Submitted:
28 July 2024
Posted:
31 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
- (1)
- Strategic U.S. national priorities;
- (2)
- eDNA technology readiness to support decision-making;
- (3)
- Emerging technologies within the strategic framework; and,
- (4)
- Implementation and adoption: from research to resource management.
2. Plenary Talk: Workshop introduction and the roll-out of the U.S. National Aquatic eDNA Strategy (Day 1)
- (1)
- “Coordinate Across Sectors to Facilitate Integration of Aquatic eDNA into Decision Making. This involves engaging cooperative mechanisms to align and promote standards and best practices for eDNA workflows across sectors, contributing technical readiness recommendations for priority applications and locales, and unifying communication strategies to enhance scientific literacy and data interpretation across all sectors;
- (2)
- Build Capacity, Infrastructure, and the Research Enterprise Needed to Employ Aquatic eDNA Technology at Scale by improving human capacity with training and education, meeting technical demands through infrastructure development — ranging from national sample repositories to interoperable data management structures, and supporting research and development to bolster and transition eDNA science into sustained operations; and
- (3)
- Advance Coordinated Aquatic eDNA Observations to Aid Assessments in U.S. Waters by harmonizing the extensive collection and delivery of eDNA data needed for robust and trustworthy metrics about aquatic health across the nation, resulting in an eDNA network to support national priorities and actions and to inform decisions that promote aquatic life and resilient ecosystems.”
- (a)
- Identifying priority sites and applications for aquatic eDNA sampling,
- (b)
- Implementing technological advances to build operational capacity, and
- (c)
- Operationalizing biological resource data for societal benefit”.
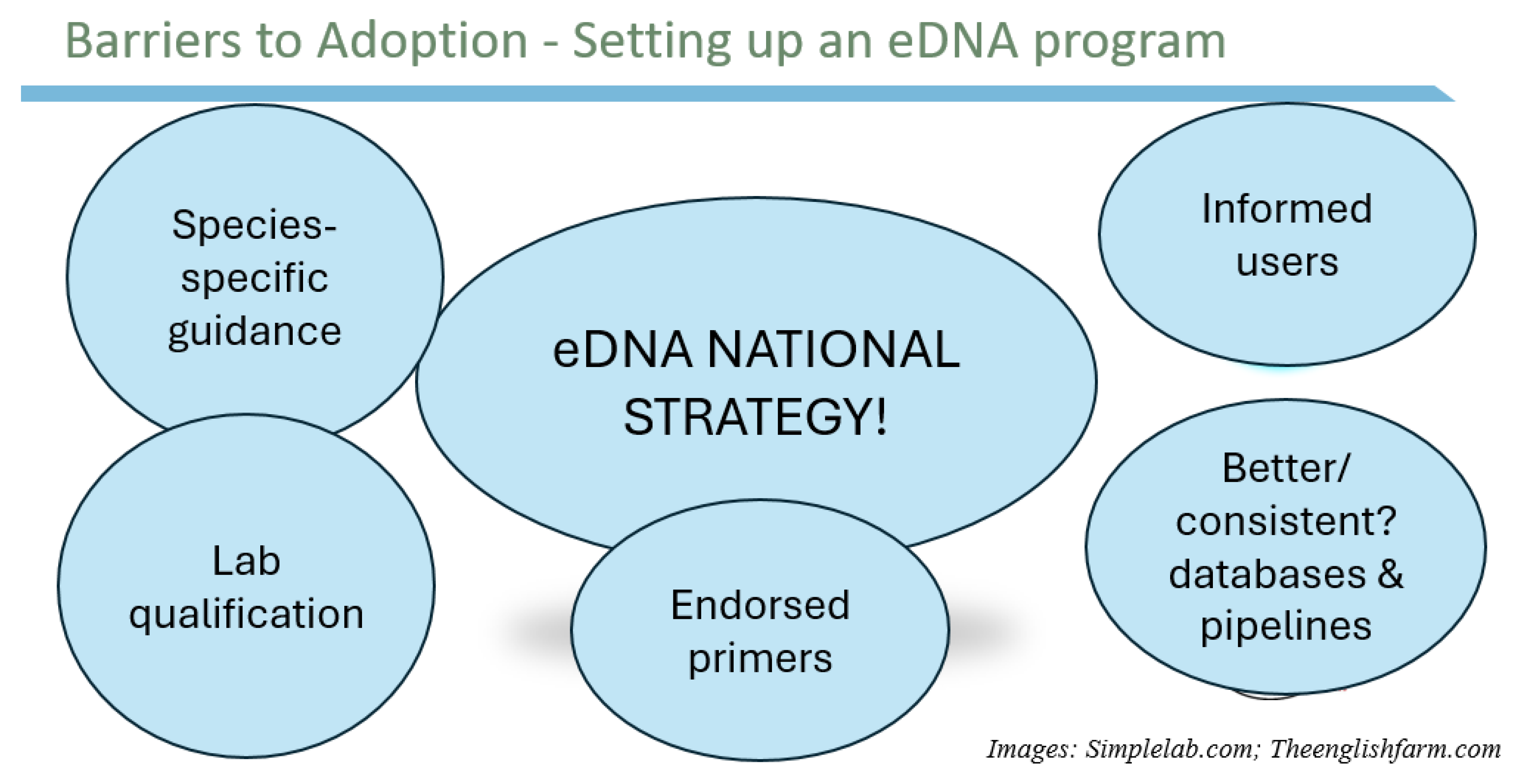
3. Strategic National Priorities for Aquatic eDNA, Adoption, and Alignment for Implementation
3.1. Strategic National Priorities for Marine and Aquatic eDNA
- (a)
- Improving field, laboratory and informatics infrastructure;
- (b)
- Better, faster, scalable sampling devices for all habitats;
- (c)
- Clean, high-throughput capacity (in labs and sequencing facilities);
- (d)
- Curated, voucher-based reference libraries;
- (e)
- Long-term biorepositories for verification and reuse; and,
- (f)
- Coordination of data workflows, computational resources, and information management”.
- (1)
- “Data infrastructure and interoperability, including data storage, analysis, visualization, and sharing of data and metadata – including bioinformatic tools, portals, and database for ASVs (amplicon sequence variants);
