Introduction
Exhaled volatile biomarkers carry information about the state of human health (Davis et al., 2021). The importance of the development of short-turnaround-time, random-access, point-of-care devices for the diagnosis of SARS-CoV-2 infections cannot be overemphasized. These tests might be very important for mass screening, real-time patient management, and infection control decisions (Tang et al., 2020).
Many of the current metabolomics studies aim to identify specific biomarkers. Different specific statistical and machine learning strategies were used to create multi-metabolite biomarker models (Xia, et al., 2012).
The advantages of using metabolite biomarkers (speed, reproducibility, quantitative accuracy, low cost, non-invasiveness, and small sample volume) combined with the remarkable success of newborn screening programmes have inspired many metabolomics researchers to pursue biomarker studies for other diseases (Chace, 2001; Wilcken et al., 2003).
Many techniques were applied for volatile biomarker quantification, such as real-time, online Proton Transfer Reaction time-of-flight mass spectrometry (PTR-MS) to perform a metabolomic analysis of expired air from patients with acute respiratory distress syndrome (ARDS) (Grassin-Delyle et al., 2021). A characteristic breathprint for COVID-19 has been identified in a multivariate analysis. In addition, PTR-MS and selected ion flow tube (SIFT)-MS, mass spectrometry with Ionic Molecule Reactions (IMR-MS), Ion Mobility Spectrometry (IMS), and Field Asymmetric Ion Mobility Spectrometry (FAIMS) have also been used to diagnose various diseases using VOC metabolites (Beliza’rio et al., 2021).
These techniques enabled researchers to establish links between specific VOC molecular biomarkers and various diseases. However, the methods described are developed mainly for laboratory conditions; they are expensive, not suitable for clinical environments, require a long time to process data, or require highly-trained personnel to operate and analyse data.
Here we are using a GC-IMS developed for clinical environments, enabling fast diagnostics and making it affordable for wider users (Ruszkiewicz et al., 2020). The GC-IMS technique is similar to GC-MS, but it is operated at atmospheric pressure. The GC-IMS delivers selectivity and sensitivity sufficient to identify molecular biomarkers. A novel data processing method based on Machine Learning (ML) algorithms has been especially developed and tested in this study.
Experimental
Participants Profile
Data were collected at the Medical University of South Carolina (MUSC). Two cohorts of COVID-19 positive participants were studied, including hospitalised patients with chronic symptoms.
For the trials, consenting participants in the trial were a mixture of COVID-19-infected patients in wards and non-COVID-19-infected participants. Eligibility criteria were defined as follows: The minimum age for participation was 16 years old. All ethnicities and genders were included. Exclusion criteria were: those below age 16, those unable to consent, ITU patients on a ventilator, and pregnant women. The breakdown of participants is shown in
Table S1;
Supplementary Materials. The median age was 60.5;
Supplementary Materials.
For the trial, the age of participants ranged from 24 to 99. The median age was 60.5. More information on the age distribution and ethnicity is shown in the
Supplementary Materials.
For the analysis, some cases were excluded, for the following reasons:
- I.
The data quality rating was Q2 or Q3. This excluded 9 from the main dataset (see section on data quality).
- II.
The Covid-19 PCR swab test was negative, but the patient was symptomatic (these were tagged as “suspected” Covid-19). This accounted for 1 case in the.
Data Collection
The breath data were sampled in clinical environments using a BreathSpec
© GC-IMS developed by G.A.S. mbH (Germany), Ruszkiewicz
et al., (2020). The breath samples were acquired by having the patient blow into a specially designed breath sampler (
Supplementary Materials).
The GC-IMS records volatile biomarkers from a breath sample. A raw spectrum comprises a 2D dataset that is defined by the GC retention time and the IMS drift time. A typical spectrum of a breath sampler contains a vast number of individual molecular VOC biomarkers (Ruszkiewicz et al., 2020). Extended libraries of both the retention times and the drift times are available for most chemicals.
