1. Introduction
Cancer is a fatal disease and a significantly global health concern due to its poor prognosis and the overexpression of genes in various cancer cells. Traditional clinical treatments are often associated with long-term physical and mental burdens, as well the development of resistance and resurgence [
1]. Consequently, extensive research has focused on developing novel strategies to improve treatment outcomes and reduce these burdens, specially targeting the overexpressed genes.
One promising approach attracting the health care community’s attention is Small Interfering RNAs (siRNA)-mediated gene silencing, a promising type of RNA-based therapeutics. The effectiveness of RNA-based gene therapy hinges on the intracellular functionality of therapeutic RNA within target cells. Categorization by their biochemical mechanisms, these therapies require careful consideration of clinically significant drug delivery method [
2]. The Food and Drug Administration (FDA) approved the first siRNA drug 20 years after the first report of RNAi in eukaryotic cells Recently, FDA and the European Medicines Agency (EMA) approved siRNA therapeutics include Patisiran for hereditary transthyretin-mediated amyloidosis (hATTR), Givosiran for acute hepatic porphyria, Lumasiran for primary hyperoxaluria type 1, and Inclisiran for hypercholesterolemia [
3]. Additionally, the FDA has accepted a new drug application for Vutrisiran, targeting polyneuropathy associated with hATTR amyloidosis [
2,
3].
siRNA has been delivered using various drug delivery approaches, including polymers [
4], aptamers [
5], lipids nanoparticles [
6], viral carriers [
7], and dendrimers [
8,
9]. However, these techniques still have limitations, such as cargo-carrying capacity, low delivery efficiency, the risk of mutation, high cytotoxicity, and lack of target specificity [
10]. A possible alternative siRNA delivery system is afforded by the use of appropriate Cell-penetrating peptides (CPPs).
CPPs containing cationic amino acids such as lysine and arginine (R) and hydrophobic residues such as tryptophan (W), alanine, or phenylalanine have been used for siRNA delivery [
11,
12,
13]. The acylation of CPPs with suitable fatty acyl chains has enhanced siRNA delivery by providing a hydrophobic region to entrap relatively large biomolecules. Furthermore, the positively charged residues, such as R, of CPPs can interact with the negatively charged backbone of siRNAs [
14]. Various combinations of R and W in CPPs have shown promise in intracellular siRNA delivery [
15,
16]. Previous data indicated that oleyl-conjugated CGRKR peptides can deliver siRNA into tumor cells due to enhanced hydrophobicity [
17,
18]. A follow-up study from our laboratories demonstrated the application of histidine (H) and R in CPPs with an oleyl chain in delivering siRNA with promising cellular uptake and gene silencing efficiency in triple-negative breast cancer cells compared to standard transfecting agents, e.g., lipofectamine [
18].
It has been observed that unsaturated fatty acids also enhance membrane permeability. The presence of R residue provides strong binding interactions to siRNA, whereas H residue helps create a proton sponge effect in endosomes, aiding in endosomal escape. H residue was selected due to its ability to utilize its imidazole ring as an endosome destabilization mechanism, facilitating the delivery of siRNA into the cytosol. The imidazole ring of histidine acts as a weak base, acquiring a cationic charge when the local pH environment drops below pH 6 [
19]. The addition of W residues enhances the amphiphilic properties of peptides. Therefore, we hypothesize that fatty acylation of (WRH)
n peptide with an oleyl chain (n = 1-4) (
Figure 1) could enhance siRNA delivery. The use of linear WRH peptides with fatty acyl chains has not yet been explored in drug delivery. To the best of our knowledge, there has been no previous report demonstrating the use of linear peptides constituting oleic acid with W, R, and H residues for siRNA delivery in MDA-MB-231 and SK-OV-3 cells. Therefore, we designed linear (WRH)
n peptides (n = 1-4) with or without oleic acid chains.
In this study, we synthesized a siRNA delivery system using Oleyl-WRH peptides using solid-phase peptide synthesis and evaluated their cytotoxicity, cellular uptake of peptide:siRNA complexes, and their STAT3 gene silencing efficiency. It is believed that amino acids with hydrophobic properties, like W, can generate a hydrophobic region and provide amphiphilicity to the peptide. Our studies will generate data to provide the feasibility of using appropriate oleyl-WRH peptides for siRNA delivery.
3. Materials and Methods
3.1. Materials
The amino acid building blocks Fmoc-His(Trt)-OH, Fmoc-Arg(Pbf)-OH, and Fmoc-Trp(Boc)-OH were obtained from AAPPTec LLC (Louisville, KY, USA). The resin H-His-(Trt)-Cl-2-chlorotrityl (loading capacity = 0.724 mmol/g) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Coupling reagents, including HCTU, DMF, DIPEA, trifluoroacetic acid (TFA), acetic acid, triisoproylsilane (TIS), and piperidine were purchased from Sigma-Aldrich Chemical Co. (Milwaukee, WI, USA) and used without further purification. F′-siRNA (Catalog #: AM4635-AM4636) was obtained from Ambion Inc. (Austin, TX, USA). All cell biology reagents were procured from Wilken Scientific (Pawtucket, RI, USA) or Fisher Scientific (Hanover Park, IL, USA).
The chemical structures of the peptides were confirmed by analyzing their masses using a high-resolution matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometer (model # GT 0264, Bruker Inc., Fremont, CA, USA) in positive ion mode with α-cyano-4-hydroxycinnamic acid as the matrix. Crude peptide purification was carried out using a reverse-phase high-performance liquid chromatography (RP-HPLC) system (Shimadzu LC-20AP, Canby, OR, USA) equipped with a Waters XBridge BEH130 preparative column (10 µm, 110 Å, 21.2 × 250 mm), employing a gradient elution of acetonitrile and water with 0.1% trifluoroacetic acid (TFA, v/v) at a flow rate of 6 mL/min, and monitored at 220 nm.