- (2)
- Training – provide eDNA workforce training and proficiency enhancement;
- (3)
- Sampling – seek automated solutions, low-cost sampling solutions, and better filtration for dilute targets and turbid conditions;
- (4)
- Accreditation – develop process and guidance for Accreditation/Certification/Validation/Quality Analysis-Quality Control (QA-QC);
- (5)
- Continued Research and Development – continued assay development and study topics to include: fate and transport, eRNA, comparisons with standard monitoring, parameterize uncertainty, and population information from eDNA,
- (6)
- Hubs – establish technology hubs and centers of excellence to accelerate technology development and provide inclusive training,
- (7)
- Reference materials – develop standard reference materials,
- (8)
- Reference sequences – expand the database of genetic markers for vouchered species of interest. Generate and maintain reference sequence libraries,
- (9)
- Lab facilities – develop dedicated lab facilities for eDNA analysis,
- (10)
- Biorepositories – deposit and maintain voucher samples and their tissues and DNA in well-referenced museum facilities, and
- (11)
- Partnerships, including with industry for analytical scale-up, cost reduction (economy of scale), standardization, and market development.”
3.2. Broader Adoption – Models and Lessons Learned for eDNA
- (a)
-
Rare species, to
- (1)
- Inform Endangered Species Act compliance, and
- (2)
- Allow research on the species without capture or take;
- (b)
-
Invasive and nuisance species,
- (1)
- Examples: nutria, dreissenid (zebra and quagga) mussels, to verify local eradication,
- (2)
- Example: to develop tools to evaluate impacts of nuisance species, such as toxic algae;
- (c)
-
Biodiversity assessment, to
- (1)
- Efficiently track species assemblages in response to habitat restoration or species reintroduction; and,
- (d)
-
Pathogen monitoring, to
- (1)
- Establish pathogen presence and prevalence.
- (2)
- Example: Ceratonova shasta near salmon hatcheries”.
3.3. Alignment for Impactful eDNA Research and Implementation
- (1)
- “Reinforce cooperation on international standards,
- (2)
- Develop national standards, or
- (3)
- Let others do this first.”
- (1)
- Promote standardization of eDNA methods,
- (2)
- Reduce ecological survey time and costs,
- (3)
- Create widely available software for modeling regional biodiversity,
- (4)
- Raise end-user proficiency regarding the use of eDNA methods, and
- (5)
- Promote mainstream eDNA best practices in management, policy, and regulations”.