Machine Learning Algorithm
Currently, the breath diagnostics paradigm is based on the identification of specific and non-specific biomarkers associated with a disease (Amman and Smith, 2013). Often, a disease generates a few specific biomarkers and many non-specific biomarkers common to many different diseases and conditions. For example, for the COVID-19 VOC 11, non-specific markers are identified by Ruszkiewicz et al. (2020). These VOC markers are not unique and are present in many breath samples associated with ketosis, gastrointestinal effects, and inflammatory processes. Therefore, the presence of a non-specific marker cannot be used for diagnostics.
For the COVID-19 diagnostics, as for many other diseases, variations in the signal intensity of selected markers enable one to find a statistically significant association with the decrease. These pattern variations were used for the diagnostic.
While it seems to be possible to manually analyse peaks in the spectra to identify COVID-19 markers, in reality, this is very difficult or often practically impossible due to the large number of spectra obtained. It might be beneficial to develop an automatic procedure to analyse and classify the entire spectrum. Therefore, in this study, instead of identifying biomarkers and their variation patterns, a new approach is suggested. It is based on a unique ML algorithm advantage, enabling one to identify patterns in the entire dataset without identification of the individual VOC biomarkers. It is expected to work much faster than a traditional approach based on the identified biomarkers.
Machine learning algorithms used here have been developed to find diagnostic features automatically in large datasets by utilising Deep Neural Networks (DNN). The dataset obtained in this trial was analysed by these algorithms (DeepBreath, Deeplight Technologies Ltd.). These algorithms are designed to work with the 2D spectra output by the GC-IMS or similar instruments, e.g., GC-MS or GC-GC.
In operation, a breath biomarker spectrum and a set of clinical data are introduced to the trained software (DeepBreath), where the information flows through several layers of neural networks (Cover and Thomas, 2006). During this information flow, more and more features relevant to positive and negative elements are accumulated. And finally, the flow of information is coming out of the neural network with predominantly positive or negative diagnostic results. After training, a single GC-IMS spectra is processed in 5 to 10 seconds.