Hanks’ Balanced Salt Solution (HBSS), fetal bovine serum (FBS), penicillin (10,000 U/mL), and streptomycin (10 mg/mL) were supplied by Life Technologies (Grand Island, NY, USA). Western blot materials, including 10% Mini-PROTEAN® TGX Stain-Free Protein Gels, Clarity Western ECL Substrate, Trans-blot® Turbo TM Cassettes, and Trans-Blot® Turbo Mini PVDF Transfer Packs, were obtained from Bio-Rad (Hercules, CA, USA). 4,6-Diamidino-2-phenylindole (DAPI) was provided by Vector Laboratories (Burlingame, CA, USA). Phosphate-buffered saline (PBS, 20x concentration), sulforhodamine 101 (Texas Red), and other chemicals were purchased from VWR (Radnor, PA, USA). Scrambled negative control siRNA (Catalogue no. AM4635), Alexa Fluor 488-labeled negative control siRNA (Catalogue no. 1027292), and siRNA targeting STAT3 (Catalogue no. SI02662338) were acquired from Thermo Fisher. Monoclonal antibodies against STAT3 (Mouse, Catalogue no. 9139S) and GAPDH (Mouse, Catalogue no. 97166S) were supplied by Cell Signaling Technology, Inc. (Danvers, MA, USA). Mouse IgG HRP-conjugated antibody was purchased from R&D Systems (Catalogue no. HAF007).
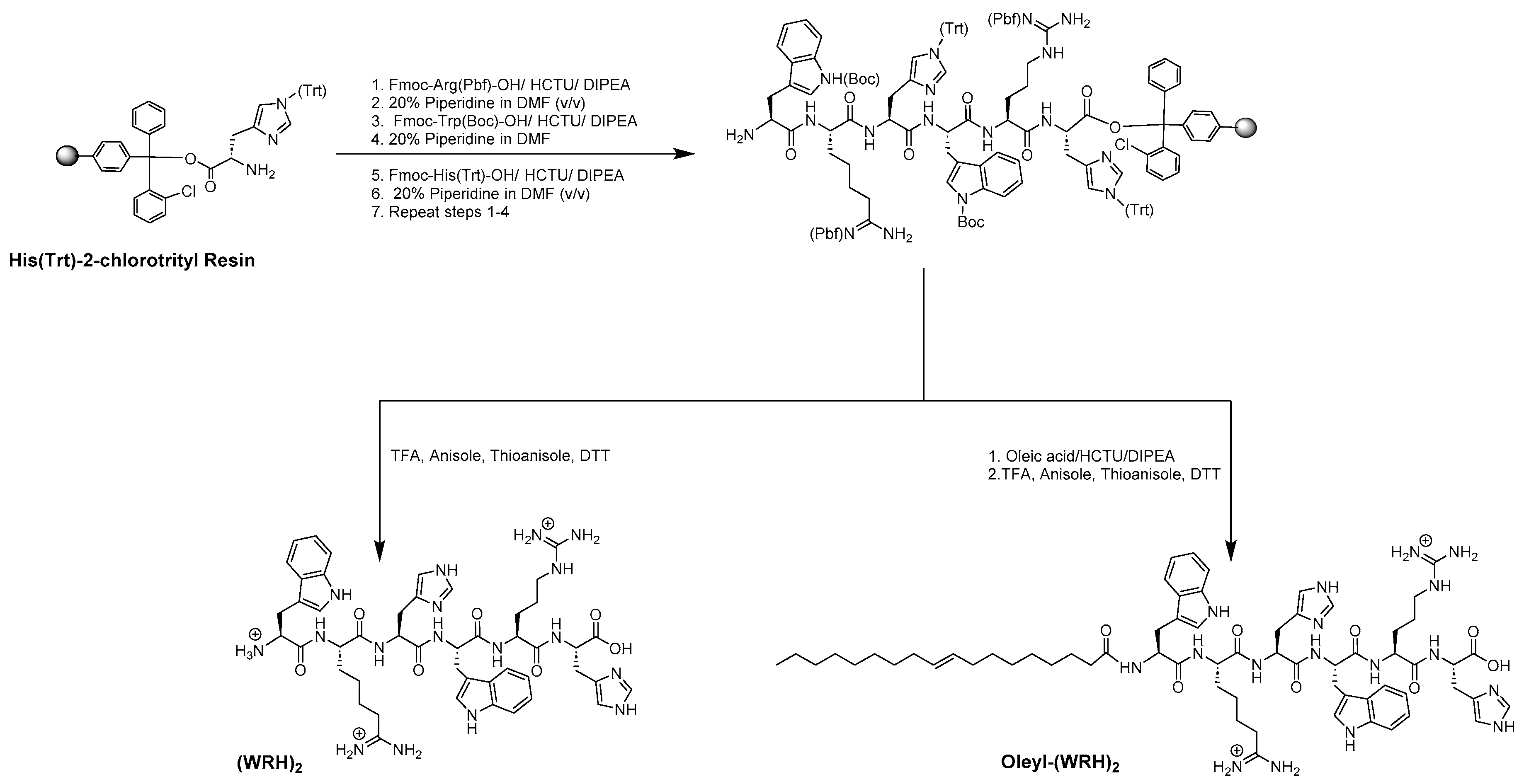
3.2. Solid-Phase Peptide Synthesis of Unmodified and Oleyl-Conjugated Peptides
The peptides were synthesized via Fmoc/tBu solid-phase peptide synthesis, following the procedures detailed in previously reported methods [
18], exemplified in
Scheme 1. In summary, the synthesis utilized preloaded H-His-(Trt)-Cl-2-chlorotrityl resin (loading capacity = 0.724 mmol/g) as the solid support. Peptides containing alternating tryptophan, arginine, and histidine residues were synthesized on a 0.50 mmol scale, following the specified peptide sequence. Subsequently, complete cleavage was achieved by incubating with a freshly prepared cleavage cocktail reagent R (trifluoroacetic acid (TFA): thioanisole: anisole: dithiothreitol (DTT) (92:5:2:3; v/v/v/v)) for 4 hours to obtain non-oleyl peptides.
