1. Introduction
Return to sport following a musculoskeletal injury focuses on reestablishing normal biomechanical patterns and if done correctly decreases reinjury risk by 85% [
1]. When performing tasks that require movement, muscles work together in tandem to execute the movement and produce the required force [
2]. Current rehabilitation techniques rely upon visual assessments and palpations during slow controlled movements as well as patient reports to monitor the recovery of muscle function [
3]. Thus, current exams are subjective and heavily dependent upon the clinician’s experience and patient feedback. To achieve more quantitative assessment, state-of-the-art return to sport protocols utilize kinetic measures involving the utilization of sensors such as force plates, dynamometers, and 3D motion capture [
4,
5]. These are extensions of routine clinical assessments; however, these measures require large equipment, specialized personnel, post-hoc analysis, and are generally not found outside of biomechanical laboratories.
Force plates have become a main component for monitoring sports performance testing and neuromuscular fatigue [
4,
6,
7,
8,
9]. The increased portability of force plates in recent years has made them standard practice in the sports performance field [
10,
11]. Specifically, isometric squat testing is a monitoring tool commonly used to assess the rate of force development in athletic populations [
12,
13,
14]. Previous literature has demonstrated a strong correlation between isometric squat testing and other dynamic performance tasks (e.g., jump testing) [
13,
15].
However, force plates only consider ground reaction forces that are produced from multiple muscle groups working together. How individual muscles are activated and contribute to force production cannot be determined from the use of force plates. This level of information is needed during return to sport protocols to ensure that an athlete is not compensating during a functional task, resulting in improper form or inefficient movement [
16,
17].
Muscle level information can be acquired using surface electromyography (sEMG) to monitor activation patterns during dynamic movement [
4,
18,
19]. However, sEMG has limitations, including complex outputs, noise influences, muscle cross-talk, and the inability to derive signals from deep muscle tissue [
20,
21]. Mechanomyography (MMG), a technique to measure mechanical activity of the muscle using specific transducers, can also be used to obtain muscle level information [
22,
23,
24]. Previous research has shown that MMG can monitor muscle function during activity but is limited as signals are influenced by movement artifacts [
25]. Together sEMG and MMG have been combined to assess muscle function information during dynamic tasks [
22,
25,
26].
Ultrasound imaging is an attractive method to image and characterize muscle tissues [
27,
28]. However, current ultrasound-based examinations are not dynamic and are performed subjectively in highly controlled environments where the patient is stationary. Dynamic ultrasound imaging allows one to characterize muscle contraction, velocity, pennation angle, and fascicle length changes, which can be inform muscle function [
29,
30,
31,
32,
33]. Kamatham et al., has shown the application of ultrasound metrics in predicting muscle force during isometric hand contractions [
34]. Additional studies have also shown changes in ultrasound measures, such as pennation angle and echogenicity, over the duration of long periods of intense exercise [
35].
Although the aforementioned studies have shown promising results, implementation in clinical settings is challenging as current clinical systems are not optimized for dynamic imaging. Furthermore, clinical ultrasound systems are hindered by bulky transducers that require hand-held operation or custom strapping mechanisms to maintain acoustic coupling during dynamic movement, typically from a single anatomical site. In such settings, even the slightest transducer movement can cause skin contact loss and/or changes in the transducer pressure, resulting in inaccurate muscle function characterization [
36]. To be feasible for imaging in dynamic and clinical settings ultrasound systems must be able to overcome these limitations.
Considering the challenges of acquiring dynamic ultrasound images using traditional ultrasound imaging transducers, wearable ultrasound technology research and development has accelerated in recent years. Current applications of wearable ultrasound technology have mainly focused on cardiovascular and pulmonary function, exoskeleton, and prosthesis control [
37,
38,
39,
40,
41,
42,
43]. Wearable ultrasound holds great promise for simultaneously imaging different muscle groups contributing to force production during movement to assess muscle function in a comprehensive and systematic fashion. However, this requires the development of a miniaturized system enabling distributed imaging of multiple muscle groups simultaneously.
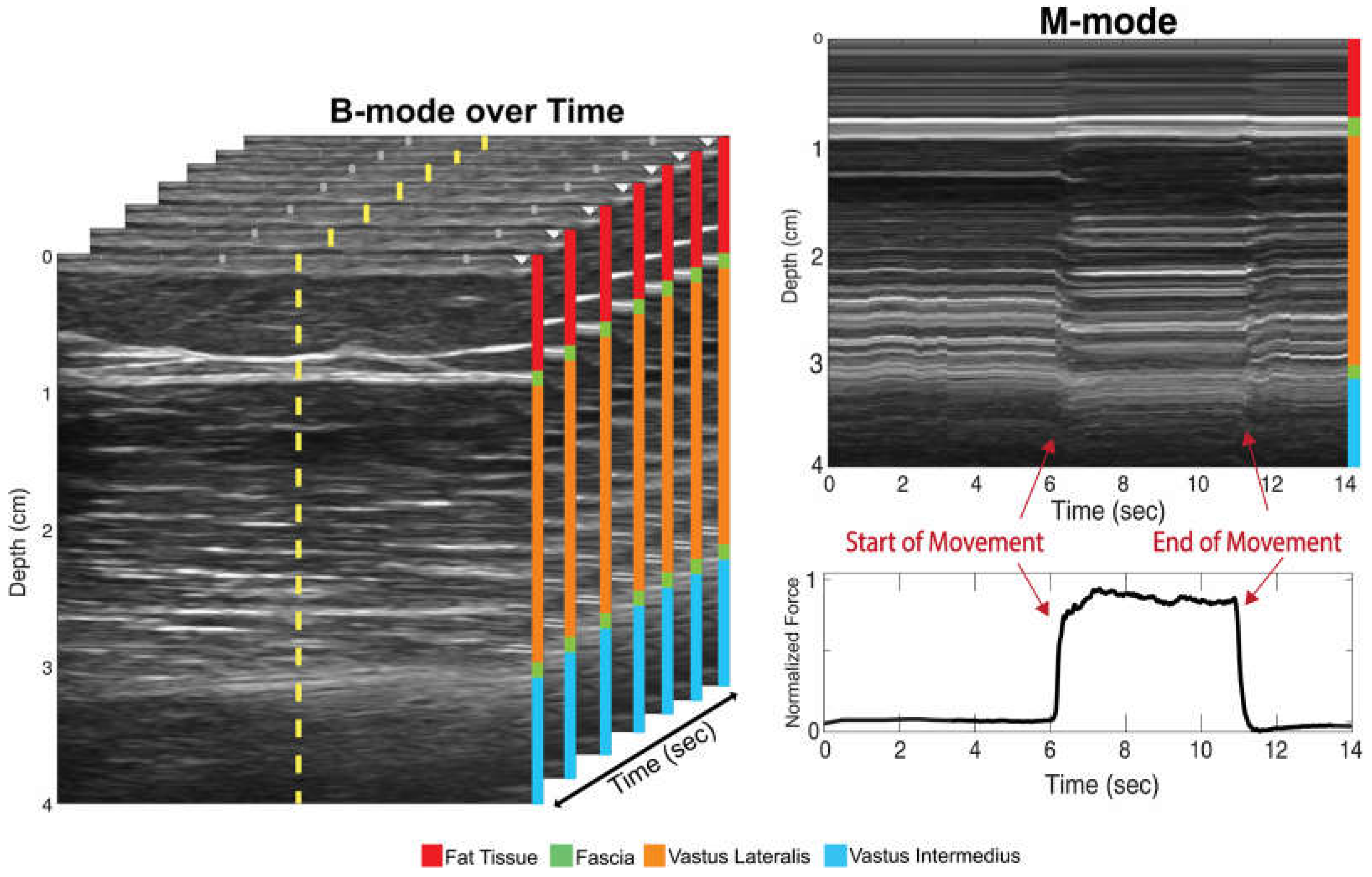
Researchers have previously shown that the use of motion mode (M-mode) ultrasound imaging can be a viable alternative to traditional B-mode imaging to investigate time-varying muscle deformation [
44]. M-mode involves imaging a single ultrasound scanline over time, allowing muscle motion to be monitored more efficiently [
33,
45]. Utilizing M-mode imaging allows for simpler electronics, smaller device footprints, lower computational requirements and the incorporation of multiple imaging sensors.
