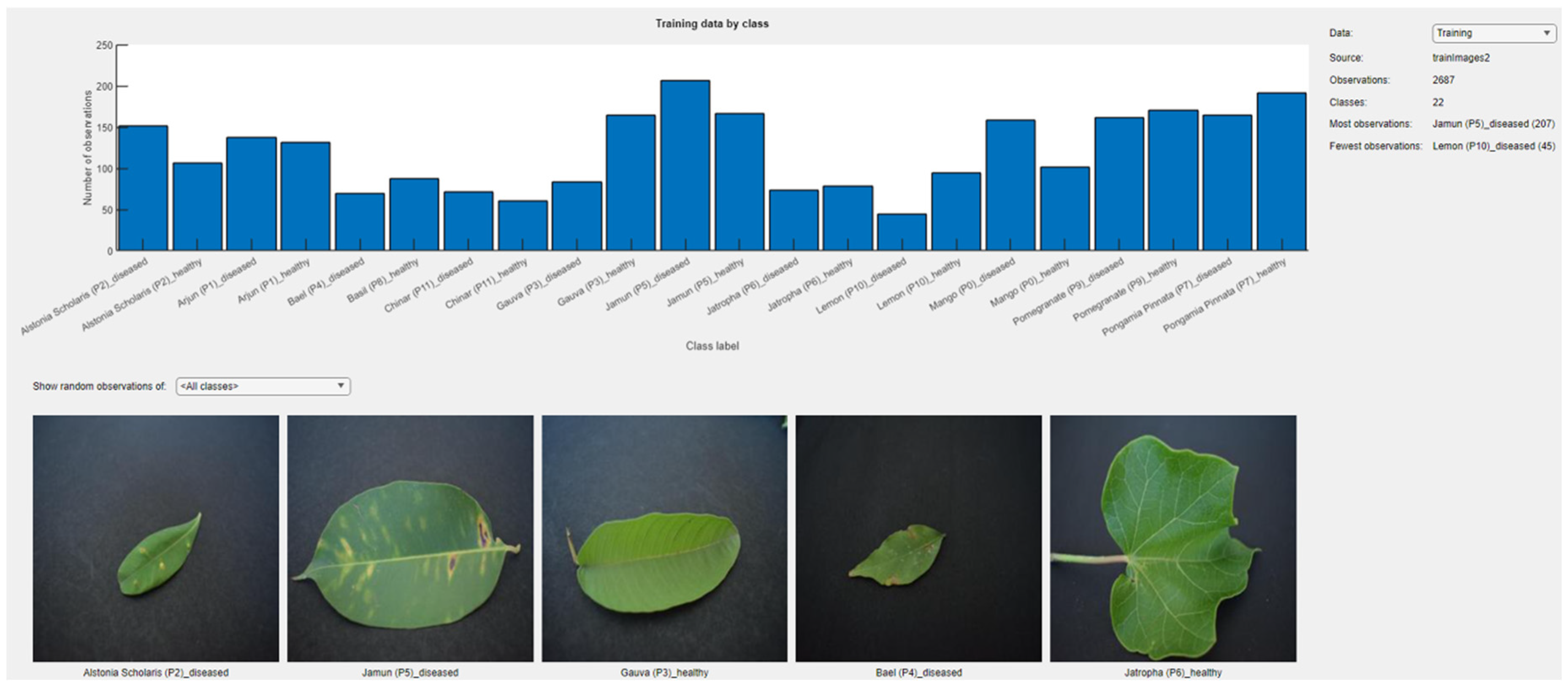
The dataset used in this study is A Database of Leaf Images [
33] available on the Mendeley Data source. It consists of images of 12 different plant diseases. The dataset’s summary is given in
Table 2 while its class samples could be seen in
Figure 2. The dataset was chosen for this study owing to previously conducted study justifying its superior quality and high significance for deep learning application, through its GLCM metrics rigorous analyses and concurrence with deep learning train-test results as in [
34]. To better understand the class samples labelled in series, the sequential numbering utilized for this studies is given as in
Table 3.
2.2. Dataset Preprocessing
The dataset’s raw images were 4000x6000x3 pixels in size. The photographs were scaled to comply with the input size specifications of the pretrained models VGG16 and Darknet53. The input size requirements for VGG16 and Darknet53 are 224x224x3 and 256x256x3, respectively. This scaling was necessary to guarantee that the convolutional layers of these models could process the images efficiently and without experiencing any dimensionality problems.
MATLAB R2023b was used for all preprocessing operations and the ensuing model training. This MATLAB edition is ideal for managing big datasets and intricate neural network designs because it offers sophisticated tools and functions for deep learning, image processing, and integration with pretrained models.
2.3. Model Architecture
The proposed hybrid model combines the VGG16 and Darknet53 architectures, as to be shown. The final layers of both networks are concatenated to form a more comprehensive feature representation, followed by fully connected layers, a softmax layer, and a classification layer. To further understand the concept, the respective models are initially described as follows:
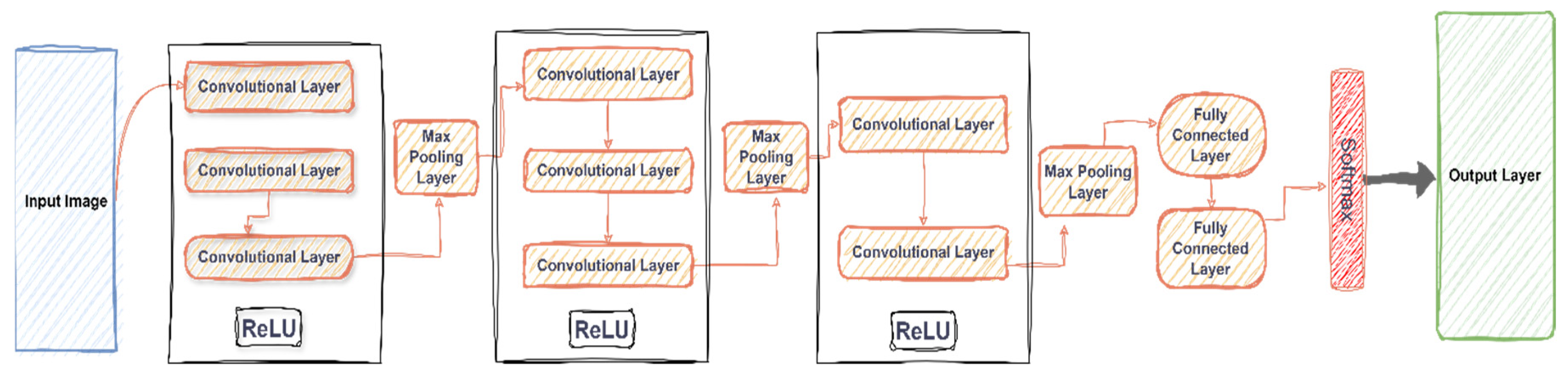
2.3.1. VGG16
VGG16 is a widely used CNN architecture known for its simplicity and depth, consisting of 16 weight layers. It is adept at extracting hierarchical features from images. VGG16 was introduced in the year 2015 as an advanced convolutional neural network model by Simonyan and Zisserman [
35] from the University of Oxford. Using ImageNet, a dataset containing over 14 million images divided into 1000 classes, the model attains 92.7% top 5 test accuracy. By sequentially replacing the large kernel-sized filters (11 and 5 in the first and second convolutional layers, respectively) with multiple 3×3 kernel-sized filters, it proves to be an enhanced version of AlexNet. In this study, VGG16 and Darknet53 were selected to develop HybridPlantNet23 due to its superb accuracy that it showed with the same selected dataset herein, confirmed from another study conducted by the authors, which is yet to be published by Springer Nature as a book chapter under the series name “Proceedings of Science and Technology”. A general concept of how VGG-16 works is provided in
Figure 3.