For the oleyl-conjugated peptides, following the conjugation of the last amino acid and Fmoc deprotection steps, N-terminal acylation with oleic acid was carried out in the presence of HCTU/DIPEA. The resin was thoroughly washed with DMF. Fmoc deprotection was achieved using piperidine (20% v/v) in DMF, and the Kaiser test confirmed completion of the coupling process with all free amino groups reacted. Finally, all peptide conjugates were cleaved using a freshly prepared cleavage cocktail reagent R (TFA: thioanisole: anisole: DTT, 92:5:2:3; v/v/v/v) for 4 h.
All crude peptides were precipitated using cold diethyl ether, centrifuged, and subsequently purified using reverse-phase high-performance liquid chromatography (RP-HPLC) on a Shimadzu LC-20AP Prominence system. The purification utilized a gradient elution system consisting of water with 0.1% TFA and acetonitrile (CH
3CN) with 0.1% TFA (v/v) (5% to 95% CH
3CN over 60 min) on a C18 column (00G-4436-P0-AX, Gemini Prep C18, 10 μm particle size). Fractions were collected and analyzed using MALDI-TOF mass spectrometry. Fractions containing the expected compounds were pooled and lyophilized to obtain solid peptide powders. Analytical HPLC was conducted to confirm the purity (>95%) of the most effective compounds used in biological assays.
Table 1 outlines the manual peptide synthesis steps.
(WRH)1: MALDI-TOF (m/z) C23H33N9O4 Calculated: 499.2645, Found: 499.4015 [M+2H]+; (WRH)2: MALDI-TOF (m/z) C46H62N18O7 Calculated: 978.5038, Found: 978.4187 [M+2H]+; (WRH)3: MALDI-TOF (m/z) C69H90N27O10 Calculated: 1456.7358, Found: 1456.5664 [M+H]+; (WRH)4: MALDI-TOF (m/z) C92H119N36O13 Calculated: 1935.9752, Found: 1935.6767 [M+H]+; Oleyl-(WRH)1: MALDI-TOF (m/z) C41H65N9O5 Calculated: 763.5098, Found: 763.6974 [M+2H]+; Oleyl-(WRH)2: MALDI-TOF (m/z) C64H94N18O8 Calculated: 1242.7491, Found: 1242.9278 [M+2H]+; Oleyl-(WRH)3: MALDI-TOF (m/z) C87H122N27O11 Calculated: 1720.9812, Found: 1720.6871 [M+H]+; Oleyl-(WRH)4: MALDI-TOF (m/z) C112H152N36O14 Calculated: 2201.2278, Found: 2201.7647 [M+2H]+.
3.3. Complex Formation of Oleyl Conjugated Peptides and Scrambled siRNA
The complexation of oleyl-conjugated peptides and siRNA involved physically mixing them together. Appropriate volumes of oleyl-conjugated peptides and scrambled siRNA solutions were combined in Hank’s Balanced Salt Solution (HBSS) buffer to form the complexes. These complexes were formed through ionic interactions between the positively charged arginine residues of the conjugates and the negatively charged phosphate groups of siRNA.
The final concentration of siRNA complexes remained constant, while the concentration of the conjugates was incrementally adjusted to achieve various N/P ratios. The N/P ratio, where N represents the moles of ionizable nitrogen in the delivery agent and P represents the moles of phosphate groups in siRNA, was calculated using the formula:
3.4. Dynamic Light Scattering
The peptide-siRNA complexes were analyzed using dynamic light scattering (DLS) methodology. The Malvern Nano ZS Zetasizer (Westborough, MA, USA) was employed to measure the hydrodynamic diameter and surface charge of the complexes at 25°C. Zeta potential measurements were conducted at 40 V using disposable folded capillary cells (DTS1070). The instrument was calibrated with a transfer standard, and disposable cuvettes were utilized for determining the hydrodynamic diameter.
A specific N/P ratio was selected for the DLS experiments, maintaining a final siRNA concentration of 50 nM within the complexes. Each measurement was performed in triplicate, with each run consisting of 20 scans. Zeta potential values were calculated using the Smoluchowski model, and all results met the instrument's quality standards.
Particle size analysis was conducted at identical N/P ratios using consistent concentrations of siRNA and oleyl-conjugated peptides. Each sample was analyzed in triplicate with an automatic attenuator setting. The results were validated against the instrument's quality standards and reported as the mean ± SD from three independent experiments.
3.5. Gel Retardation Assay for Binding Affinity
Gel electrophoresis was employed to assess the binding affinity of the peptides with scrambled siRNA. The synthesized peptides were mixed with siRNA at N/P ratios ranging from 0 to 60, maintaining a final siRNA concentration of 36 nM in the complexes. Complex formation occurred in 1X PBS buffer at room temperature. The negative control (N/P ratio 0) lacked peptides entirely.
Freshly prepared 1% agarose gel was stained with 0.5 μg/mL ethidium bromide. Prior to loading into the wells, each complex received 5 μL of 6X purple gel loading dye. Gel electrophoresis was conducted at 70 V and 400 mA for 20 min in TAE buffer (2 M Tris base, 1 M glacial acetic acid, 0.5 M sodium EDTA, pH 8.3). Band visualization utilized the ChemiDoc XRS+ system with CCD high-resolution imaging (Bio-Rad Imager), and band intensities (indicative of unbound siRNA) were quantified using Image LabTM software. The experiment was performed in triplicate, and the percentage of binding (%Binding) was calculated using the formula:
3.6. Protection of siRNA against Enzymatic Degradation
In the harsh biological environment of the human body, various endonucleases rapidly degrade nucleic acids upon entry. Here, we investigated the ability of oleyl-conjugated peptides to protect siRNA from these endonucleases. Peptide-siRNA complexes were prepared at N/P ratios ranging from 0 to 60, with a final siRNA concentration of 36 nM. Fetal bovine serum (FBS) served as a positive control, while Hank’s Balanced Salt Solution (HBSS) served as the negative control, representing 100% intact siRNA.