We have developed a
Simultaneous Musculoskeletal Assessment with Real Time Ultrasound (SMART-US) device that uses novel ultrasound imaging technology to acquire four channels of ultrasound M-mode data concurrently [
46,
47,
48]. In this study, our research objective was to investigate whether ground reaction force during an isometric squat task can be predicted from M-mode ultrasound data from our novel distributed system using a machine learning algorithm. The three following hypotheses were tested: 1) M-mode ultrasound imaging can predict ground reaction force during isometric squats, 2) force predictions using distributed SMART-US sensor are comparable to those obtained from traditional clinical ultrasound systems, and 3) a distributed SMART-US imaging device will have higher accuracy than a single-channel image acquisition system. Our results indicate support for all these three hypotheses.
2. Materials and Methods
Five able-bodied female subjects (24 ± 3.4 years) participated in the study. All participants were former National Collegiate Athletic Association Division I athletes, in good physical health, and recreationally active. They reported no lower body injuries, had the risks and benefits explained before testing, and signed an institutionally approved informed consent form.
Before testing began, the dominant leg was determined by asking the participants which leg they used to kick a ball [
49]. The participants' dominant leg was then scanned with a clinical ultrasound transducer (eSaote BioSound MyLab 25, Biosound Esaote, Inc., Indianapolis, IN, USA). Static B-mode images were acquired at four locations: Rectus Femoris, Vastus Lateralis, Medial Hamstring, and Vastus Medialis. Locations of clinical images were marked using a custom template for wearable sensor placement later in the protocol. After scanning was concluded, participants completed a standardized 15-minute dynamic exercise warm-up.
Following the warmup, participant conducted two rounds of three isometric squats at maximum voluntary contraction on a force plate (AccuPower; AMTI, Watertown, MA, USA) acquired at 1600 Hz with a custom data acquisition system utilizing LabVIEW (National Instruments, Austin, TX, USA). Two rounds of non-randomized ultrasound imaging conditions were conducted with at least 10 minutes of rest in between rounds. Participants were cued using verbal commands from researchers on when to start and end the movement. Participants executed a maximum voluntary contraction squat for approximately 5 seconds when cued. All data were time-synched using a custom keyboard that enabled the researchers to start and stop the data collection on each machine simultaneously.
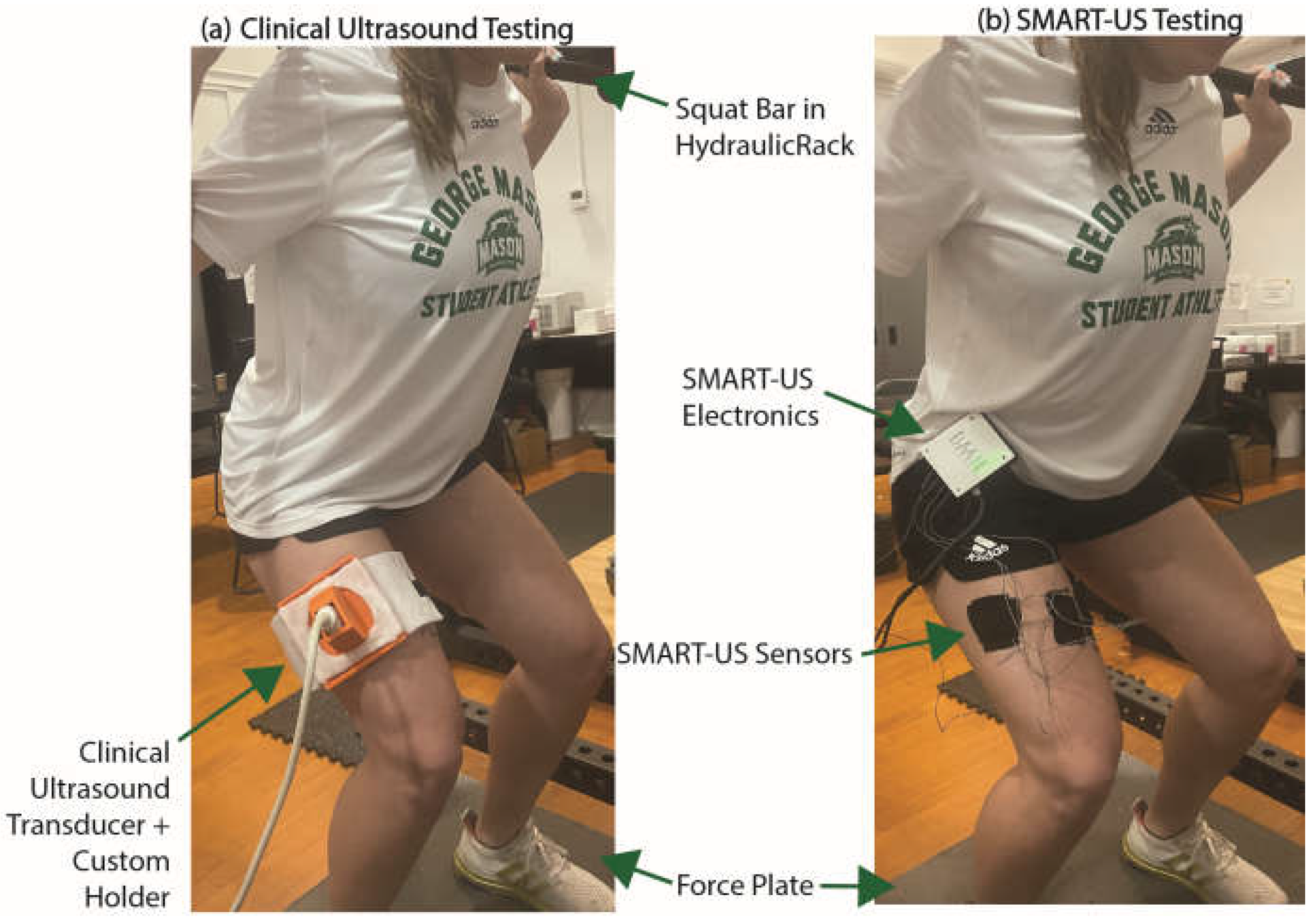
2.1. Round 1 - Clinical Ultrasound Testing
A clinical two-dimensional ultrasound system (eSaote BioSound MyLab 25, Biosound Esaote, Inc., Indianapolis, IN, USA) with a 5-cm linear transducer (frequency, 7.5 MHz; axial resolution < 0.5 mm) was used for round one of testing. The clinical system was fitted to the participant’s dominant vastus lateralis using a custom 3D-printed holder (
Figure 1a).