3.4. Emerging Technologies within the Strategic Framework
3.5. Emerging eDNA and eRNA Topics
3.6. eDNA Fate and Transport, and eDNA Data Models
- (a)
- “How long has the eDNA been there and where did it originate from?
- (b)
- Can sampling reconstruct the properties of source emission?
- (c)
- What about continuous source emissions vs. intermittent releases (e.g., skin cells vs. excrement)?, and
- (d)
- What are the model requirements for eDNA prediction?”
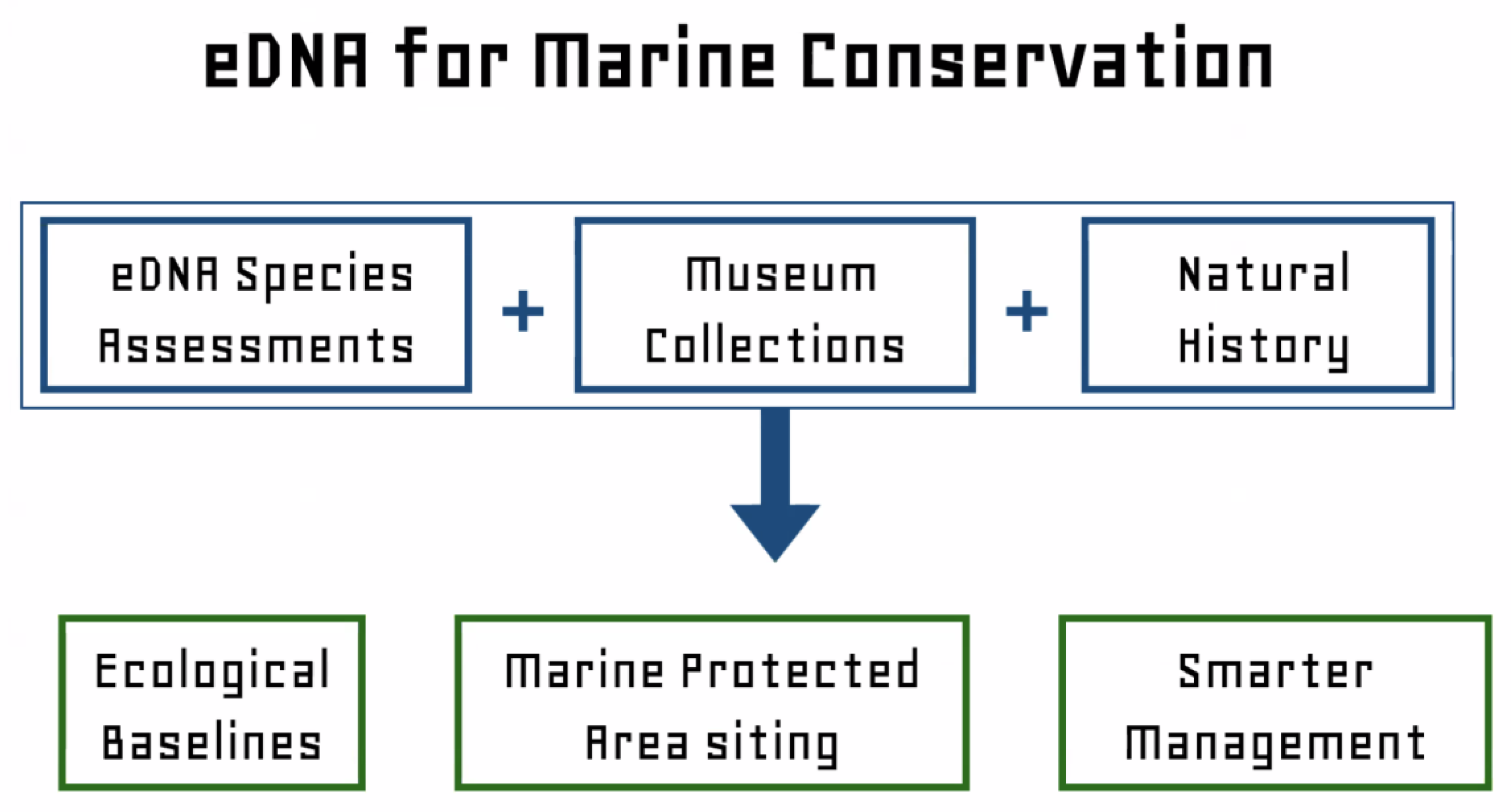
4. Capstone Talk by Dr. Enric Sala (Pristine Seas Program), and a Panel of Agency and Industry Representatives about the use of eDNA
5. Conclusions and Summary
Funding
Acknowledgments
Conflicts of Interest
References
- Third National Marine Environmental DNA Workshop. June 3 – 5, 2024. Available online: https://secwww.jhuapl.edu/EventLink/Event/300 (accessed on 18 June 2024).