Each peptide-siRNA complex at different N/P ratios was exposed to a 25% (v/v) FBS solution in HBSS and then incubated at 37°C for 24 h. After incubation, the heparin competition assay was employed to dissociate the peptide-siRNA complex and assess the amount of intact siRNA remaining after exposure to FBS. Specifically, 5 μL of a 2:3 mixture (v/v) of heparin (5% solution in normal saline) and ethylenediaminetetraacetic acid (0.5 mM) was added to each complex and incubated for 10 min. Subsequently, 5 μL of 6X purple gel loading dye was added to each mixture, and samples were analyzed using 1% ethidium bromide gel electrophoresis at 70 V and 400 mA for 20 minutes. Gels were visualized under UV illumination, and band intensities were quantified using Image LabTM software.
3.7. Cell Culture and In Vitro Cytotoxicity Assay
The cell lines selected for this study included MDA-MB-231 (ATCC No. HTB-26, triple-negative breast cancer cell line), MCF-7 (ATCC No. HTB-22, breast adenocarcinoma cells), HEK-293 (ATCC No. CRL-1573, normal kidney cells), and SK-OV-3 (ATCC No. HTB-77, human epithelial ovary adenocarcinoma cells).
MDA-MB-231, MCF-7, and SK-OV-3 cell lines were cultured in DMEM media supplemented with 10% fetal bovine serum (FBS 10%) and a penicillin-streptomycin solution (10,000 units of penicillin and 10 mg of streptomycin per mL in 0.9% NaCl, 1%). All cell lines were maintained in a humidified atmosphere containing 5% CO2 and 95% air at 37°C. They were handled under sterile conditions within Biological Safety Cabinets throughout the study.
The cytotoxicity of the synthesized oleyl-conjugated peptides complexed with scrambled siRNA was evaluated in vitro using the aforementioned cell lines. Peptide-siRNA complexes were prepared at N/P ratios ranging from 10 to 100.
For the assay, 10,000 cells per 0.1 mL were seeded into each well of a 96-well plate using a multichannel pipette and allowed to adhere for 24 h in a standard cell culture incubator. Cell health and confluency were assessed after the initial 24 h period. Subsequently, varying concentrations of freshly prepared peptide-siRNA complexes were added to each well in triplicate and incubated for 48 hours at 37°C in a humidified atmosphere with 5% CO2.
% Cell Viability = [(Average OD values of the samples treated with the compound)- (OD value of culture medium)]/ [(Average OD values of no treatment) -( OD value of culture medium)] x 100.
3.8. Cellular Internalization of siRNA
Cellular uptake of oleyl-conjugated peptide-siRNA complexes was assessed using flow cytometry in live cells. Alexa Fluor 488-labeled scrambled siRNA was used to form complexes with oleyl-conjugated peptides. This investigation was conducted in MDA-MB-231 and SK-OV-3 cell lines.
Approximately 500,000 cells were seeded per well in a six-well plate and allowed to adhere for 24 hours. Subsequently, cells were treated with conjugate-siRNA complexes and incubated for an additional 24 h. After treatment, cells were washed three times with 1X PBS, trypsinized, and centrifuged at 1000 rpm (96× g) for 5 min at 4°C. The cell pellet was washed three more times with PBS to remove the residual medium. Finally, cells were resuspended in PBS for flow cytometric analysis using the Alexa Fluor 488 channel to quantify mean fluorescence intensity.
Mean fluorescence intensity in treated cells was calculated for each sample, referencing the signal calibration with non-treated cells serving as the negative control. Alexa Fluor 488-labeled scrambled siRNA acted as an additional negative control, while lipofectamine-siRNA-treated cells served as the positive control.
3.9. Cellular Uptake Study Using Confocal Microscopy
In light of previous studies demonstrating the use of fixed or live cells for imaging siRNA cellular uptake, confocal microscopy was employed to corroborate data acquired from flow cytometry. Approximately 400,000 MDA-MB-231 cells were seeded on coverslips in 6-well plates and allowed to adhere for 24 h. During this period, cell confluency and health were assessed under a standard microscope.
Peptide-siRNA complexes were prepared with a final siRNA concentration of 36 nM and administered to the cells. Non-treated cells served as the negative control, while lipofectamine-siRNA complexes were employed as the positive control. After 24 h of incubation with the complexes, cells on coverslips were washed thrice with 1X PBS and fixed with 3.7% formaldehyde solution for 30 min at room temperature.
Cell membranes were stained using Texas Red (TR) Phalloidin (Invitrogen) (1:250 in HBSS), and DAPI was used to stain cell nuclei. Images of fixed cells were captured using a Nikon A1R confocal microscope system equipped with a 60X objective and various filters for Alexa-488 (FITC), TR, and DAPI fluorescence channels.
3.10. Protein Silencing Effect of siRNA (Western Blot)
STAT-3 was chosen as the model protein due to its significant overexpression in various types of cancer, including breast cancer. The aim of this study was to assess the silencing efficiency of STAT-3 siRNA delivered via oleyl-conjugated peptides by evaluating STAT-3 expression in cells treated with peptide-siRNA complexes. Non-treated cells served as the negative control, while cells treated with lipofectamine (20 μg/mL) complexed with STAT-3 siRNA (50 nM) served as the positive control.
To evaluate STAT-3 expression, Western blotting was performed. Approximately 500,000 MDA-MB-231 cells were seeded in T-25 flasks and incubated under standard growth conditions at 37°C. After 24 h, cells were treated with peptide-siRNA complexes for 48 hours, with a final siRNA concentration of 50 nM. Cell lysates were prepared using RIPA buffer according to standard protocols, followed by sonication in intervals to ensure complete cell lysis. The lysates were then centrifuged at 12,000 rpm (RCF = 13,870× g) at 4°C for 15 minutes, and the supernatant was transferred to pre-cooled microtubes.