Images were collected using a USB-based video grabber (DVI2USB 3.0, Epiphan Systems, Inc., Palo Alto, CA) at 50 Hz during the isometric squat. Ultrasound settings remained constant across participants (Depth: 4 cm, Gain: 50).
After data acquisition, the two-dimensional ultrasound image sets were imported into a custom MATLAB (MATLAB 2022b, MathWorks Inc., Natick, MA, USA) script and transformed into an M-mode image. This was accomplished by taking a singular scanline at the center of the two-dimensional image over time (
Figure 2).
2.2. Round 2 – SMART-US Testing
A wearable, custom designed ultrasound system with multiple distributed imaging sensors, was strapped to the participant’s waist (
Figure 1b). Additional device details can be found in previous publications [
48] but a summary of the capabilities is included in this section. The SMART-US system can capture four channels of M-modes at a frame rate of 50 Hz and a depth of 4 cm using PZT transducers with a center frequency of 4.25 MHz. The system uses a frequency sweep with known sweep rate (10 ms long) to encode depth as frequency, such that shallow depths are low frequency components and deeper muscles are higher frequency components in a multi-component sinusoidal signal. Image processing consists of performing a fast Fourier transform (FFT) on the 10 ms window to acquire the frequency components and thus the muscle interfaces at where the transducers were placed. Transducers were placed in custom 3D-printed housings on the dominant leg in the following locations: vastus lateralis (at the center of the clinical transducer from round one), rectus femoris, medial hamstring, and vastus medialis (
Figure 1b). Sensor placement for the vastus lateralis was determined using a custom 3D printed template, which marked the center of the clinical ultrasound transducer, ensuring placement was the same across collections.
2.3. Modeling
A ridge regression model was used for machine learning analysis. Ridge regression estimates coefficients of linear models that include a linearly correlated predictor. Specifically, this model is used to address the problem of multicollinearity by utilizing a penalty term:
where
k is the ridge parameter
(k = 5), and
l is the identity matrix. Smaller positive values of
k improve the conditioning and reduce the variance of model estimates, resulting in a smaller mean squared error [
50].
The ridge function used in MATLAB solves the following equation for a given l value:
where
N is the number of observations,
yi is the response at observation
i,
xi is the data with a vector of length
p at observation
i, is a nonnegative regularization parameter,
is a scalar, and
is a vector the length of
p.
2.3.1. Feature Extraction
Image features were extracted from M-mode images using a mean depth calculation. Mean depth was used to track the muscle interface as it expands or contracts during the specified task. The calculation is a weighted average, that weighs each pixel by the pixel intensity at that corresponding depth. This in turn results in a frequency, and in this case depth, where most of the energy is centered in the window. Features sections are calculated by taking a moving depth window of 25 pixels over each amplitude line (A-line) of the image, with a window overlap of 50%. To calculate each window's mean depth, the window's pixel intensities (
A) are first multiplied by the corresponding pixel depth (
D). The sum of the multiplied components is then divided by the sum of the pixel intensities in that window (Equation (3)).
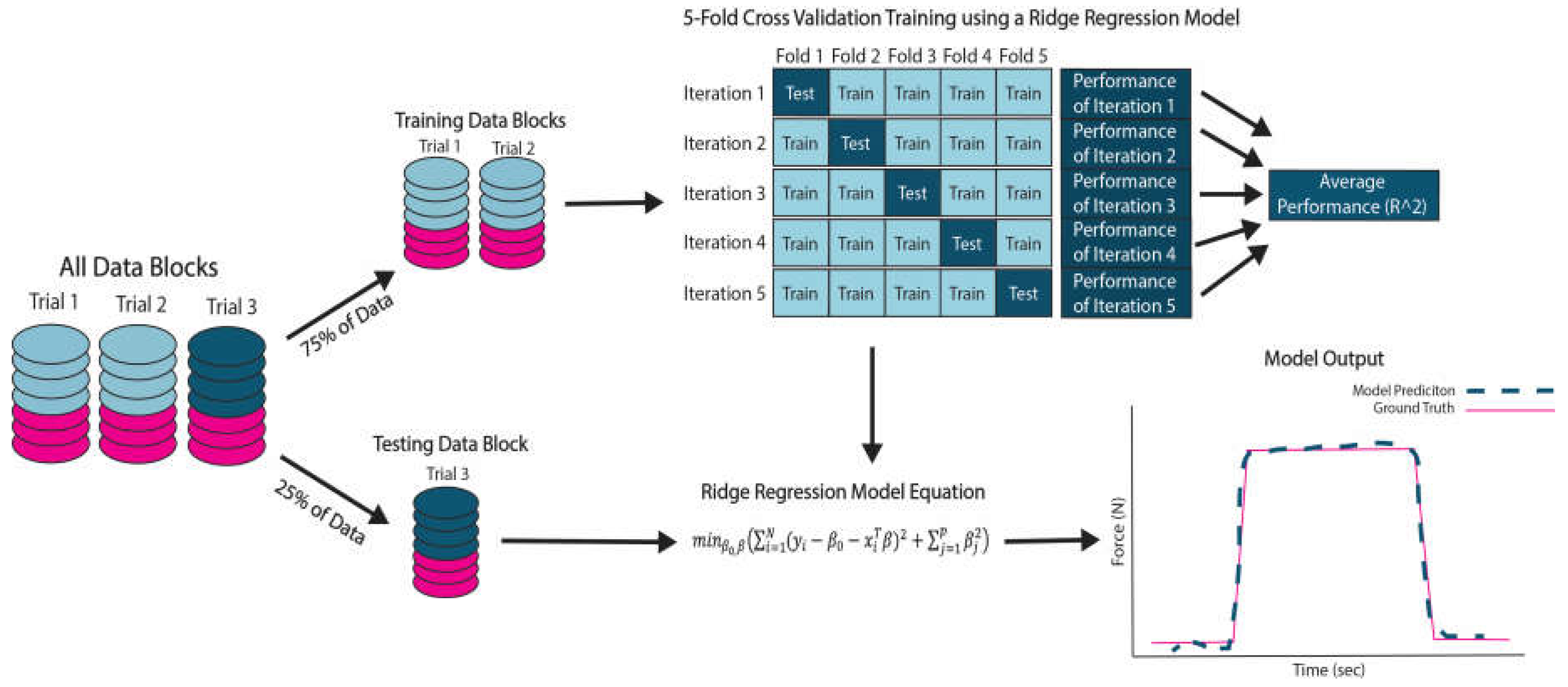
The extracted features of each column in the M-mode image were then converted to Z-scores. A principal component analysis (PCA) was run the resulting Z-scores to obtain the most significant features. The features that explained 99% of the variance were retained for further analysis. Data containing all features was split into training and testing sets to validate and test the machine learning models (
Figure 3). The acquired M-mode images were put into six categories for analysis and comparison (
Table 1)
.