- U.S. National Aquatic Environmental DNA Strategy. A report by the Interagency Working Group on Biodiversity of the Subcommittee on Ocean Science and Technology Committee on Environmental of the National Science and Technology Council. By Prabhakar, A. et al. June 2024. Available online: https://www.whitehouse.gov/wp-content/uploads/2024/06/NSTC_National-Aquatic-eDNA-Strategy.pdf (accessed on 18 June 2024).
- Stepien, C.A; Theroux, S.; Weisberg, S. Commentary: The Second National Workshop on Marine eDNA: A workshop to accelerate the incorporation of eDNA science into environmental management applications. Environmental DNA 2022, 6(1), E 379. Available online: https://onlinelibrary.wiley.com/doi/10.1002/edn3.379 (accessed on 18 June 2024).
- Kelly, R. P.; Lodge, D.M.; Lee, K.N.; et al. Perspective: Toward a national eDNA strategy for the United States. Environmental DNA 2023. 6(1), e42. [CrossRef]
- Capitol Hill Ocean Week (CHOW), June 4–6, 2024, Washington, D.C. Available online: https://capitolhilloceanweek.app.swapcard.com/event/capitol-hill-ocean-week?trck=https%3A%2F%2Fapi.swap-card.com%2Ftrack%2Fclick%2FO4f14bbfFZryqzbgZ8QKzQ%3D~TkDWETnoBMneCuGXRD2YAjwxiCWU_JPCiHqVn3soA_mPVTvV21cyL23aDRqqkvZj (accessed on 18 June 2024).
- Johns Hopkins University. Applied Physics Laboratory (APL). Available online: Climatetrace.org (accessed on 18 June 2024).
- U.S. National Ocean Biodiversity Strategy. A report by the Interagency Working Group on Biodiversity of the Subcommittee on Ocean Science and Technology Committee on Environmental of the National Science and Technology Council. By Prabhakar, A. et al. June 2024. Available online: https://www.whitehouse.gov/ostp/news-updates/2024/06/03/nstc-the-national-ocean-biodiversity-strategy (accessed on 18 June 2024).
- U.S. National Strategy for a Sustainable Ocean Economy. A report by the Interagency Working Group on Biodiversity of the Subcommittee on Ocean Science and Technology Committee on Environmental of the National Science and Technology Council. By Prabhakar, A. et al. June 2024. https://www.whitehouse.gov/ostp/news-updates/2024/06/03/national-strategy-for-a-sustainable-ocean-economy/ (accessed on 18 June 2024).
- Monterey Bay Aquarium Research Institute (MBARI), Monterey, California. Environmental Sampler Processor (ESP). https://www.mbari.org/technology/environmental-sample-processor-esp/ (accessed on 18 June 2024).
- Sepulveda, A.J., Huegh, A., Gage, J.A., et al. Integrating environmental DNA results with diverse data sets to improve biosurveillance of river health. Front. Ecol. Evol., 2021, 9. [CrossRef]
- Ocean Biomolecular Observing Network (OBON), the United Nations Decade of Ocean Science for Sustainable Development (2021–2030). https://oceandecade.org/actions/ocean-biomolecular-observing-network-obon/ (accessed on 18 June 2024).
- Stepien, C.A., Lance, R.F., Klymus, K.E., and Hunter, M.E. Commentary: 6th Annual eDNA Technical Exchange Workshop. Environmental DNA. 2023. https://onlinelibrary.wiley.com/doi/epdf/10.1002/edn3.466 (accessed on 18 June 2024). [CrossRef]
- Morisette, J., Burgiel, S., Brantley, K., et al. (U.S. National Invasive Species Council eDNA Task team). Strategic considerations for invasive species managers in the utilization of environmental DNA (eDNA): steps for incorporating this powerful surveillance tool. Management of Biological Invasions. 2021, 12(3), 747–775. https://www.reabic.net/journals/mbi/2021/3/MBI_2021_Morisette_etal.pdf (accessed on 18 June 2024).