The total protein concentration in the lysates was determined using a BSA assay. Briefly, a working reagent (50:1 A: B) was added to standards and unknown samples in triplicate in a 96-well plate. After mixing and incubating for 30 minutes at 37°C with 5% CO2, absorbance at 562 nm was measured using a Spectra MAX M5 microplate reader to quantify protein levels.
Approximately 15 μg of protein from each sample was loaded onto a 10% Mini-PROTEAN® TGX Stain-Free protein gel, and electrophoresis was carried out in SDS buffer at 200 V for 30 minutes. The separated proteins were transferred to a Mini PVDF membrane using a Trans-Blot® Turbo system. The membrane was blocked with 5% BSA for 3 h, followed by overnight incubation at 4°C with primary antibody (1:1000 in TBS-T). After washing the membrane with TBS-T, it was incubated with a secondary HRP-linked antibody (1:1000 in TBS-T) for 1 h, followed by additional washing steps.
Protein bands were visualized using an ECL Detect Kit and imaged with a ChemiDoc imager. Band intensities were quantified using Image LabTM software.
3.10. Statistical Analysis
Experiments were conducted in triplicate, and results are presented as mean ± standard deviation unless stated otherwise. Data from flow cytometry and western blotting experiments were analyzed using standard one-way analysis of variance (ANOVA) with Tukey’s multiple comparison test. The effects of varying N/P ratios on particle size and zeta potential were evaluated using two-way ANOVA with Tukey's multiple comparison test. A p-value > 0.05 was considered non-significant (ns). Statistical significance for all analyses was set at p < 0.05.
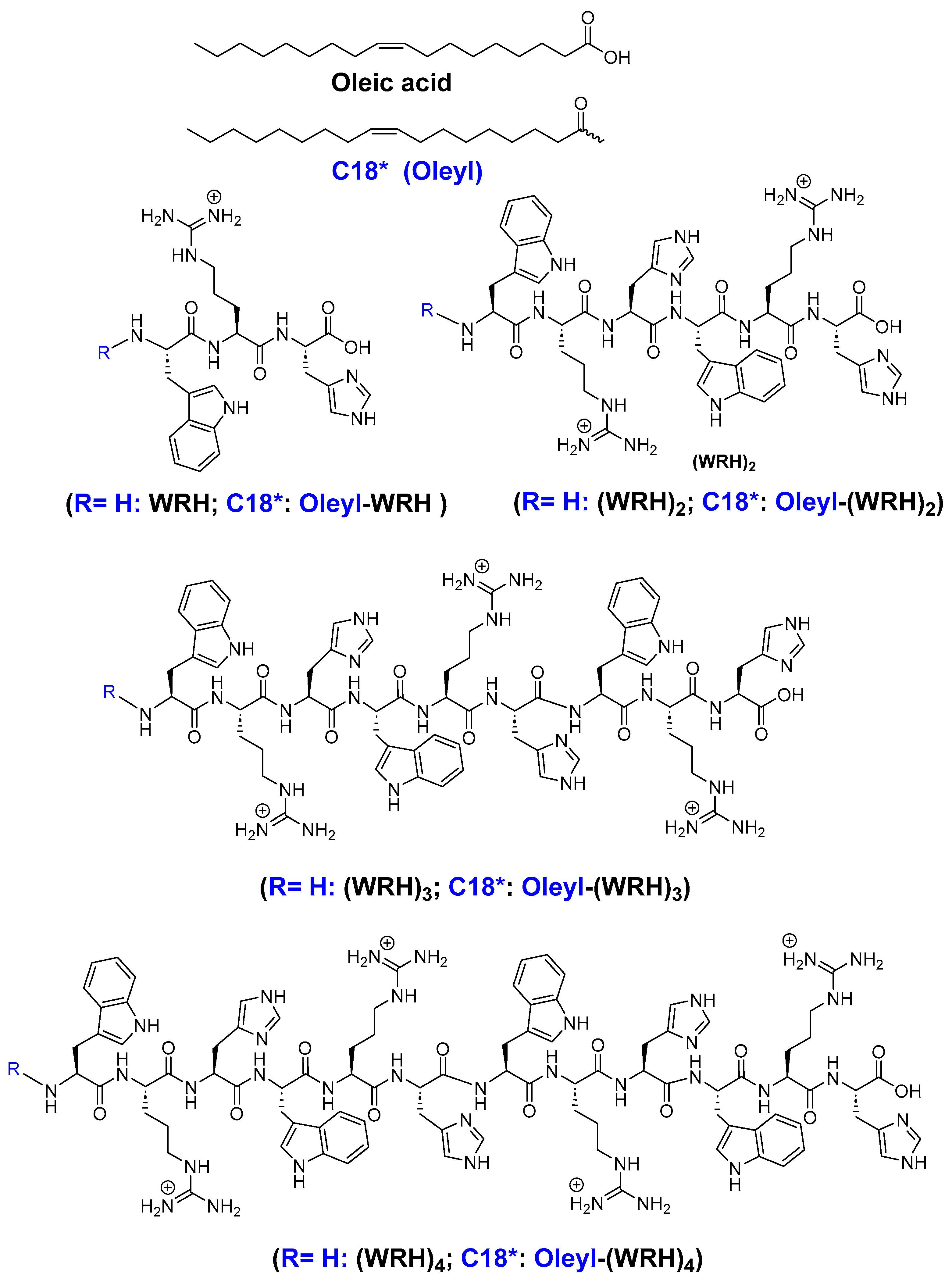
Figure 1.
Structure of synthesized peptides.
Figure 1.
Structure of synthesized peptides.
Scheme 1.
A synthetic scheme of (WRH)2 and Oleyl-(WRH)2 peptides.