2.3.2. Model Validation
Trials 1 and 2 were used for 5-fold cross-validation to evaluate the model’s performance in making predictions compared to the ground truth, which is force plate data obtained during the movement. Each round of cross-validation involves randomly partitioning all feature data into five subsets of equal size, with four subsets used for training and the remaining for testing. The process is repeated five times to ensure that each subset is used once as the testing set with the other subsets used to train the model. The participants average cross-validation error, reported as R
2, was used to evaluate performance (
Figure 3).
2.3.3. Model Testing
The third trials were reserved for model testing. The predicted force was filtered before the R2 calculation using a moving averaged filter with a window size of 25 pixels. The R2 value for each tested trial was calculated and reported.
2.4. Statistical Analysis
The coefficient of determination (R2) was the primary outcome measure used to evaluate model performance and determine the accuracy of the predicted force outputs compared to the ground-truth force measures. An R2 coefficient closer to 1 was indicative of a higher model accuracy.
Data were assessed for normality via Levene’s test. If data did not violate the Levene’s test, then results were compared using a two-sample t-test. If data violated Levene’s test (p<0.05), non-parametric statistics were performed. A Kruskal-Wallis test was used to determine if model conditions were significant during training. If significant post-hoc Tukey’s honestly significant difference test was performed to determine where significance occurred (p<0.05). All statistical procedures were performed in MATLAB 2023b.
3. Results
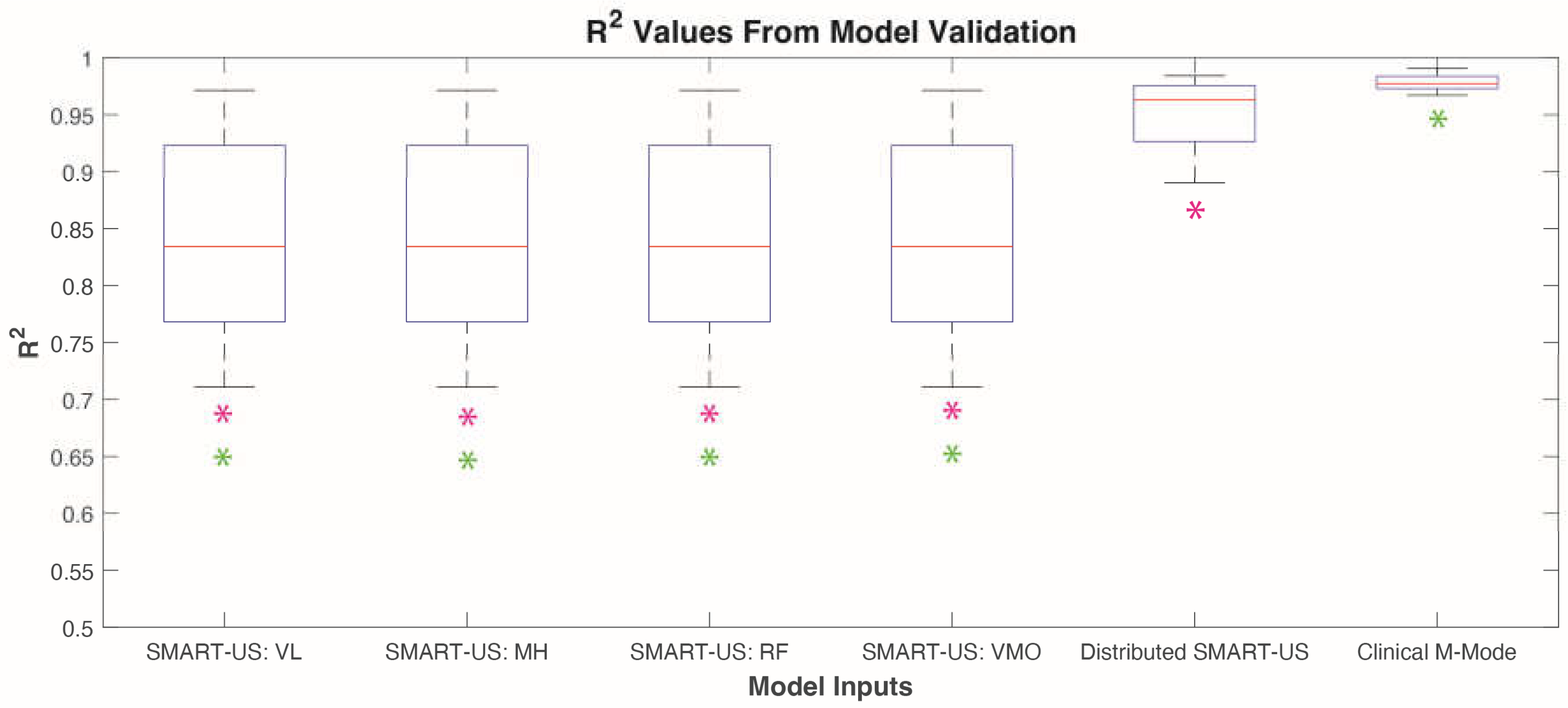
3.1. Model Validation
The six categories of M-mode image data were analyzed independently for cross-validation analysis using the method outlined in
Figure 3. Results showed that the
Clinical M-mode performed the best with an R
2 value of 0.98±0.01 (
Figure 4). The next best was the
Distributed SMART-US model as the inputs with an R
2 of 0.95±0.03. Finally, for force prediction using an individual SMART-US sensor, the results were as follows:
SMART-US: VL = 0.85±0.09,
SMART-US: MH = 0.82±0.10,
SMART-US:RF = 0.85±0.07,
SMART-US:VMO = 0.74±0.14. Model validation results violated Levene’s test. Therefore, a Kruskal-Wallis test was used to indicate significant difference between model conditions (p<0.0001). A post hoc Tukey’s test showed that the
Clinical M-mode model and the
Distributed SMART-US were both significantly different from the
SMART-US: VL,
SMART-US: MH,
SMART-US:RF,
SMART-US:VMO (p<0.0001) (
Figure 4). There was no significant difference between the
Clinical M-mode and
Distributed SMART-US models.