- National Early Detection and Rapid Response (EDRR) Framework, USGS, 2023. https://www.usgs.gov/tools/national-early-detection-and-rapid-response-edrr-framework (accessed on 18 June 2024).
- Langhammer, P.F.; Bull, J.W.; Bicknel, J.E.; et al. The positive impact of conservation action. Science, 2024, 384(6694): 453–458., DOI: 10.1126/science.adj6598. Available online: https://www.science.org/doi/10.1126/science.adj6598 (accessed on 18 June 2024). [CrossRef]
- Southern eDNA Society. Society of Australian/New Zealand environmental DNA researchers and end-users. Available online: https://sednasociety.com (accessed on 18 June 2024).
- Stepien, C.A.; Schultz, H.K.; McAllister, S.M.; et al. Evaluating mtDNA metabarcoding markers to identify zooplankton and ichthyoplankton in the Salish Sea: Resolution in comparison to morphology and detection of rare, threatened, or invasive species. DNA, 2024, 4, 1–33. https://www.mdpi.com/2673-8856/4/1/1 (accessed on 18 June 2024). [CrossRef]
- California Department of Water Resources (DWR) environmental DNA strategy. Available online: https://resources.ca.gov/-/media/DWR-Website/Web-Pages/What-We-Do/Science/Files/eDNA_Strategy.pdf (accessed on 18 June 2024).
- International eDNA Standardization Task Force. A hub for eDNA standardization development. Available online: https://iestf.global (accessed on 18 June 2024).
- Scholes, R.J.; Walters, M.; Turak, E.; et al. Building a global observing system for biodiversity. Current Opinion in Environmental Sustainability, 2012, 4(1), 139–146. [CrossRef]
- iTrack-DNA. Available online: https://itrackdna.ca (accessed on 18 June 2024).
- Abbott, C.; Coulson, M; Gagné, N.; et al. Guidance on the use of targeted environmental DNA (eDNA) analysis for the management of aquatic invasive species and species at risk. DFO Can. Sci. Advis. Sec. Res. Doc. 2021, 019,. iv + 42 p. Available online: https://waves-vagues.dfo-mpo.gc.ca/library-bibliotheque/40960791.pdf (accessed on 18 June 2024).
- Ocean Bioinformatic Information System. OBIS. Intergovernmental Oceanographic Commission (IOC), June 11, 2024, Available online: https://manual.obis.org and https://www.gbif.us/post/2024/gbif-obis-joint-action-plan/ (accessed on 18 June 2024).
- California Cooperative Oceanic Fisheries Investigations (CalCOFI). Available online: https://calcofi.org (accessed on 19 June 2024).
- GenBank, N.I.H. (National Institutes of Health), National Library of Medicine, National Center for Biotechnology Information (NCBI). Available online: https://www.ncbi.nlm.nih.gov/genbank/ (accessed on 18 June 2024).
- Global Biodiversity Information Facility. Available online: https://www.gbif.org. Prototype metabarcoding tool at: (https://edna-tool.gbif-uat.org/). (accessed on 18 June 2024).
- Darwin Core Standard. Available online: https://dwc.tdwg.org/terms/ (accessed on 18 June 2024).
- NASA (National Aeronautics and Space Administration), PACE (Plankton, Aerosol, ocean Cloud Ecosystem). Available online: https://pace.oceansciences.org/home.htm (accessed on 18 June 2024).
- Preston, C.; Yamahara, K.; Pargett, D.; et al. Autonomous eDNA collection using an uncrewed surface vessel over a 4200-km transect of the eastern Pacific Ocean, Environmental DNA, 2024, 6(1), e468. [CrossRef]
- Stevens, J.D.; Parsley, M.B. Environmental RNA applications and their associated gene targets for management and conservation. Environmental DNA, 2023, 5(2), 227–239. [CrossRef]
- Parsley, M.B.; Goldberg, C.S. Environmental RNA can distinguish life stages in amphibian populations, Molecular Ecology Resources, 2024, 24(4), e13857 . [CrossRef]
- eDNA Explorer Platform. Available online: https://www.ednaexplorer.org (accessed on 18 June 2024).
- Smithsonian Institution, Life on a Sustainable Planet and Ocean DNA Initiative. https://naturalhistory.si.edu/exhibits/sant-ocean-hall/ocean-dna (accessed on 18 June 2024).