Scheme 1.
A synthetic scheme of (WRH)2 and Oleyl-(WRH)2 peptides.
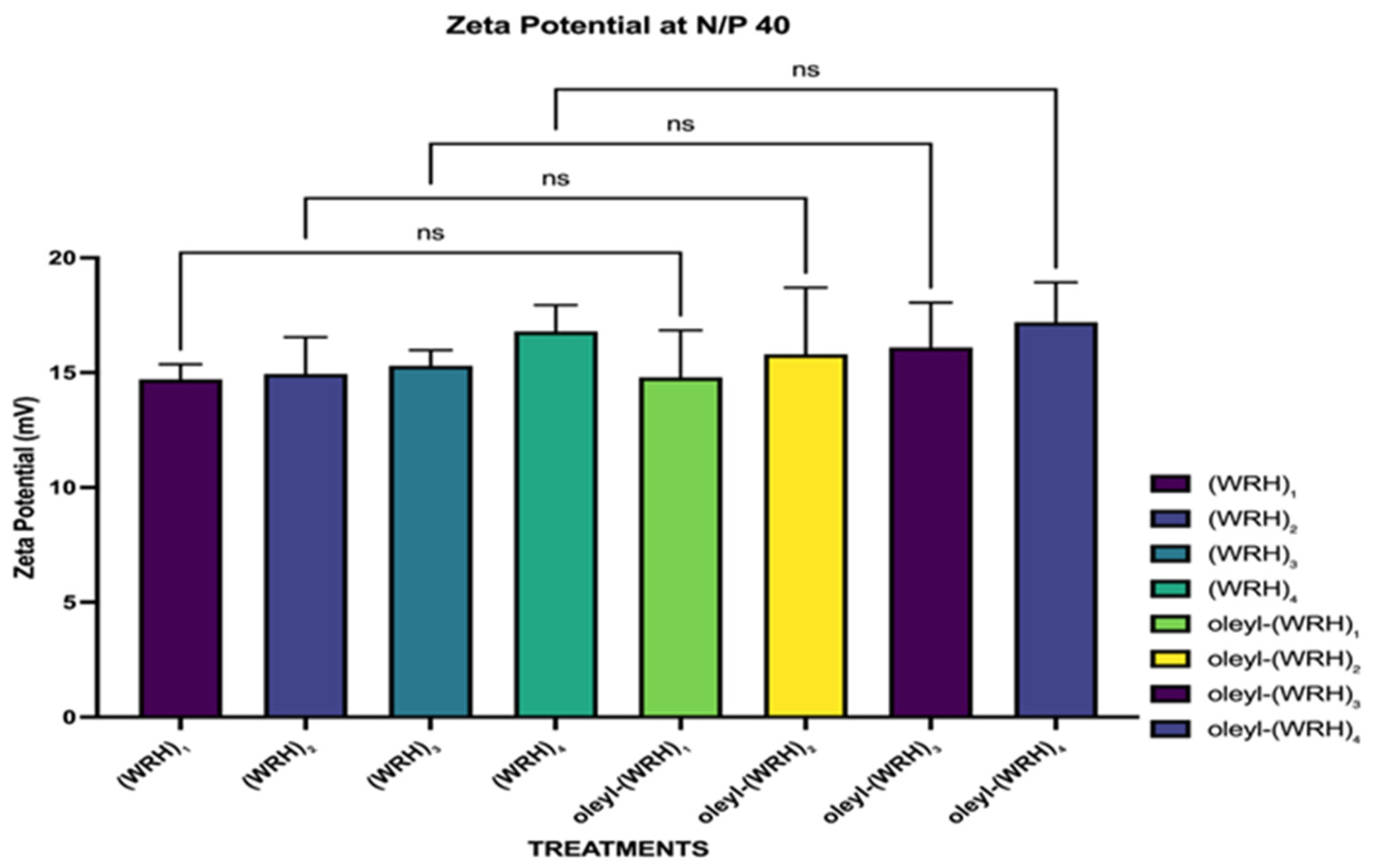
Figure 2.
Zeta potential values of peptide: siRNA complexes at N/P ratio 40. Two-way ANOVA was performed using Turkey’s multiple comparisons test to determine the effect of changing the N/P ratio on zeta potential. ns = non-significant.
Figure 2.
Zeta potential values of peptide: siRNA complexes at N/P ratio 40. Two-way ANOVA was performed using Turkey’s multiple comparisons test to determine the effect of changing the N/P ratio on zeta potential. ns = non-significant.