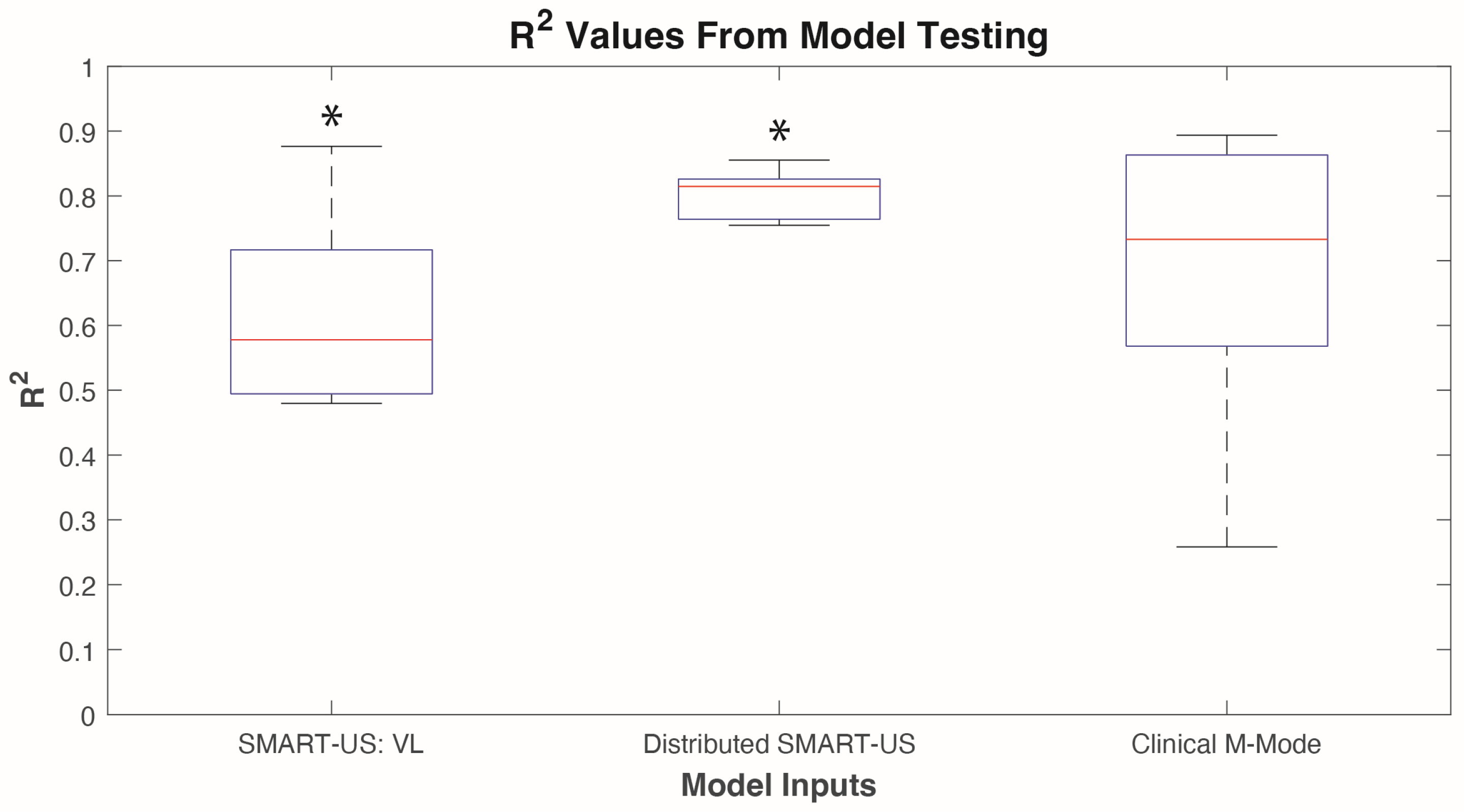
3.3. Model Testing
Online testing using the last trial was conducted on the two models that were not significantly different in the cross-validation (i.e., Distributed SMART-US and Clinical M-mode). The SMART-US: VL was also included in model testing, as the sensor was placed in the same location as the Clinical M-mode sensor. Model testing data did not violate Levene’s test (p = 0.66) and a two- sampled t-test was used to inidcate signifcance between model types. Results showed that the Clinical M-mode model had an average R2 of 0.680.25, SMART-US: VL model had an average R2 of 0.0.16, and Distributed SMART-US mode had an average R2 of 0.800.04 (Table 2). Two-sample t-test indicated that only SMART-US: VL vs. Distributed SMART-US models were statistically significant from eachother (p<0.05).
4. Discussion
As demonstrated in this research a novel distributed wearable ultrasound system can accurately predict force during a maximal isometric squat task. Three hypotheses were tested: 1) M-mode ultrasound imaging can be used to predict force during a maximal isometric squat task by utilizing a machine learning algorithm, 2) force predictions from a novel wearable ultrasound system are comparable to those obtained from traditional unportable clinical ultrasound systems, and 3) a novel distributed ultrasound device will have higher prediction accuracy than traditional single anatomical site imaging modalities due to its ability to image multiple muscles that contribute to force production.
Model validation results indicated that a ridge regression model using
Clinical M-mode was able to predict force production during an isometric squat task with an R
2 value of 0.98±0.01. These results are comparable to previous literature using machine learning models that utilize M-mode imaging metrics to predict force during isometric hand dynamometry tasks [
34]. The current study expands upon the previous literature by utilizing a novel distributed imaging system, allowing for analysis of force development from a whole muscle group and focusing on lower body isometric movements.
Validation results comparing the Distributed SMART-US model and the Clinical M-mode model were not statistically significant, providing evidence that a Distributed SMART-US model can be used as a reliable alternative to predicting force during isometric squats. The Distributed SMART-US model and the Clinical M-mode model were significantly different from the individual SMART-US sensor models. This can be attributed to the Distributed SMART-US model’s multi-sensing modality and the Clinical M-mode models higher imaging resolution.
Testing results also found no statistical significance in model accuracies when comparing force prediction from the Distributed SMART-US and the Clinical M-mode models. Even though it was not statistically significant, the Distributed SMART-US model showed higher precision and robustness across all participants than the Clinical M-mode model, demonstrating its advantage.
The precision of the
Distributed SMART-US model could be attributed to its multi-anatomical imaging nature. Muscles work together to produce a force or movement. When performance of one of the muscles in the group is affected, another muscle will recruit and activate stronger [
51,
52]. By having multiple sensors placed over the quadriceps and hamstring, multiple muscle forces in the
Distributed SMART-US model can be accounted for. Whereas the
Clinical M-mode model only examines the force contribution from a single muscle. Additionally, bulky strapping mechanisms, that were used for transducer placement (
Figure 1a), can restrict muscle movement during exercise and influenced the
Clinical M-mode model’s variability.
In previous studies single anatomical site acquisition was identified when comparing sEMG, ultrasound, and dynamometry fatigue assessments [
53]. A study conducted by Varol et al. found weak associations among sEMG, ultrasound, and dynamometry systems and identified the necessity to measure all three to provide synergistic information in fatigue assessments [
53]. A distributed sensing system, which can provide deep muscle information from multiple anatomical sites, could provide more accurate information about muscle group fatigue during a dynamic task.
sEMG has been used commonly in monitoring muscle fatigue as well as in control paradigms for prosthesis and exoskeletons. [
7,
14,
19]. Previous research has shown that while sEMG may outperform ultrasound-based machine learning models to predict different levels of force in a grip task, ultrasound imaging can be a more robust and accurate measure when muscle fatigue is incorporated into the protocol [
54,
55]. Additionally, Zhang et al. showed that machine learning models improved in accuracy when ultrasound features were added in addition to sEMG to predict ankle dorsiflexion moment [
56]. They theorized that this was due to ultrasound’s ability to directly visualize muscle activity, which is unobtainable in sEMG devices.
Furthermore, sEMG can monitor multiple anatomical locations simultaneously to probe synergistic muscle activation patterns, however this use case has been cautioned against in previous literature. Tweedle et al. showed that muscle activation measured using ultrasound signals occurred 98 milliseconds before muscle activation was detected by both sEMG and fine wire EMG in the vastus lateralis and biceps brachii [
57]. Hence, the use of ultrasound imaging for detection of muscle activation is recommended due to its ability to visualize deep-seated muscle tissue, which can allow for a more accurate interpretation of results.
A previously discussed the use of ultrasound technology for monitoring muscle function has increased in recent years. Current research on the implementation of miniaturized wearable ultrasound technology has included multiple use cases ranging from prosthesis control to monitoring physiological signals [
40,
42,
48,
55,
58].