- National Geographic Society, Pristine Seas Program. Available online: https://www.nationalgeographic.org/society/our-programs/pristine-seas/ (accessed on 18 June 2024).
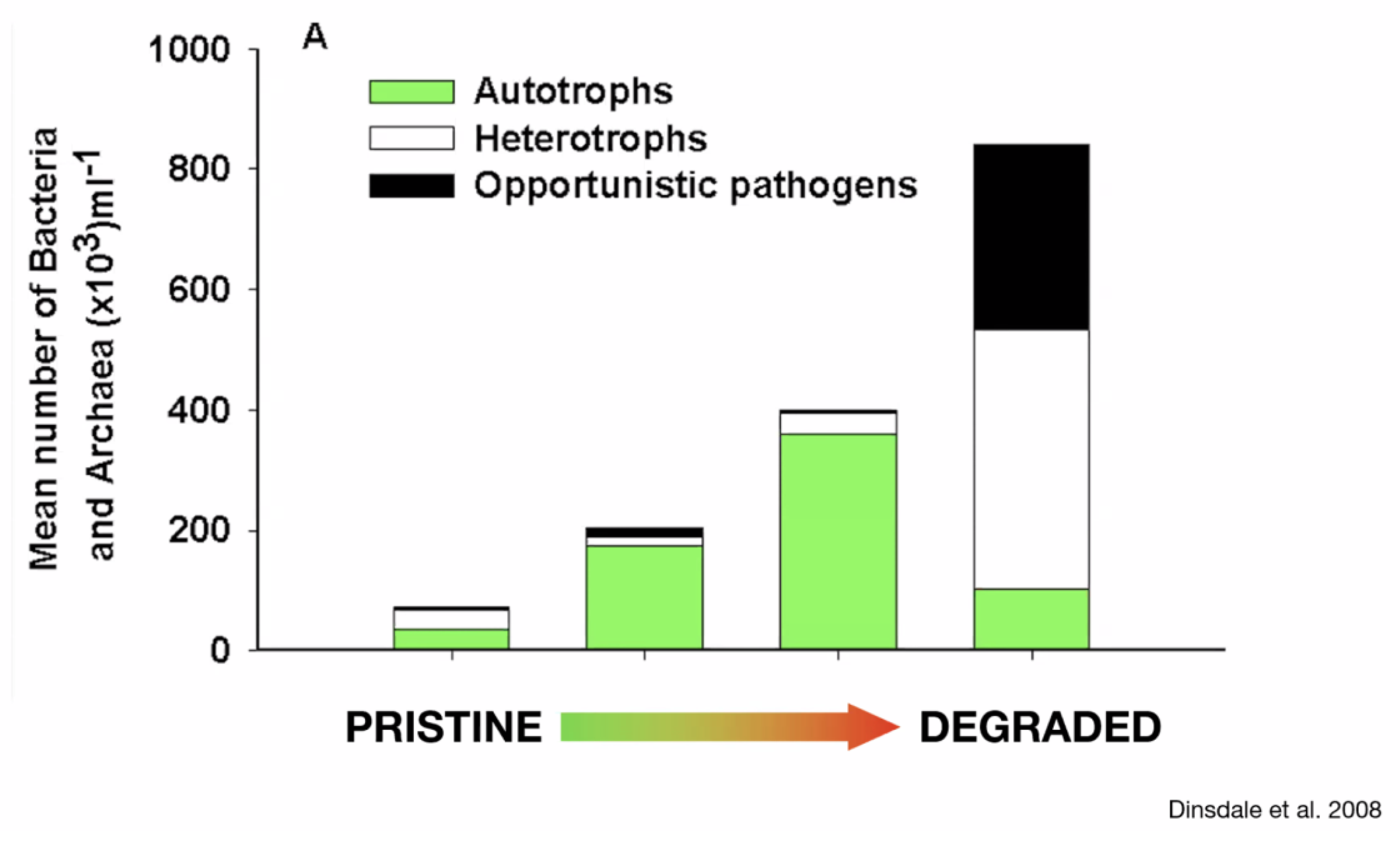
- Dinsdale, E.A.; Pantos, O.; Smriga, S., et al. Microbial ecology of four coral atolls in the Northern Line Islands. PLoS One, 2008, 3(2), e1584. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