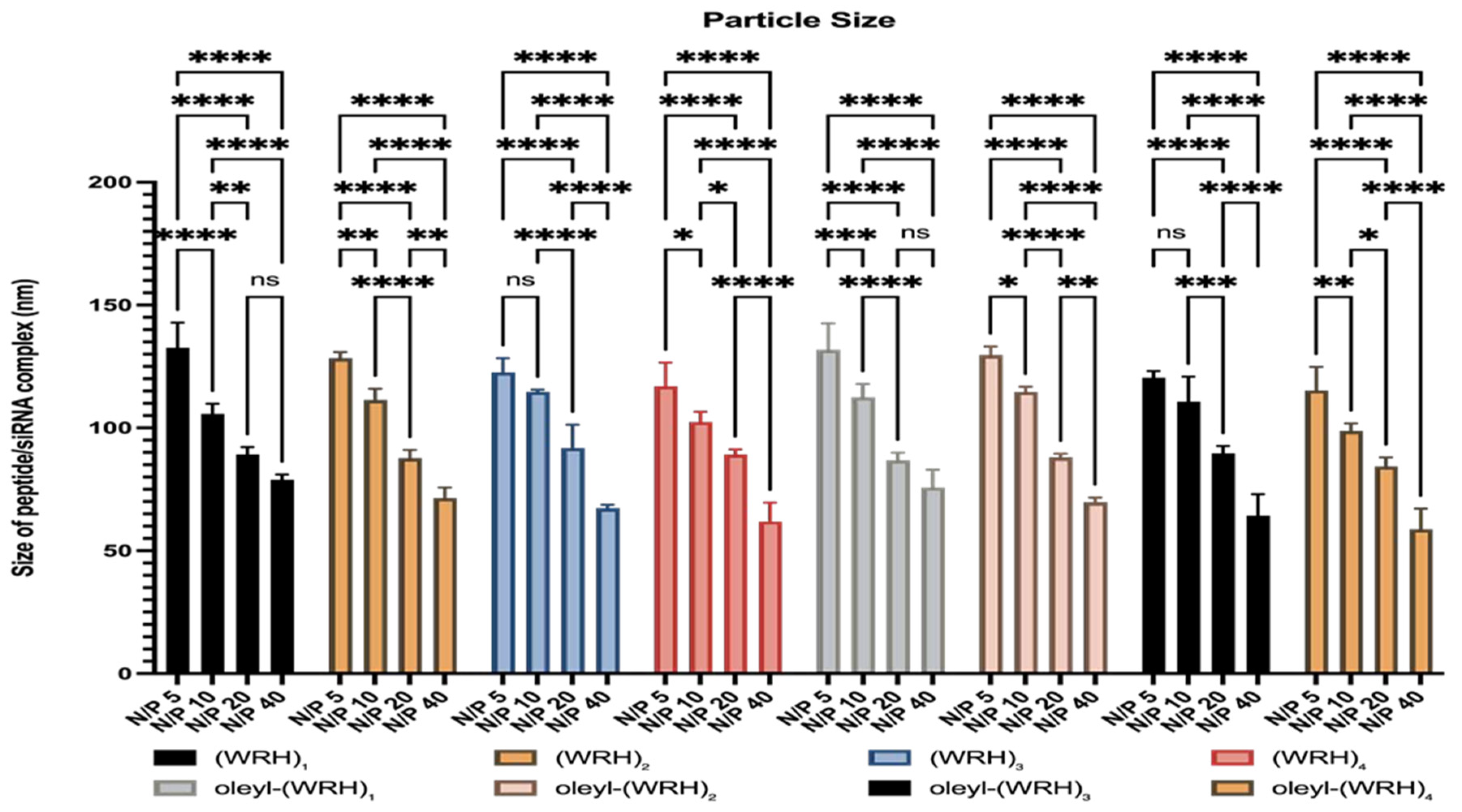
Figure 3.
Particle size evaluation of peptides (WRH)1 to oleyl-(WRH)4 at different N/P ratios. Two-way ANOVA was performed using Tukey’s multiple comparisons test to determine the effect of changing the N/P ratio on the particle size. ns = non-significant (p-value > 0.05), * p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, and **** p-value < 0.0001. Error bars represent standard deviation (n = 3).
Figure 3.
Particle size evaluation of peptides (WRH)1 to oleyl-(WRH)4 at different N/P ratios. Two-way ANOVA was performed using Tukey’s multiple comparisons test to determine the effect of changing the N/P ratio on the particle size. ns = non-significant (p-value > 0.05), * p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, and **** p-value < 0.0001. Error bars represent standard deviation (n = 3).
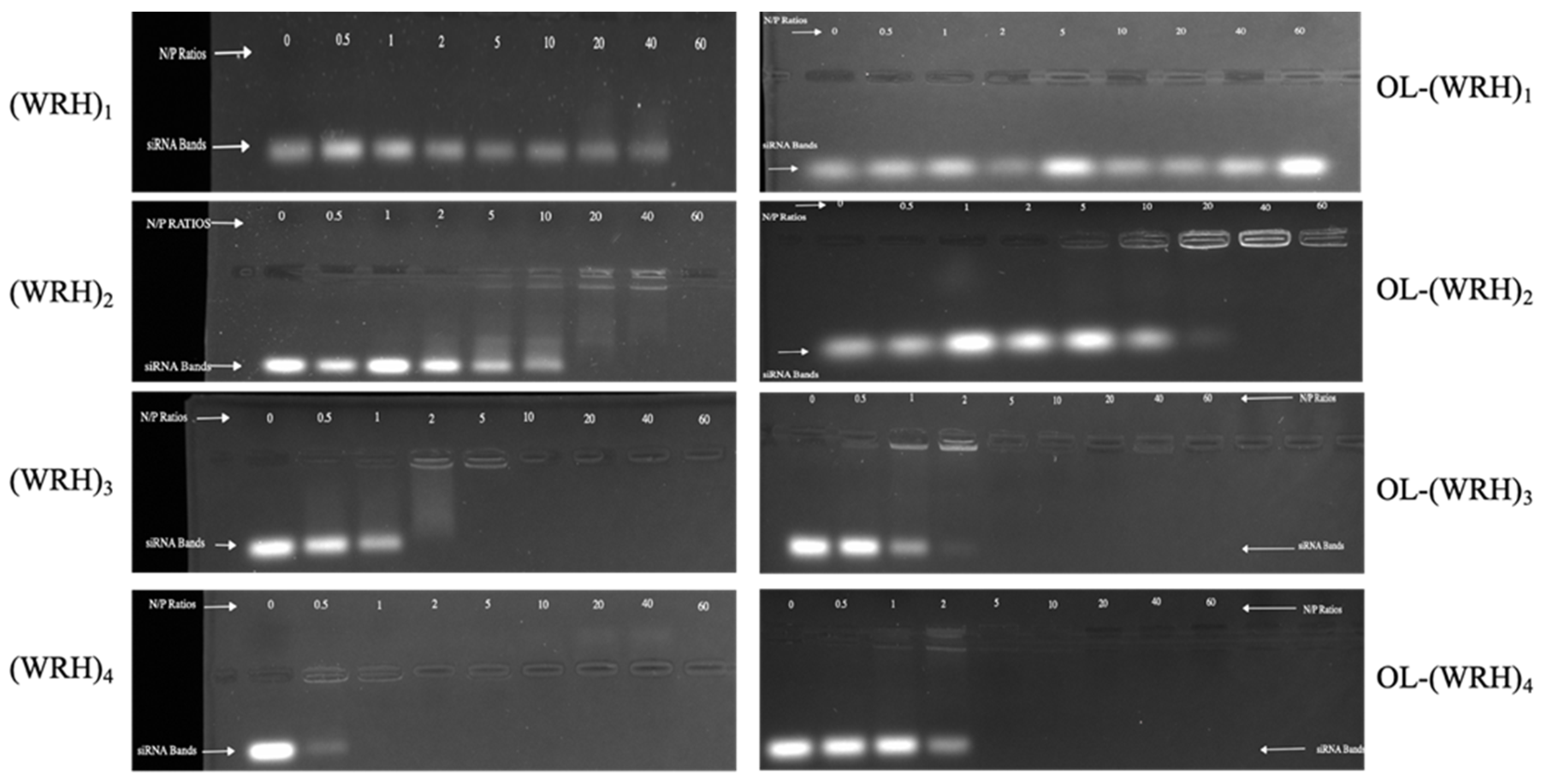
Figure 4.