Results and Discussion
Then the model was trained using a proportion of the dataset, keeping the remainder for testing. During the testing, for each participant, a score between zero and one was generated by the ML algorithms to indicate the likelihood of a negative (zero) or positive (one) diagnosis. The score depends on how well the features in the spectra match with the trained model. A well-matched spectrum will give a score close to the RT-PCR diagnosis for that participant, i.e., close to zero for a COVID-19 “negative participant” and close to 1 for a COVID-19 “positive participant.”
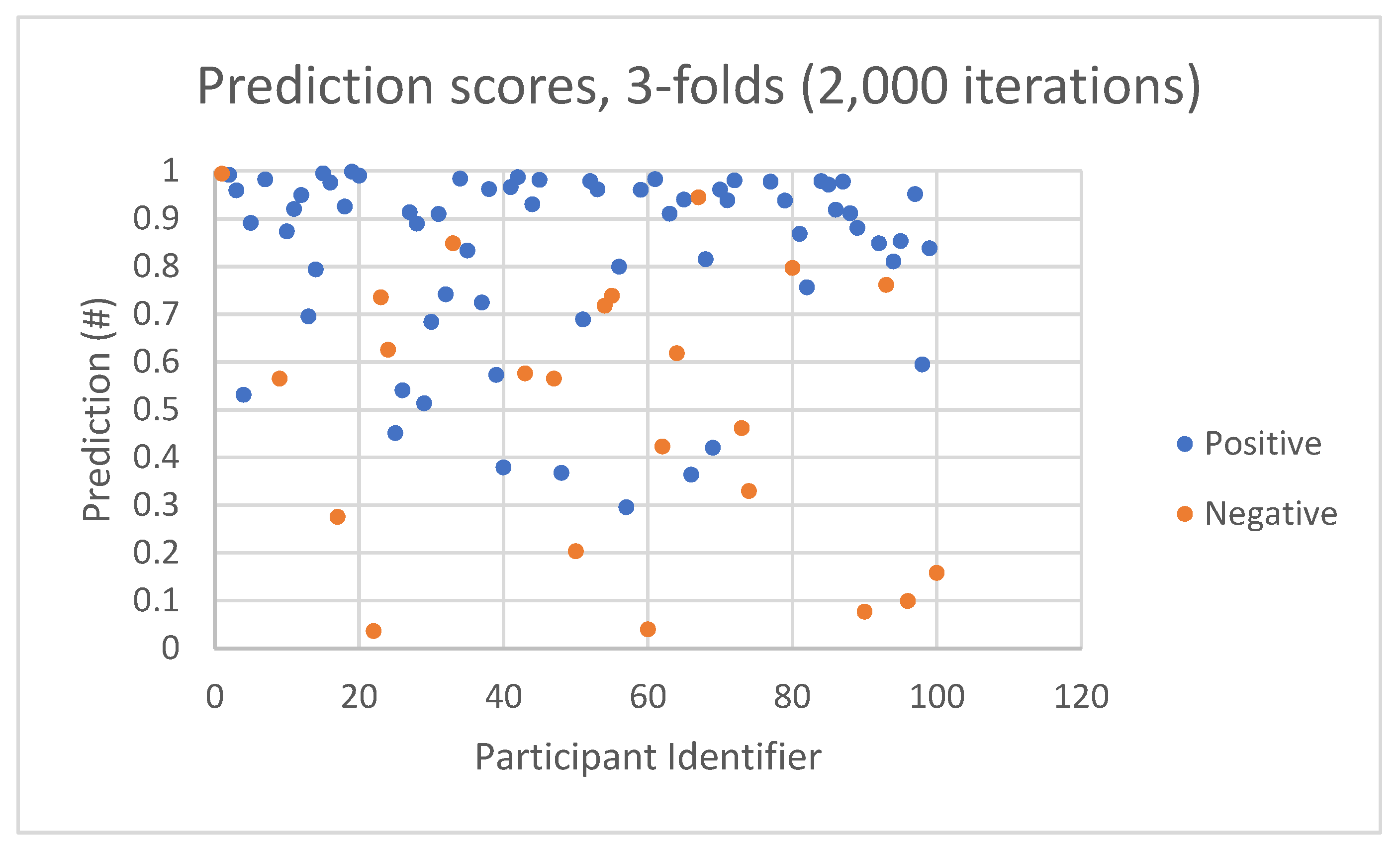
An example of the prediction score (
Figure 4) shows two pluralities of positive and negative dots. Each dot represents a participant with a COVID-19 diagnostic prediction score in the range of 0 to 1. Ideally, one can expect dots to be fully separated and located near the perdition score 1 for “positive participant” and near the perdition score 0 for “negative participant.” In reality, two important factors have to be taken into account:
Figure 4.
An example of prediction scores by participant identifiers.
Figure 4.
An example of prediction scores by participant identifiers.
In
Figure 4, the majority of positive prediction scores (blue dots) are in the range of 0.8 to 1. However, a small number of positive prediction scores are spread over 0.3 to 0.8. The negative prediction scores (orange dots) are spread over a wider range from 0.04 to 0.95.
It should be noted that the prediction score is not a full Bayesian probability and is shown as an example. The prediction score is influenced by the randomization of data for training and testing of the ML algorithms. Importantly, the prediction score shows the statistical nature of the ML algorithms. These graphs enable one to quantify the accuracy, sensitivity, and selectivity of the diagnostics without using conventional statistical models (Bzdok
et al., 2018); see
Supplementary Materials.
A ROCAUC of 84.1 was obtained, with a 95% CI of 1.3 (
Table 5). This gives a precision of 93.6% (±5.7%) and an accuracy of 78.4% (±4.8%). These results seem to indicate that there is a solid difference between COVID-19 positive and negative participants in testing unknown participants using ML algorithms trained on known diagnostics.