For example, Yang et al., presented a wearable multi-channel A-mode ultrasound system that can be used for in vivo muscle deformation detection and virtual prosthesis control demonstrating a task completion rate of 100% and path efficiency of 93.30% [
58]. Lin et al. developed a wearable ultrasound patch that can be used to continuously monitor physiological signals over a 12-hour period [
42]. These studies focused on utilizing wearable ultrasound systems for upper body tasks or monitoring physiological signals (i.e., heart rate, blood pressure) at a single location [
42,
58]. To our knowledge, this is the first study utilizing a novel distributed wearable ultrasound system for lower limb imaging and force prediction during an isometric task.
Results from the current research can potentially improve the understanding of complex muscle mechanisms responsible for force output. This technology enables muscle function to be analyzed in real-time, providing feedback on neuromuscular performance and recovery during activity that is currently unavailable [
59].
The incorporation of machine learning is useful for monitoring individual muscle force development and how it relates to overall forces exerted during a task. The potential for this research and SMART-US ranges from rehabilitation to prosthesis and exoskeleton control and can provide biofeedback warnings about compensatory movements that could increase the risk of injury.
Limitations of the current study include utilization of single force plate. Single force plate collection could have affected machine learning models as force production was the total force produced between both limbs. However, imaging modalities were only placed on the participants dominants limb, therefore machine learning models did not consider the nondominant side when predicting force. Additionally, a fundamental limitation is that machine learning models were subject-specific, and one model cannot be generalized to the whole group. Moreover, validation and testing on different performance tasks are needed to discern the importance of sensor location. The image resolution of the clinical and SMART-US systems was not compared, and the difference may have impacted feature extraction and quality. Furthermore, clinical transducer pressure and strapping was variable across participants, potentially inhibiting muscle movement during exercise and influencing results.
Future work to address the aforementioned would include 1) expanding sample size and including different population types, 2) developing a semi-real-time algorithm that can determine if muscles are engaging or disengaging during isometric tasks, and 3) utilizing SMART-US for more complex dynamic performance testing. Additionally, collecting longitudinal data during a rehabilitation period can shed light on how muscle groups react to injury types, locations, and treatments. This type of knowledge can inform clinical decision-making and improve rehabilitation outcomes.
5. Conclusions
In conclusion, this study has demonstrated that a distributed wearable SMART-US system can predict ground reaction force using a linear ridge regression model. This research has the potential to be expanded into understanding muscle function patterns during recovery after musculoskeletal injuries and can potentially help personalize patient care. The overall results of this study prove the feasibility of wearable ultrasound as a force estimation device, which can create new possibilities in the field of injury and recovery by utilizing information obtained at a muscle level.
Author Contributions
Conceptualization, E.L.K., S.S., and P.V.C.; methodology, E.L.K., M.T.J., Q.W., S.S., and P.V.C.; software, E.L.K., and A.B.; validation, E.L.K., S.P., A.B., and M.M.; formal analysis, E.L.K., S.P., A.B., and M.M.; investigation, E.L.K.; resources, M.T.J., SS and P.V.C.; data curation, E.L.K.; writing—original draft preparation, E.L.K.; writing—review and editing, M.T.J., Q.W.,A.B., S.P., S.S., and P.V.C.; visualization, E.L.K.; supervision, P.V.C..; project administration, E.L.K.; funding acquisition, P.V.C, SS. All authors have read and agreed to the published version of the manuscript.
Funding
The research is supported in part by the following grants: Medical Technology Enterprise Consortium – Military Performance Advancement Initiative (MTEC-MPAI) under Grant W81XWH-15-9-0001, NIH 5U01EB027601, US DoD W81XWH2010817, W81XWH2010190 and W81XWH-22-9-0016. E.L.K acknowledges graduate fellowship funded by the NSF grant DGE: 1922598.
Institutional Review Board Statement
This study was conducted according to the Declaration of Helsinki guidelines and the procedures were approved by the Institutional Review Boards at George Mason University (#1989591-2) for studies involving humans.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Data is available from the corresponding authors upon reasonable request.
Conflicts of Interest
Drs. Chitnis and Sikdar hold multiple patents on wearable ultrasound technology and applications. The other authors have no conflicts to disclose.
References
- Grindem, H.; Snyder-Mackler, L.; Moksnes, H.; Engebretsen, L.; Risberg, M.A. Simple Decision Rules Can Reduce Reinjury Risk by 84% after ACL Reconstruction: The Delaware-Oslo ACL Cohort Study. Br. J. Sports Med. 2016, 50, 804–808. [Google Scholar] [CrossRef] [PubMed]
- Hug, F.; Tucker, K. Muscle Coordination and the Development of Musculoskeletal Disorders. Exerc. Sport Sci. Rev. 2017, 45, 201. [Google Scholar] [CrossRef] [PubMed]
- Bohannon, R.W. Manual Muscle Testing: Does It Meet the Standards of an Adequate Screening Test? Clin. Rehabil. 2005, 19, 662–667. [Google Scholar] [CrossRef]
- Taborri, J.; Keogh, J.; Kos, A.; Santuz, A.; Umek, A.; Urbanczyk, C.; van der Kruk, E.; Rossi, S. Sport Biomechanics Applications Using Inertial, Force, and EMG Sensors: A Literature Overview. Appl. Bionics Biomech. 2020, 2020, e2041549. [Google Scholar] [CrossRef] [PubMed]
- Undheim, M.B.; Cosgrave, C.; King, E.; Strike, S.; Marshall, B.; Falvey, É.; Franklyn-Miller, A. Isokinetic Muscle Strength and Readiness to Return to Sport Following Anterior Cruciate Ligament Reconstruction: Is There an Association? A Systematic Review and a Protocol Recommendation. Br. J. Sports Med. 2015, 49, 1305–1310. [Google Scholar] [CrossRef]
- De Fazio, R.; Mastronardi, V.M.; De Vittorio, M.; Visconti, P. Wearable Sensors and Smart Devices to Monitor Rehabilitation Parameters and Sports Performance: An Overview. Sensors 2023, 23, 1856. [Google Scholar] [CrossRef] [PubMed]
- Alba-Jiménez, C.; Moreno-Doutres, D.; Peña, J. Trends Assessing Neuromuscular Fatigue in Team Sports: A Narrative Review. Sports 2022, 10, 33. [Google Scholar] [CrossRef]
- Merrigan, J.J.; Stone, J.D.; Thompson, A.G.; Hornsby, W.G.; Hagen, J.A. Monitoring Neuromuscular Performance in Military Personnel. Int. J. Environ. Res. Public. Health 2020, 17, 9147. [Google Scholar] [CrossRef]
- Merrigan, J.J.; Stone, J.D.; Martin, J.R.; Hornsby, W.G.; Galster, S.M.; Hagen, J.A. Applying Force Plate Technology to Inform Human Performance Programming in Tactical Populations. Appl. Sci. 2021, 11, 6538. [Google Scholar] [CrossRef]
- Ferguson, J.; Gibson, N.V.; Weston, M.; McCunn, R. Reliability of Measures of Lower-Body Strength and Speed in Academy Male Adolescent Soccer Players. J. Strength Cond. Res. 2024, 38, e96. [Google Scholar] [CrossRef]
- Cabarkapa, D.; Philipp, N.M.; Cabarkapa, D.V.; Fry, A.C. Position-Specific Differences in Countermovement Vertical Jump Force-Time Metrics in Professional Male Basketball Players. Front. Sports Act. Living 2023, 5, 1218234. [Google Scholar] [CrossRef] [PubMed]
- Bazyler, C.; Beckham, G.; Sato, K. The Use of the Isometric Squat as a Measure of Strength and Explosiveness. J. Strength Cond. Res. Natl. Strength Cond. Assoc. 2014, 29. [Google Scholar] [CrossRef]
- Ishida, A.; Bazyler, C.D.; Suarez, D.G.; Slaton, J.A.; White, J.B.; Stone, M.H. The Difference between Several Neuromuscular Tests for Monitoring Resistance-Training Induced Fatigue. J. Sports Sci. 2023, 41, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Subbu, R.; Weiler, R.; Whyte, G. The Practical Use of Surface Electromyography during Running: Does the Evidence Support the Hype? A Narrative Review. BMJ Open Sport Exerc. Med. 2015, 1, e000026. [Google Scholar] [CrossRef] [PubMed]
- Bishop, C.; Jordan, M.; Torres-Ronda, L.; Loturco, I.; Harry, J.; Virgile, A.; Mundy, P.; Turner, A.; Comfort, P. Selecting Metrics That Matter: Comparing the Use of the Countermovement Jump for Performance Profiling, Neuromuscular Fatigue Monitoring, and Injury Rehabilitation Testing.