An RGB image with a fixed size of 224 by 224 is the input for the convolution layer 1. The image is fed through a series of convolutional layers, where filters are applied with a little receptive field of 3x3. This is the smallest size that can effectively capture left/right, up/down, and center concepts. As a linear transformation of the input channels, one of the configurations additionally makes use of 1×1 convolution filters. The spatial padding of the convolution layer input is set to preserve the spatial resolution after the convolution, meaning that for 3x3 convolution layers, the padding is 1 pixel. The convolution stride is fixed at 1 pixel. The five max-pooling layers, which come after parts of the convolutional Layers, handle spatial pooling. Using stride 2, max-pooling is applied to the 2x2 pixel window. The stack of convolutional layers is followed by three Fully-Connected (FC) layers, each with 4096 channels. The third FC layer, which conducts 1000-way ILSVRC classification, has 1000 channels in total. The convolutional layers vary in depth and architecture. The softmax layer comes in last. Every network has the same configurations for the completely connected tiers. Every buried layer has non-linearity in rectification (ReLU). Notedly, all but one of the networks lack Local Response Normalization (LRN); this normalization results in higher computation and memory usage but does not enhance performance on the ILSVRC dataset [
36,
37,
38,
39].
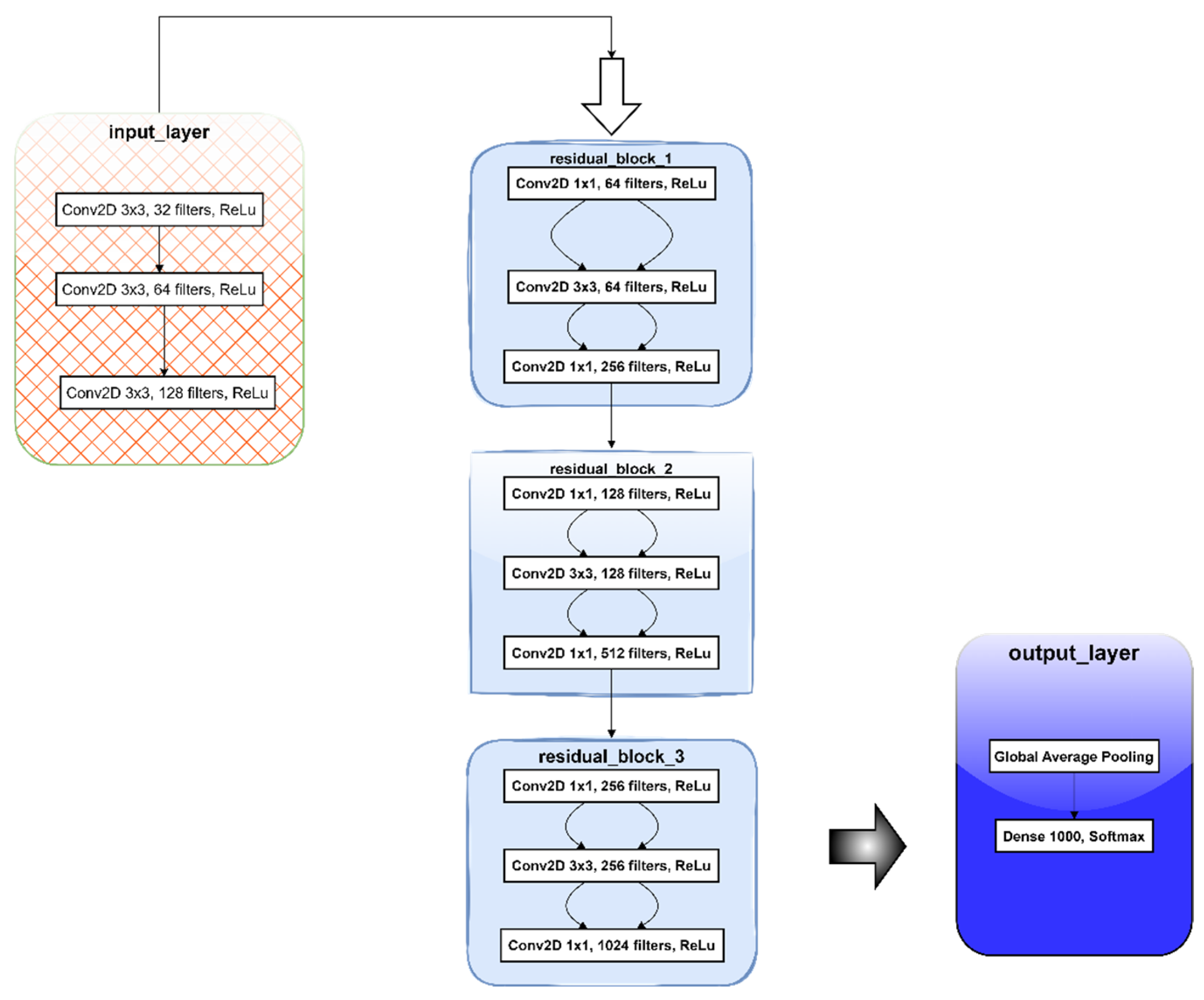
2.3.2. Darknet53
Darknet-53 is a convolutional neural network (CNN) architecture that serves as the backbone for the YOLOv3 (You Only Look Once, version 3) object detection system. It is an advancement over earlier iterations of YOLO, created especially to improve the precision and effectiveness of object identification activities [
40]. The advantages of this model include improved accuracy, efficiency, and versatility. A schematic of the model’s general concept is provided in
Figure 3.
Darknet-53, as the name implies, has 53 convolutional layers that are deeper than the 19 layers that make up Darknet-19, the foundation of YOLOv2. The network can learn more intricate features and improves detection accuracy with the increased depth. A combination of 1x1 and 3x3 convolutional filters is used by the network. While the 1x1 filters are used for dimension reduction, which lowers computational complexity, the 3x3 filters are utilized to capture spatial characteristics. The residual connections in Darknet-53 are modeled after ResNet (Residual Networks). With a shortcut link that adds the block’s input to its output, each residual block is made up of two convolutional layers. By making these connections, the vanishing gradient issue is lessened and the network is able to train more efficiently. Bottleneck layers with 1x1 convolutions are employed within residual blocks to minimize the number of input channels, thus assisting in lowering the computational cost without sacrificing performance [
41,
42,
43].