A review of the accuracy of COVID-19 reference methods reported false negative rates ranging from 2% to 29% (or sensitivity 71–98%) based on negative RT-PCR tests that were positive on repeat testing (Arevalo-Rodriguez et al., 2020). The repeat RT-PCR testing is likely to underestimate the true rate of false negatives because of the virus load that is influenced by the stage of disease (Sethuraman et al., 2020) and degree of viral multiplication (Wölfel et al., 2020). The probability of having a negative RT-PCR test result is also influenced by the number of days since exposure (Kucirka et al., 2020), which can also be attributed to variation in the virus load. The average viral load in some respiratory specimens is 105 copies per mL, and maximal virus loading is found at circa 108 copies per mL. The viral loads in throat swab and sputum samples peaked around 5–6 days after symptom onset, ranging from around 103 to 7·108 copies per mL according to To et al. (2020) and Health Information and Quality Authority (2020). There are also false positives in PCR tests due to the ability to detect RNA fragments in inactivated viruses.
Given the allowance for PCR accuracy being below 100%, we have to consider that the breath diagnostic results are being influenced by the accuracy of PCR reference methods. This is less important for symptomatic cases, but it is expected to be more important for asymptomatic cases, where we know PCR accuracy is expected to be far less than 100%. The ROCAUC 84.1% is an indication of the strong predictive power of the GC-IMS with the DeepBreath ML data processing algorithms to discriminate between positive and negative cases of COVID-19 and, therefore, facilitate a fast diagnosis of this infection.
The data analysis has been undertaken without identification of the biomarkers. All features, such as molecular VOC, were included in the training and testing algorithms. The strong prediction score confirms the assumption that the ML algorithms automatically select predictive features and analyse them. This approach is faster than conventional, requires fewer resources, and can constantly improve performance by adding new data to the training and testing rounds.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Acknowledgements
G. Silvestri for supplying the raw GC-IMS spectra.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Amman A, Smith D. Volatile biomarkers. Amsterdam: Elsevier BV; 2013.
- Arevalo-Rodriguez I, Buitrago-Garcia D, Simancas-Racines D, et al. (2020). False-negative results of initial RT-PCR assays for covid-19: a systematic review. medRxiv 20066787. 2020. [CrossRef]
- Barker, M., & Rayens, W. (2003). Partial least squares for discrimination. Journal of Chemometrics, 17(3), 166–173. [CrossRef]
- Beauchamp J, Davis C, Pleil J. (2020). Breathborne biomarkers and the human volatilome. Elsevier Science, 2nd Edition; 722p. ISBN: 9780128199671.
- Beliza’rio J. E., J Faintuch and M G Malpartida (2021). Breath Biopsy and Discovery of Exclusive Volatile Organic Compounds for Diagnosis of Infectious Diseases. Frontiers in Cellular and Infection Microbiology; 10. [CrossRef]
- Bourgon, R., Gentleman, R., & Huber, W. (2010). Independent filtering increases detection power for high-throughput experiments. Proceedings of the National Academy of Sciences of the United States of America, 107(21), 9546–9551. [CrossRef]
- Breiman, L. (2001). Random forests. Machine Learning, 45(1), 5–32.
- Bzdok D, Altman N, Krzywinski M. (2018). Statistics versus Machine Learning. Nature Methods; 15:233-234.
- Chace, D. H. (2001). Mass spectrometry in the clinical laboratory. Chemical Reviews, 101(2), 445–477. [CrossRef]
- Cortes, C., & Vapnik, V. (1995). Support-vector networks. Machine Learning, 20(3), 273–297.
- Cover, T. M. and Thomas, J. A. Elements of Information Theory. John Wiley & Sons, 2nd edition, 2006.
- Davis C. E., M. Schivo, N. J. Kenyon (2021). A breath of fresh air; the potential for COVID-19 breath diagnostics. EBioMedicine, 63. [CrossRef]
- Eriksson, L., Johansson, E., Kettaneh-Wold, N., & Wold, S. (2001). Multi- and Megavariate Data Analysis Principles and Applications. Dublin: Umetrics Academy.