- Buckthorpe, M. Recommendations for Movement Re-Training After ACL Reconstruction. Sports Med. 2021, 51, 1601–1618. [Google Scholar] [CrossRef] [PubMed]
- Piech, J.; Bajorek, W.; Płonka, A.; Kuchciak, M.; Bobula, G. Lateralization Value of Functional Movement Rating in Volleyball Players’ Injury Prevention. J. Phys. Educ. Sport 2020, 20, 1475–1480. [Google Scholar] [CrossRef]
- Powers, C.M.; Landel, R.; Perry, J. Timing and Intensity of Vastus Muscle Activity During Functional Activities in Subjects With and Without Patellofemoral Pain. Phys. Ther. 1996, 76, 946–955. [Google Scholar] [CrossRef]
- Kotov-Smolenskiy, A.M.; Khizhnikova, A.E.; Klochkov, A.S.; Suponeva, N.A.; Piradov, M.A. Surface EMG: Applicability in the Motion Analysis and Opportunities for Practical Rehabilitation. Hum. Physiol. 2021, 47, 237–247. [Google Scholar] [CrossRef]
- Felici, F.; Del Vecchio, A. Surface Electromyography: What Limits Its Use in Exercise and Sport Physiology? Front. Neurol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.J.; Acosta-Sojo, Y. sEMG: A Window Into Muscle Work, but Not Easy to Teach and Delicate to Practice—A Perspective on the Difficult Path to a Clinical Tool. Front. Neurol. 2021, 11. [Google Scholar] [CrossRef]
- Woodward, R.B.; Stokes, M.J.; Shefelbine, S.J.; Vaidyanathan, R. Segmenting Mechanomyography Measures of Muscle Activity Phases Using Inertial Data. Sci. Rep. 2019, 9, 5569. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; Sundaraj, K.; Ahmad, R.B.; Ahamed, N.U. Mechanomyogram for Muscle Function Assessment: A Review. PLOS ONE 2013, 8, e58902. [Google Scholar] [CrossRef] [PubMed]
- Orizio, C.; Gobbo, M. Mechanomyography. In Wiley Encyclopedia of Biomedical Engineering; Akay, M., Ed.; Wiley, 2006 ISBN 978-0-471-24967-2.
- Correa, M.; Projetti, M.; Siegler, I.A.; Vignais, N. Mechanomyographic Analysis for Muscle Activity Assessment during a Load-Lifting Task. Sensors 2023, 23, 7969. [Google Scholar] [CrossRef] [PubMed]
- Linderman, S.E.; Scarborough, D.M.; Aspenleiter, R.; Stein, H.S.; Berkson, E.M. Assessing Quadriceps Muscle Contraction Using a Novel Surface Mechanomyography Sensor during Two Neuromuscular Control Screening Tasks. Sensors 2023, 23, 6031. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.-V.; Wu, W.-T.; Özçakar, L. Ultrasound Imaging and Rehabilitation of Muscle Disorders: Part 1. Traumatic Injuries. Am. J. Phys. Med. Rehabil. 2019, 98, 1133. [Google Scholar] [CrossRef]
- Whittaker, J.L.; Stokes, M. Ultrasound Imaging and Muscle Function. J. Orthop. Sports Phys. Ther. 2011, 41, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zheng, Y.P.; Chen, X.; Huang, Q.H. Assessment of Muscle Fatigue Using Sonomyography: Muscle Thickness Change Detected from Ultrasound Images. Med. Eng. Phys. 2007, 29, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Yang, X.; Yin, G.; Guo, J. Skeletal Muscle Fatigue State Evaluation with Ultrasound Image Entropy. Ultrason. Imaging 2020, 42, 235–244. [Google Scholar] [CrossRef]
- Huang, Z.-H.; Ma, C.Z.-H.; Wang, L.-K.; Wang, X.-Y.; Fu, S.-N.; Zheng, Y.-P. Real-Time Visual Biofeedback via Wearable Ultrasound Imaging Can Enhance the Muscle Contraction Training Outcome of Young Adults. J. Strength Cond. Res. 2022, 36, 941–947. [Google Scholar] [CrossRef]
- Majdi, J.A.; Acuña, S.A.; Chitnis, P.V.; Sikdar, S. Toward a Wearable Monitor of Local Muscle Fatigue during Electrical Muscle Stimulation Using Tissue Doppler Imaging. Wearable Technol. 2022, 3, e16. [Google Scholar] [CrossRef]
- Naranjo-Cinto, F.; Pecos-Martín, D.; Cuenca-Zaldivar, J.N.; Achalandabaso-Ochoa, A.; Quintero-Pérez, J.; Bierge-Sanclemente, P.; García-Escudero, M.; Fernández-Carnero, S. M-Mode Ultrasound Behavior of Rectus Femoris and Vastus Intermedius during Contraction with Anthropometric Correlations: Cross-Sectional Study. Appl. Sci. 2023, 13, 2589. [Google Scholar] [CrossRef]
- Kamatham, A.T.; Alzamani, M.; Dockum, A.; Sikdar, S.; Mukherjee, B. Sparse Sonomyography-Based Estimation of Isometric Force: A Comparison of Methods and Features. IEEE Trans. Med. Robot. Bionics 2022, 4, 821–829. [Google Scholar] [CrossRef]
- Sjoerdsma, M.; Caresio, C.; Tchang, B.; Meeder, A.; van de Vosse, F.; Lopata, R. The Feasibility of Dynamic Musculoskeletal Function Analysis of the Vastus Lateralis in Endurance Runners Using Continuous, Hands-Free Ultrasound. Appl. Sci. 2021, 11, 1534. [Google Scholar] [CrossRef]
- Heres, H.M.; Sjoerdsma, M.; Schoots, T.; Rutten, M.C.M.; van de Vosse, F.N.; Lopata, R.G.P. Image Acquisition Stability of Fixated Musculoskeletal Sonography in an Exercise Setting: A Quantitative Analysis and Comparison with Freehand Acquisition. J. Med. Ultrason. 2020, 47, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, Z.; Hettiarachchi, N.; Yan, J.; Liu, H. A Wearable Ultrasound System for Sensing Muscular Morphological Deformations. IEEE Trans. Syst. Man Cybern. Syst. 2021, 51, 3370–3379. [Google Scholar] [CrossRef]
- Yang, X.; Liu, Y.; Yin, Z.; Wang, P.; Deng, P.; Zhao, Z.; Liu, H. Simultaneous Prediction of Wrist and Hand Motions via Wearable Ultrasound Sensing for Natural Control of Hand Prostheses. IEEE Trans. Neural Syst. Rehabil. Eng. 2022, 30, 2517–2527. [Google Scholar] [CrossRef]