By normalizing the output of each layer, batch normalization, when applied after each convolutional layer, aids in stabilizing and speeding up the training process. The network can learn more intricate representations by introducing non-linearity through the use of the Leaky Rectified Linear Unit (Leaky ReLU) activation function. By permitting a little, non-zero gradient while the unit is dormant, the leaky version aids in the prevention of dead neurons. The model employs global average pooling at the final stage of the convolutional layers in place of fully linked layers. As a result, a feature vector for every class is produced, bringing the spatial dimensions down to 1x1. Depending on the particular application, the resultant feature vector from global average pooling is then processed through fully connected layers to carry out classification or regression tasks [
41,
42,
44].
Darknet-53’s main responsibility in YOLOv3 is to extract features from the input pictures [
40]. Remaining connections in the deeper network facilitate the capture of more abstract and detailed data, which are essential for precise object detection.
Figure 4.
Schematic of Darknet53 Model’s architecture.
Figure 4.
Schematic of Darknet53 Model’s architecture.
2.3.3. HybridPlantNet23
HybridPlantNet23 is an enhanced model ensembled with the purpose of precisely identifying and categorizing plant diseases. In order to attain excellent performance in recognizing plant illnesses, this model carefully integrates the strengths of two potent convolutional neural network (CNN) architectures: Darknet53 and VGG-16. Using a large dataset of pictures of different plant species in both healthy and disease conditions, the VGG-16 and Darknet53 ensemble approach in HybridPlantNet23 trains both models independently. Using its special advantages, each model gains the ability to distinguish different features from the input images. Its major architectural descriptions are presented and briefly discussed in
Table 4. As it could be seen vividly from stage number 4 of the methodology, provided in
Figure 5, the layers utilized and the arranged is presented.
2.4. Combined Datastore
To guarantee that each model received images of the right size, separate datastores were established for VGG16 and Darknet53. For this, MATLAB’s imageDatastore function was utilized, which effectively manages big image collections. Preprocessing pipelines were included in each datastore to resize images to the necessary sizes of 256x256x3 for Darknet53 and 224x224x3 for VGG16, respectively. As is often done in image preparation, these pipelines also normalized pixel values by removing the mean and dividing by the standard deviation.
To create a merged feature set, the feature vectors that were extracted from VGG16 and Darknet53’s penultimate layers were concatenated. With VGG16 being particularly good at capturing fine-grained textures and Darknet53 being particularly good at larger-scale features, this combination potentially made the most of both models’ strengths.
2.5. Training
The hybrid model is trained using 2687 images (60%) of the dataset with an initial learning rate of 0.01. The categorical cross-entropy loss function is used to handle the multi-class classification problem. The model’s selected training parameters show a carefully calculated trade-off between model accuracy and training speed. Given the depth and complexity of the combined VGG-16 and Darknet53 architectures, the model’s convergence is accelerated by the relatively high initial learning rate.
To minimize the risk of overfitting, the model was trained with 30 epochs, which is enough to teach it all the complex properties needed to detect plant diseases accurately. The conventional selection of 32 mini-batch sizes offers a fair balance between the computational efficiency of the training process and the stability of gradient estimations. For every epoch, the data was shuffled and validated to verify that the model generalizes effectively and its performance is periodically reviewed, allowing for fast modifications supposing the model begins to overfit.
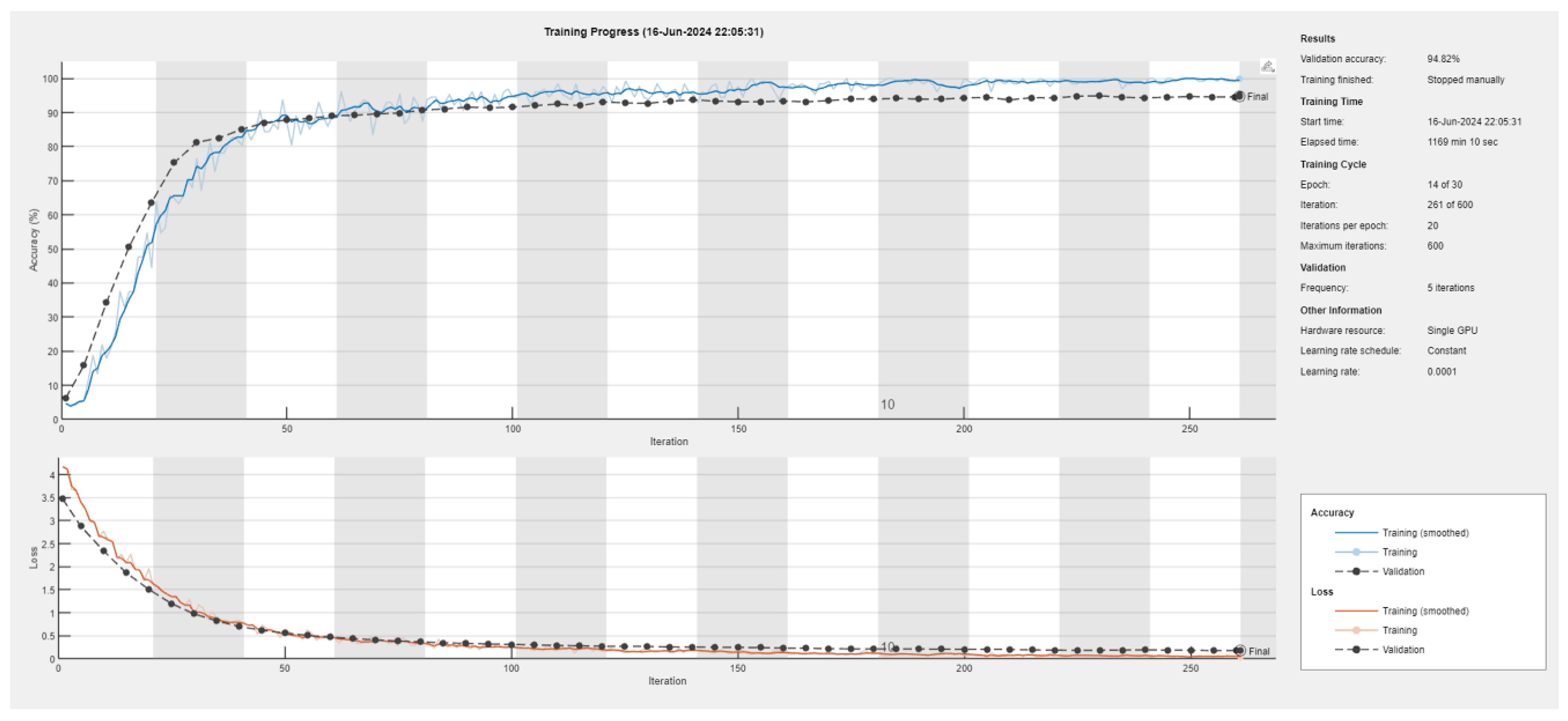
Figure 6 provides the training progress of the ensemble model. The top graph displays the training and validation accuracy over respective iterations. The blue line is representing the training accuracy. Dashed black line visible in the figure is a representation of the validation accuracy. The bottom graph further displays the training and validation loss over iterations where red line presents the training loss. The validation loss is represented by the dashed black line.