- Grassin-Delyle S., C. Roquencourt, P. Moine, G. Saffroy, Stanislas Carn, N. Heming, J. Fleuriet, H. Salvator, E. Naline, l-J Couderc, P. Devillier, E. A. Thevenot, D. Annane (2021). Metabolomics of exhaled breath in critically ill COVID-19 patients: A pilot study EBioMedicine, 63. [CrossRef]
- Jianguo Xia, David I. Broadhurst, Michael Wilson, David S. Wishart (2012). Translational biomarker discovery in clinical metabolomics: an introductory tutorial, Metabolomics. [CrossRef]
- Hackstadt, A. J., & Hess, A. M. (2009). Filtering for increased power for microarray data analysis. BMC Bioinformatics, 10, 11.
- Health Information and Quality Authority (2020). Evidence summary for SARS-CoV-2 viral load and infectivity over the course of an infection. 9 June 2020, Health Information and Quality Authority. Report. https://www.hiqa.ie/sites/default/files/2020-06/Evidence-Summary_SARS- CoV-2-duration-of-infectivity.pdf.
- Kohl, S. M., Klein, M. S., Hochrein, J., Oefner, P. J., Spang, R., & Gronwald, W. (2012). State-of-the art data normalization methods improve NMR-based metabolomic analysis. Metabolo- mics, 8(Suppl 1), 146–160.
- Kucirka L. M.; Lauer S. A.; Laeyendecker O.; Boon D.; and Lessler J. (2020). Variation in False-Negative Rate of Reverse Transcriptase Polymerase Chain Reaction–Based SARS-CoV-2 Tests by Time Since Exposure. Annals of Internal Medicine. [CrossRef]
- Ruszkiewicz DM, Sanders D, O’Brien R, Hempel F, Reed, MJ Riepe,AC, Bailie K, Brodrick E, Darnley, K Ellerkmann, R Mueller, O Skarysz A, Truss M, Wortelmann T, Yordanov S, Thomas CLP, Schaaf B & Eddleston M (2020). Diagnosis of COVID-19 by analysis of breath with gas chromatography-ion mobility spectrometry - a feasibility study. EClinicalMedicine, 29-30, 100609; (http://creativecommons.org/licenses/by-nc-nd/4.0/).
- Saeys, Y., Inza, I., & Larranaga, P. (2007). A review of feature selection techniques in bioinformatics. Bioinformatics, 23(19), 2507–2517.
- Sethuraman N, Sundararaj Stanleyraj J, Ryo A. (2020). Interpreting diagnostic tests for SARS-CoV-2. JAMA 2020. [CrossRef]
- Szymanska, E., Saccenti, E., Smilde, A. K., & Westerhuis, J. A. (2012). Double-check: Validation of diagnostic statistics for PLS-DA models in metabolomics studies. Metabolomics, 8(Sup- pl 1), 3–16.
- Tang Y-W, J E. Schmitz, D H. Persing, C W. Strattonb (2020). Laboratory Diagnosis of COVID-19: Current Issues and Challenges; Journal of Clinical Microbiology, 58, p. 1-9.
- To, K.K.W., Tsang, O.T.Y., Leung, W.S., Tam, A.R., Wu, T.C., Lung, D.C., Yip, C.C.Y., Cai, J.P., Chan, J.M.C., Chik, T.S.H., Lau, D.P.L., Choi, C.Y.C., Chen, L.L., Chan, W.M., Chan, K.H., Ip, J.D., Ng, A.C.K., Poon, R.W.S., Luo, C.T.,... Yuen, K.Y. (2020). Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: An observational cohort study. Lancet Infect. Dis. 20, 565–574. [CrossRef]
- Trygg, J., Holmes, E., & Lundstedt, T. (2007). Chemometrics in metabolomics. Journal of Proteome Research, 6(2), 469–479.
- van den Berg, R. A., Hoefsloot, H. C., Westerhuis, J. A., Smilde, A. K., & van der Werf, M. J. (2006). Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genomics, 7, 142.
- Wilcken, B., Wiley, V., Hammond, J., & Carpenter, K. (2003). Screening newborns for inborn errors of metabolism by tandem mass spectrometry. New England Journal of Medicine, 348(23), 2304–2312.
- Wölfel R, Corman VM, Guggemos W, et al. (2020). Virological assessment of hospitalized patients with COVID-2019. Nature 2020. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).