- Yang, X.; Yan, J.; Chen, Z.; Ding, H.; Liu, H. A Proportional Pattern Recognition Control Scheme for Wearable A-Mode Ultrasound Sensing. IEEE Trans. Ind. Electron. 2020, 67, 800–808. [Google Scholar] [CrossRef]
- Song, P.; Andre, M.; Chitnis, P.; Xu, S.; Croy, T.; Wear, K.; Sikdar, S. Clinical, Safety and Engineering Perspectives on Wearable Ultrasound Technology: A Review. IEEE Trans. Ultrason. Ferroelectr. Freq. Control 2023, 1–1. [Google Scholar] [CrossRef] [PubMed]
- La, T.-G.; Le, L.H. Flexible and Wearable Ultrasound Device for Medical Applications: A Review on Materials, Structural Designs, and Current Challenges. Adv. Mater. Technol. 2022, 7, 2100798. [Google Scholar] [CrossRef]
- Lin, M.; Zhang, Z.; Gao, X.; Bian, Y.; Wu, R.S.; Park, G.; Lou, Z.; Zhang, Z.; Xu, X.; Chen, X.; et al. A Fully Integrated Wearable Ultrasound System to Monitor Deep Tissues in Moving Subjects. Nat. Biotechnol. 2024, 42, 448–457. [Google Scholar] [CrossRef]
- Amado-Rey, A.; GonçalvesSeabra, A.; Stieglitz, T. Towards Ultrasound Wearable Technology for Cardiovascular Monitoring: From Device Development to Clinical Validation. IEEE Rev. Biomed. Eng. 2024, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Bunce, S.M.; Hough, A.D.; Moore, A.P. Measurement of Abdominal Muscle Thickness Using M-Mode Ultrasound Imaging during Functional Activities. Man. Ther. 2004, 9, 41–44. [Google Scholar] [CrossRef]
- Dieterich, A.V.; Pickard, C.M.; Deshon, L.E.; Strauss, G.R.; Gibson, W.; Davey, P.; McKay, J. M-Mode Ultrasound Used to Detect the Onset of Deep Muscle Activity. J. Electromyogr. Kinesiol. 2015, 25, 224–231. [Google Scholar] [CrossRef]
- Tarbox, E.; Akhlaghi, N.; Dhawan, A.; Gammell, P.; Chitnis, P.; Sikdar, S. Low-Power Ultrasound Imaging Systems Using Time Delay Spectrometry. In Proceedings of the 2017 IEEE International Ultrasonics Symposium (IUS); September 2017; pp. 1–1. [Google Scholar]
- Chitnis, P.; Akhlaghi, N.; Tarbox, E.; Otto, P.; Gammell, P. (72) Inventors: Siddhartha Sikdar, Washington, DC. 24.
- Bashatah, A.; Mukherjee, B.; Rima, A.; Patwardhan, S.; Otto, P.; Sutherland, R.; King, E.L.; Lancaster, B.; Aher, A.; Gibson, G.; et al. Wearable Ultrasound System Using Low-Voltage Time Delay Spectrometry for Dynamic Tissue Imaging. IEEE Trans. Biomed. Eng. 2024, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Van Melick, N.; Meddeler, B.M.; Hoogeboom, T.J.; Nijhuis-van Der Sanden, M.W.G.; Van Cingel, R.E.H. How to Determine Leg Dominance: The Agreement between Self-Reported and Observed Performance in Healthy Adults. PLOS ONE 2017, 12, e0189876. [Google Scholar] [CrossRef]
- Hoerl, A.E.; Kennard, R.W. Ridge Regression: Biased Estimation for Nonorthogonal Problems. Technometrics 2000, 42, 80–86. [Google Scholar] [CrossRef]
- Hart, J.M.; Pietrosimone, B.; Hertel, J.; Ingersoll, C.D. Quadriceps Activation Following Knee Injuries: A Systematic Review. J. Athl. Train. 2010, 45, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Smeets, A.; Malfait, B.; Dingenen, B.; Robinson, M.A.; Vanrenterghem, J.; Peers, K.; Nijs, S.; Vereecken, S.; Staes, F.; Verschueren, S. Is Knee Neuromuscular Activity Related to Anterior Cruciate Ligament Injury Risk? A Pilot Study. The Knee 2019, 26, 40–51. [Google Scholar] [CrossRef]
- Varol, U.; Navarro-Santana, M.J.; Valera-Calero, J.A.; Antón-Ramírez, S.; Álvaro-Martínez, J.; Díaz-Arribas, M.J.; Fernández-de-las-Peñas, C.; Plaza-Manzano, G. Convergent Validity between Electromyographic Muscle Activity, Ultrasound Muscle Thickness and Dynamometric Force Measurement for Assessing Muscle. Sensors 2023, 23, 2030. [Google Scholar] [CrossRef]
- Zeng, J.; Zhou, Y.; Yang, Y.; Xu, Z.; Zhang, H.; Liu, H. Robustness of Combined sEMG and Ultrasound Modalities Against Muscle Fatigue in Force Estimation. In Proceedings of the Intelligent Robotics and Applications; Liu, X.-J., Nie, Z., Yu, J., Xie, F., Song, R., Eds.; Springer International Publishing: Cham, 2021; pp. 213–221. [Google Scholar]
- Xia, W.; Zhou, Y.; Yang, X.; He, K.; Liu, H. Toward Portable Hybrid Surface Electromyography/A-Mode Ultrasound Sensing for Human–Machine Interface. IEEE Sens. J. 2019, 19, 5219–5228. [Google Scholar] [CrossRef]
- Zhang, Q.; Iyer, A.; Kim, K.; Sharma, N. Evaluation of Non-Invasive Ankle Joint Effort Prediction Methods for Use in Neurorehabilitation Using Electromyography and Ultrasound Imaging. IEEE Trans. Biomed. Eng. 2021, 68, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- Tweedell, A.J.; Tenan, M.S.; Haynes, C.A. Differences in Muscle Contraction Onset as Determined by Ultrasound and Electromyography. Muscle Nerve 2019, 59, 494–500. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, Z.; Hettiarachchi, N.; Yan, J.; Liu, H. A Wearable Ultrasound System for Sensing Muscular Morphological Deformations. IEEE Trans. Syst. Man Cybern. Syst. 2021, 51, 3370–3379. [Google Scholar] [CrossRef]
- Paoletta, M.; Moretti, A.; Liguori, S.; Snichelotto, F.; Menditto, I.; Toro, G.; Gimigliano, F.; Iolascon, G. Ultrasound Imaging in Sport-Related Muscle Injuries: Pitfalls and Opportunities. Medicina (Mex.) 2021, 57, 1040. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).