The model’s validation accuracy was recorded as 94.82%. This high validation accuracy shows how well it performs on unseen data, hence indicating effective learning and generalization. The training process was halted manually because the model had already reached satisfactory performance levels, a recommended by experts and professionals in the field. A significant computational effort is indicated by the total training time elapsed, which is typically necessary for training deep learning models with large datasets. The model was trained for 14 epochs out of the planned 30, suggesting early stopping due to reaching satisfactory performance. Furthermore, it only completed 261 iterations out of the maximum 600 planned iterations.
To ensure frequent monitoring and adjustment of the training process, the model’s performance was validated in every 5 iterations. Single GPU and constant learning rate (0.0001) were utilized straight throughout the training phase of the model. The relatively low learning rate is necessary for careful weight adjustments, which helps in achieving fine-tuned model performance.
2.6. Performance Evaluation
Total images used to test and validate the model were 908 and 907 images, representing a 20% and 20%, respectively, of the total images. The model’s performance is evaluated using standard metrics such as accuracy, precision, recall, and F1-score, which calculated using equations 1- 4 respectively. The results are compared with those of standalone VGG16 and Darknet53 models, earlier trained before the ensemble model, to demonstrate the effectiveness of the hybrid approach.
3. Results
This section provides a concise and precise description of the results and their interpretation.
4. Discussion
The training, test and performance metrics observed in this study indicate a scientific progress and reliability of these models in plant disease classification. To further digest the results, the following itemizations will be utilized:
4.1. Single Models Classification
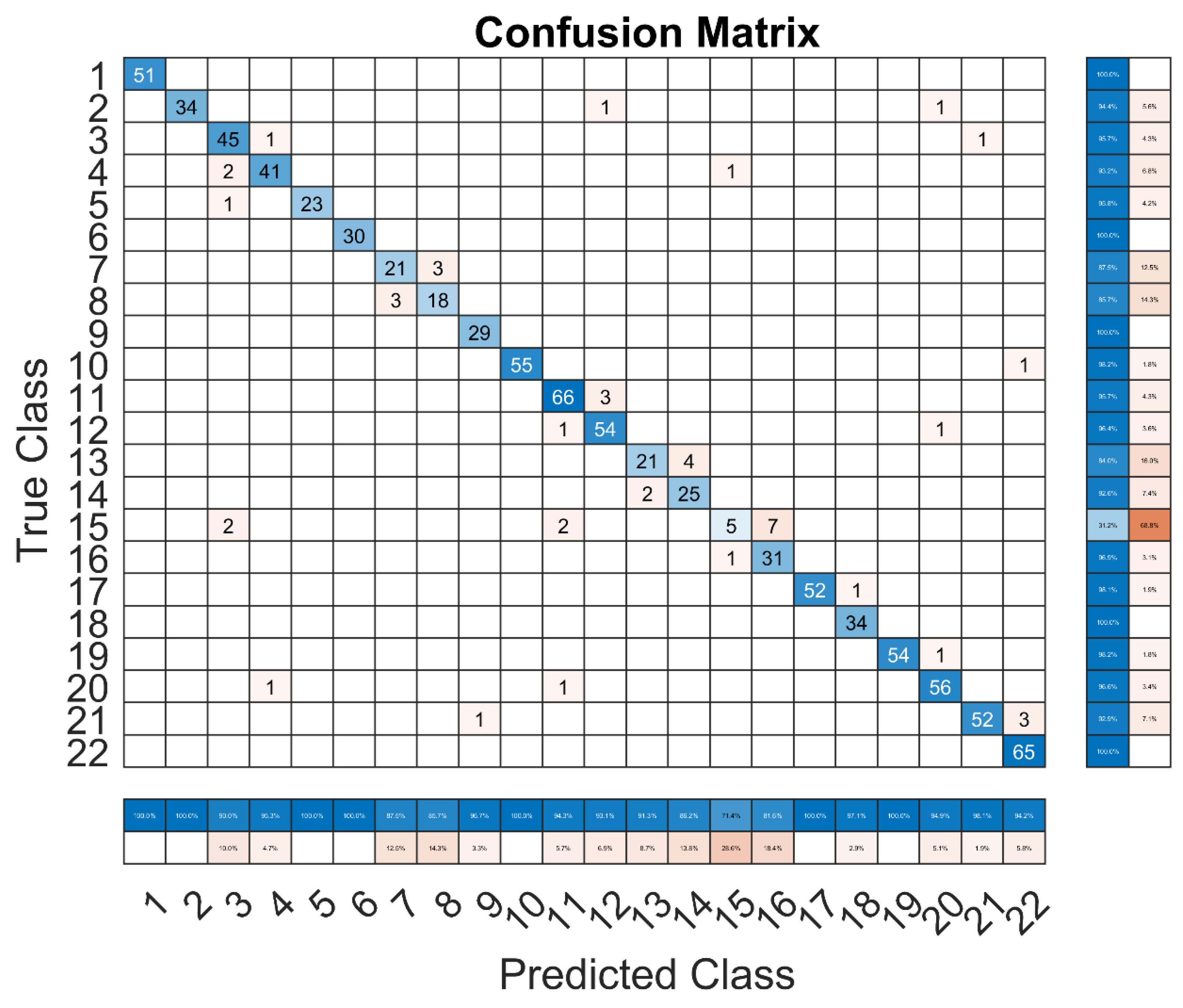
The performance across different classes is displayed in the Darknet53 model’s confusion matrix (
Figure 7). Although there are some noticeable instances where the model falls short, overall accuracy is quite great. As an example, class 15 (lemon) exhibits a notable decline in accuracy, which is reflected in the performance metrics and the confusion matrix.
Class 15, where the accuracy of the model is only 31.2%, is where it struggles the most. This implies that the model has trouble differentiating between other classes and lemons. In the dataset, class 15 had the fewest photos, suggesting that there is a class imbalance. Higher misclassification rates may result from the model’s under fitting of this class as a result of this imbalance. Moreover, Class 7 performs better with an accuracy of 87.5%, while class 8 has an accuracy of 85.7%. These classes still exhibit some misclassification, but the effect is less severe than in class 15. This suggests that even with a certain amount of misclassification, the model works fairly well on these classes overall.
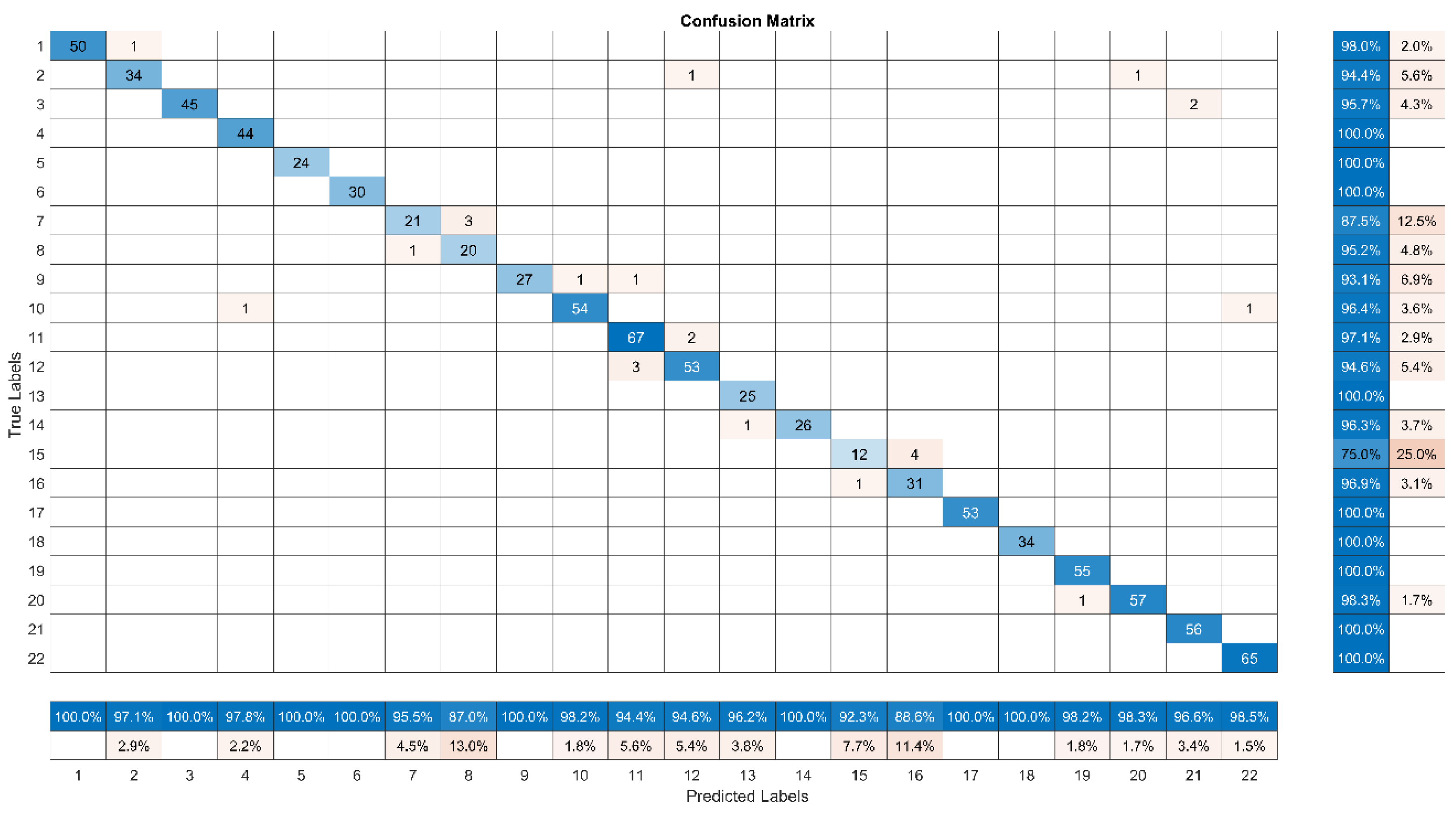
Compared to Darknet53, the VGG16 confusion matrix (
Figure 8) displays a more balanced performance across classes, as well as higher overall accuracy, precision, recall, and F1-score.
Although class 15’s accuracy is 75%, it is still significantly lower than Darknet53, indicating a considerable issue in accurately categorizing lemons. This improvement implies that VGG16’s architecture may be able to manage the class imbalance marginally better than Darknet53’s, but the problem still exists because of the dataset’s inherent imbalance. Furthermore, Class 8 accuracy of 95.2% and class 7 accuracy of 87.5% demonstrate the robust performance of VGG16. The resilience of VGG16 in managing these classes is demonstrated by these results, which are probably attributable to its deeper network and superior feature extraction skills when compared to Darknet53.
4.2. HybridPlantNet23 Classification
The training accuracy of the ensemble model rapidly increases from the start and begins to plateau around 90-100 iterations. As the validation accuracy also follows a similar trend, this implies that the model generalizes effectively well to unseen data. After 200 iterations, both the training and validation accuracies stabilized around 95%, thus indicating the efficient and excellent performance of the model. From the training and validation loss over iterations observed earlier in
Figure 6, the training loss decreases rapidly at the beginning and then gradually levels off, thereby indicating the model’s learning capability and how it adjusts its weights effectively. The validation loss, as well, follows a similar trait, mirroring the training loss closely, thereby suggesting absence of overfitting in the model.
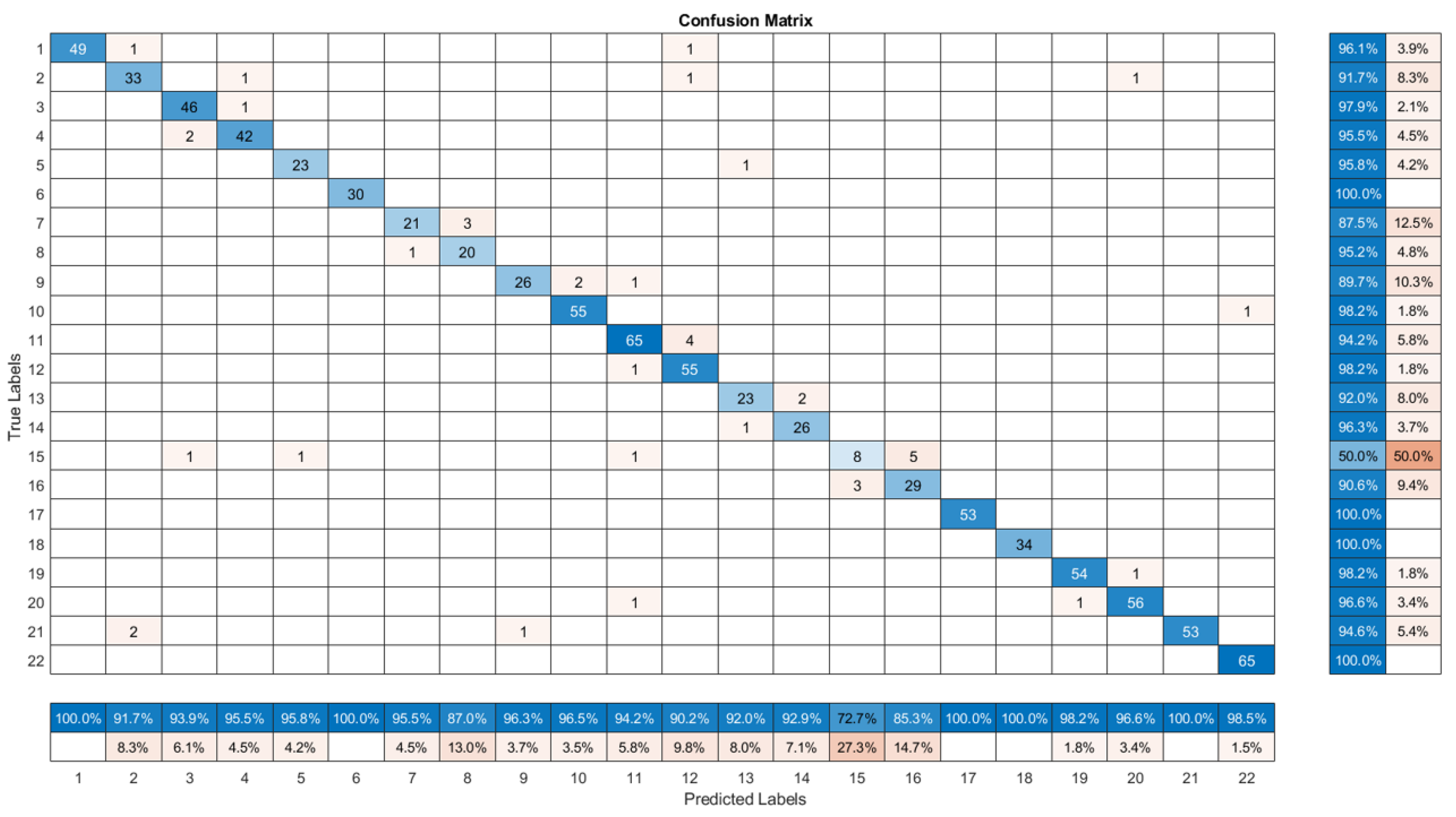
The provided confusion matrix in
Figure 9 presents a detailed view of the performance of the ensemble HybridPlantNet23 model in the act of classifying plant diseases as trained. Normally, a confusion matrix allows stakeholders to assess the model’s accuracy and identify any misclassifications accordingly. From the matrix, it could be realized that the majority of class entries are concentrated along the diagonal, which indicates the correct predictions for most classes. The vivid dark blue squares along the diagonal represent the number of correct predictions for each class. From a Class-Wise Accuracy point of view, 49 correct predictions, representing 96.1% accuracy was achieved for Class 1. In addition, 2 incorrect predictions were misclassified as other classes. Class 2 recorded 33 correct predictions, equivalent to 91.7% accuracy with only 3 misclassifications. Furthermore, class 3 correctly predicted 46 classes which is 97.9% accurate and only 1 misclassification. Moreover, class 4 has 42 correct predictions representing a 95.5% accuracy and 2 misclassifications respectively. Where class 5, class 17, class 18, and class 22, respectively recorded absolutely correct predictions with ideal (100%) accuracies, as this pattern continues for all classes, with accuracy rates ranging from 85.3% to 100%, the robustness of HybridPlantNet23 across various categories is established.
Misclassifications of the model are relatively low and scattered across different classes. With a 50% accuracy rate and 50% misclassified classes, Class 15 stand as the most significant misclassification occurs with the model. This occurs as the model struggles with this particular class, likely due to overlapping features with other classes and, more precisely, inadequate training data of the class. Furthermore, Misclassified classes frequently have misclassifications with nearby classes, suggesting that these classes may be similar. This could be true for classes 8 and 7, with 85.3% and 87.0% classification accuracy, respectively.
The overall high accuracy for most classes, in essence, indicates the model’s effectiveness in distinguishing between different plant diseases. The small number of incorrect classifications points to potential areas for model improvement—possibly through dataset expansion or architectural optimization. For the lower accuracy observed in class 15, 8 and 7, respectively, increasing their sample sizes, examining their features, and possibly implementing additional preprocessing strategies to enhance feature differentiation is recommendable for improved classification results. Moreover, the high variability within Class 7 images, is making it challenging for the model to learn a consistent pattern thus the need for some better and more standardized guidelines in plant disease datasets preparation and utilization.
4.2. Comparative Analysis and Insights
With superior accuracy of 97.25%, VGG16 appears to be the most reliable model in terms of correct predictions as seen in
Table 5. HybridPlantNet23 overtakes Darknet53 even though it falls short of VGG16 thus suggesting that: while ensemble technique improves performance, VGG16’s standalone capability is superior. Furthermore, VGG16 leads with the highest precision (96.30%), indicating the least false positives. Since HybridPlantNet23 model improves upon Darknet53’s precision, this showcases the advantage of combining features from both models. In addition, VGG16’s recall (96.96%) being the highest suggests its effectiveness in identifying true positives. HybridPlantNet23, as well, demonstrated substantial progress over Darknet53, thus indicating a better classification of actual disease cases. Even though HybridPlantNet23 improves over Darknet53 in terms of balanced performance, the F1-Score in VGG16 identifies it as the highest (96.55%). This could somewhat justify that the ensemble approach benefits from the strengths of both models.
From the results of this study, it might seem counterintuitive for single model (VGG16) to outperform HybridPlantNet23, since ensemble models are generally expected to utilize the powers of their component models to deliver higher performance. Technically, in the methodology and dataset preprocessing stage, the images were resized to the requirements of the respective pretrained models (VGG16 and Darknet53). While this effort ensures compatibility, it may introduce slight inconsistencies in feature extraction when the combined datastore is formed. This would make it necessary to select single models that are having equal or similar image size requirements thus avoiding potential inconsistencies in feature extraction. Furthermore, the ensemble model relies fully on the combined datastore, which must balance the feature extraction methods of both VGG16 and Darknet53. Hence, any misalignment or bias in feature representation may have the potential to affect the HybridPlantNet23’s performance.
For cases of ensemble modelling where one model significantly outperforms the other (as VGG16 does), the ensemble has high tendency to be dragged down by the weaker model, which in turn could displace the overall performance gain expected from ensembling. This could highlight vividly the nature and complexity of ensemble models like HybridPlantNet23. In addition, the ensemble model’s architecture plays a decisive role, such as how it weighs the classifications from each single model. On this note, it could be iterated that where the combination strategy of HybridPlantNet23 does not optimally utilize the strengths of VGG16 and Darknet53, the ensemble’s performance might not surpass VGG16.
One important point to note is the effect of data distribution and class imbalance from the dataset utilized in this study. For example, with the same dataset utilized in this study, the authors trained a Darknet19 model with a balanced dataset of 77 images each from all classes, choosing 77 as the minimal number of images in the whole dataset which was a representation from class 15 (lemon) [
34]. Despite variations with different models and data sizes, class 15 always poses a threat to the overall performance of the corresponding model (
Table 6). The significant imbalance in the number of samples per class skews the learning process. Models tend to be biased towards classes with more samples, leading to higher performance for those classes and lower performance for minority classes like class 15. Despite being an ensemble model designed to leverage the strengths of VGG16 and Darknet53, HybridPlantNet23 also suffers from this problem. Class 15’s performance is particularly poor, which drags down the overall model performance. Furthermore, the overall performance of the model in the former study is significantly lower than that attained herein, despite the fact that the former study used a ‘balanced’ dataset, whereas herein the whole dataset was used, implying that the total number of images used in this study is greater. This suggests that for effective deep learning applications, the total number of images in datasets is more significant than the class balance.
5. Conclusions
This study thoroughly assessed the results of a hybrid ensemble model called HybridPlantNet23, Darknet53, VGG16, and other deep learning models on a job of classifying plant species. It was clear from a thorough investigation of confusion matrices and a rigorous analysis of performance indicators including accuracy, precision, recall, and F1-score that VGG16 consistently beat Darknet53 and the ensemble model. Due to its better architecture, VGG16 was able to manage the dataset’s complexity more skillfully, which raised its classification accuracy in the majority of classes. Class imbalance was still a problem, though, and it continued to have an impact on students’ performance, especially in underrepresented classes like class 15.
The study’s findings emphasize how crucial it is to solve problems with data distribution and class imbalance in order to enhance model performance. Although VGG16 showed resilience in both feature extraction and classification, the ensemble model’s total performance did not improve as anticipated, where scientific insights were discussed. Future research should concentrate on dataset balancing methods such sophisticated data augmentation and synthetic data synthesis. Furthermore, investigating more complex architectures and utilizing transfer learning may improve model robustness and classification accuracy even more. These findings highlight the necessity of balanced datasets and strong model architectures, and they provide the groundwork for further advances in the classification of plant species using deep learning.
Author Contributions
Conceptualization, M.K. and S.E.; methodology, M.K., S.E., and T.C.A. A - validation, M.K., S.E., and T.C.A.; resources M.K., and S.E.; data curation, M.K. and S.E.; writing—original draft preparation, M.K., S.E., and T.C.A. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by Firat University, ADEP Project No. 23. 23.
Data Availability Statement
The data can be shared up on request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Berners-Lee, M.; Kennelly, C.; Watson, R.; Hewitt, C.N. Current global food production is sufficient to meet human nutritional needs in 2050 provided there is radical societal adaptation. Elementa. 2018, 6. [Google Scholar] [CrossRef]
- Singh, S.; Singh, B.; Singh, A.P. Nematodes: A Threat to Sustainability of Agriculture. Procedia Environ. Sci. 2015, 29, 215–216. [Google Scholar] [CrossRef]
- Strange, R.N.; Scott, P.R. Plant Disease: A Threat to Global Food Security. Annu. Rev. Phytopathol. 2005, 43, 83–116. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, X. Plant diseases and pests detection based on deep learning: a review. Plant Methods. 2021, 17, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Horst, R.K. Plant Diseases and Their Pathogens. Westcott’s Plant Dis. Handb. 2001, 65–530. [Google Scholar] [CrossRef]
- Calderón, R.; Navas-Cortés, J.A.; Zarco-Tejada, P.J. Early detection and quantification of verticillium wilt in olive using hyperspectral and thermal imagery over large areas. Remote Sens. 2015, 7, 5584–5610. [Google Scholar] [CrossRef]
- Ferentinos, K.P. Deep learning models for plant disease detection and diagnosis. Comput. Electron. Agric. 2018, 145, 311–318. [Google Scholar] [CrossRef]
- Bouguettaya, A.; Zarzour, H.; Kechida, A.; Taberkit, A.M. A survey on deep learning-based identification of plant and crop diseases from UAV-based aerial images. Cluster Comput. 2022, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Buja, I.; Sabella, E.; Monteduro, A.G.; Chiriacò, M.S.; De Bellis, L.; Luvisi, A.; Maruccio, G. Advances in plant disease detection and monitoring: From traditional assays to in-field diagnostics. Sensors. 2021, 21, 1–22. [Google Scholar] [CrossRef]
- Valle, S.S.; Kienzle, J. Agriculture 4.0 ‐ Agricultural robotics and automated equipment for sustainable crop production, Food and Agriculture Organization of the United Nations, 2020. Available online: https://www.fao.org/3/cb2186en/CB2186EN.pdf (accessed on 17 August 2024).
- FAO, International Year of Plant Health 2020 | FAO | Food and Agriculture Organization of the United Nations, FAO. 2020. Available online: https://www.fao.org/plant-health-2020/home/en/ (accessed on 14 August 2022).
- Ghobadpour, A.; Boulon, L.; Mousazadeh, H.; Malvajerdi, A.S.; Rafiee, S. State of the art of autonomous agricultural off‐road vehicles driven by renewable energy systems, in: M. Becherif, H.S. Ramadan, M.T. Benchouia, M. Khaled, M. Ramadan, M. Hilairet (Eds.), Emerg. Renew. ENERGY Gener. Autom., ELSEVIER SCIENCE BV, SARA BURGERHARTSTRAAT 25, PO BOX 211, 1000 AE AMSTERDAM, NETHERLANDS, 2019: pp. 4–13. [CrossRef]
- Yang, Y.-X.; Wen, C.; Xie, K.; Wen, F.-Q.; Sheng, G.-Q.; Tang, X.-G. Face Recognition Using the SR-CNN Model. Sensors. 2018, 18, 4237. [Google Scholar] [CrossRef]
- Mikołajczyk, A.; Grochowski, M. Data augmentation for improving deep learning in image classification problem, in: 2018 Int. Interdiscip. PhD Work. IIPhDW 2018, Institute of Electrical and Electronics Engineers Inc., 2018: pp. 117–122. [CrossRef]
- Tibdewal, M.N.; Kulthe, Y.M.; Bharambe, A.; Farkade, A.; Dongre, A. Deep Learning Models for Classification of Cotton Crop Disease Detection. Zeichen J. 2022, 8. [Google Scholar]
- Sachan, R.; Kundra, S.; Dubey, A.K. Paddy Leaf Disease Detection using Thermal Images and Convolutional Neural Networks, in: Proc. Int. Conf. Comput. Intell. Sustain. Eng. Solut. CISES 2022, Institute of Electrical and Electronics Engineers Inc., 2022: pp. 471–476. [CrossRef]
- Azlah, M.A.F.; Chua, L.S.; Rahmad, F.R.; Abdullah, F.I.; Alwi, S.R.W. Review on techniques for plant leaf classification and recognition. Computers. 2019, 8. [Google Scholar] [CrossRef]
- P. A, B.K. S, D. Murugan, Paddy Leaf diseases identification on Infrared Images based on Convolutional Neural Networks, (2022). [CrossRef]
- Lee, C.P.; Lim, K.M.; Song, Y.X.; Alqahtani, A. Plant-CNN-ViT: Plant Classification with Ensemble of Convolutional Neural Networks and Vision Transformer. Plants. 2023, 12, 2642. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Aslani, S.; Zhao, A.; Shahin, A.; Barber, D.; Emberton, M.; Alexander, D.C.; Jacob, J. A hybrid CNN-RNN approach for survival analysis in a Lung Cancer Screening study. Heliyon. 2023, 9, e18695. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y. A Sentiment Analysis of Medical Text Based on Deep Learning. 2024. Available online: http://arxiv.org/abs/2404.10503 (accessed on 26 June 2024).
- Thakur, P.S.; Sheorey, T.; Ojha, A. VGG-ICNN: A Lightweight CNN model for crop disease identification. Multimed. Tools Appl. 2023, 82, 497–520. [Google Scholar] [CrossRef]
- Lee, S.H.; Chan, C.S.; Wilkin, P.; Remagnino, P. Remagnino, Deep‐plant: Plant identification with convolutional neural networks, in: Proc. ‐ Int. Conf. Image Process. ICIP, 2015: pp. 452–456. [CrossRef]
- Su, J.; Liu, C.; Coombes, M.; Hu, X.; Wang, C.; Xu, X.; Li, Q.; Guo, L.; Chen, W.H. Wheat yellow rust monitoring by learning from multispectral UAV aerial imagery. Comput. Electron. Agric. 2018, 155, 157–166. [Google Scholar] [CrossRef]
- Tan, J.W.; Chang, S.W.; Abdul-Kareem, S.; Yap, H.J.; Yong, K.T. Deep Learning for Plant Species Classification Using Leaf Vein Morphometric. IEEE/ACM Trans. Comput. Biol. Bioinforma. 2020, 17, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Chen, Z.; Yang, M.; Zhang, R.; Cui, Y. A multiscale fusion convolutional neural network for plant leaf recognition. IEEE Signal Process. Lett. 2018, 25, 853–857. [Google Scholar] [CrossRef]
- Kaya, A.; Keceli, A.S.; Catal, C.; Yalic, H.Y.; Temucin, H.; Tekinerdogan, B. Analysis of transfer learning for deep neural network based plant classification models. Comput. Electron. Agric. 2019, 158, 20–29. [Google Scholar] [CrossRef]
- Pearline, S.A.; Kumar, V.S.; Harini, S. A study on plant recognition using conventional image processing and deep learning approaches, in: J. Intell. Fuzzy Syst., IOS Press, 2019: pp. 1997–2004. [CrossRef]
- Riaz, S.A.; Naz, S.; Razzak, I. Multipath Deep Shallow Convolutional Networks for Large Scale Plant Species Identification in Wild Image, in: Proc. Int. Jt. Conf. Neural Networks, 2020. [CrossRef]
- Litvak, M.; Divekar, S.; Rabaev, I. Urban Plants Classification Using Deep-Learning Methodology: A Case Study on a New Dataset. Signals. 2022, 3, 524–534. [Google Scholar] [CrossRef]
- Arun, Y.; Viknesh, G.S. Leaf Classification for Plant Recognition Using EfficientNet Architecture, in: Proc. IEEE 2022 4th Int. Conf. Adv. Electron. Comput. Commun. ICAECC 2022, 2022. [CrossRef]
- Beikmohammadi, A.; Faez, K.; Motallebi, A. SWP-LeafNET: A novel multistage approach for plant leaf identification based on deep CNN. Expert Syst. Appl. 2022, 202. [Google Scholar] [CrossRef]
- Chouhan, S.S.; Kaul, A.; Singh, U.P.; Jain, S. A Database of Leaf Images: Practice towards Plant Conservation with Plant Pathology. 2020. [Google Scholar] [CrossRef]
- Kabir, M.; Unal, F.; Akinci, T.C.; Martinez-Morales, A.A.; Ekici, S. Revealing GLCM Metric Variations across a Plant Disease Dataset: A Comprehensive Examination and Future Prospects for Enhanced Deep Learning Applications. Electron 2024, 13, 2299. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large‐scale image recognition, in: 3rd Int. Conf. Learn. Represent. ICLR 2015 ‐ Conf. Track Proc., International Conference on Learning Representations, ICLR, 2015. Available online: https://arxiv.org/abs/1409.1556v6 (accessed on 28 June 2024).
- Ahmad, A.; El Gamal, A.; Saraswat, D. Toward Generalization of Deep Learning-Based Plant Disease Identification Under Controlled and Field Conditions. IEEE Access. 2023, 11, 9042–9057. [Google Scholar] [CrossRef]
- Bi, C.; Xu, S.; Hu, N.; Zhang, S.; Zhu, Z.; Yu, H. Identification Method of Corn Leaf Disease Based on Improved Mobilenetv3 Model. Agronomy. 2023, 13, 300. [Google Scholar] [CrossRef]
- Wang, G.; Sun, Y.; Wang, J. Automatic Image-Based Plant Disease Severity Estimation Using Deep Learning. Comput. Intell. Neurosci. 2017, 2017. [Google Scholar] [CrossRef]
- Sarwar, F.; Griffin, A.; Rehman, S.U.; Pasang, T. Detecting sheep in UAV images - ScienceDirect. Comput. Electron. Agric. 2021, 187, 106219. [Google Scholar] [CrossRef]
- Won, J.H.; Lee, D.H.; Lee, K.M.; Lin, C.H. An Improved YOLOv3‐based Neural Network for De‐identification Technology, in: 34th Int. Tech. Conf. Circuits/Systems, Comput. Commun. ITC‐CSCC 2019, Institute of Electrical and Electronics Engineers Inc., 2019. [CrossRef]
- Wang, H.; Zhang, F.; Wang, L. Fruit classification model based on improved darknet53 convolutional neural network, in: Proc. ‐ 2020 Int. Conf. Intell. Transp. Big Data Smart City, ICITBS 2020, Institute of Electrical and Electronics Engineers Inc., 2020: pp. 881–884. [CrossRef]
- Rachburee, N.; Punlumjeak, W. An assistive model of obstacle detection based on deep learning: YOLOv3 for visually impaired people. Int. J. Electr. Comput. Eng. 2021, 11, 3434–3442. [Google Scholar] [CrossRef]
- Hamidon, M.H.; Ahamed, T. Detection of Tip-Burn Stress on Lettuce Grown in an Indoor Environment Using Deep Learning Algorithms. Sensors. 2022, 22, 7251. [Google Scholar] [CrossRef]
- Vasavi, S.; Priyadarshini, N.K.; Harshavaradhan, K. Invariant Feature-Based Darknet Architecture for Moving Object Classification. IEEE Sens. J. 2021, 21, 11417–11426. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).