Representative agarose gel images of gel retardation assay for evaluating the binding affinity of the non-oleyl conjugated and oleyl-conjugated peptides/siRNA complexes at different N/P ratios using scrambled siRNA.
Figure 4.
Representative agarose gel images of gel retardation assay for evaluating the binding affinity of the non-oleyl conjugated and oleyl-conjugated peptides/siRNA complexes at different N/P ratios using scrambled siRNA.
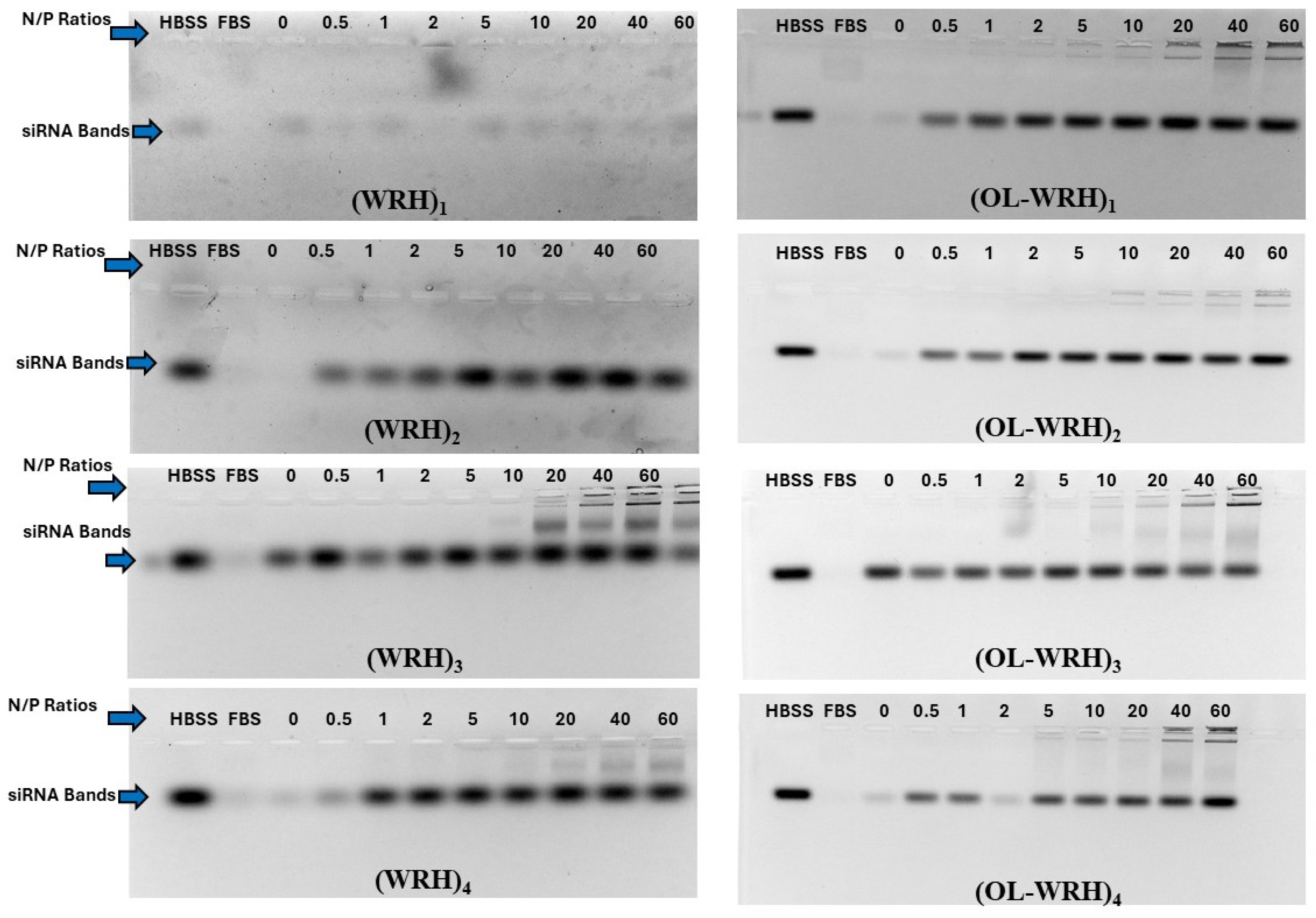
Figure 5.
Representative agarose gel images of gel retardation assay for evaluating the serum stability of the non-oleyl conjugated and oleyl-conjugated peptides/siRNA complexes at different N/P ratios using scrambled siRNA. Heparin sulfate dissociated siRNA from the peptide/siRNA complexes (FBS 25%, Incubation time 24 h). The band representing negative control (scrambled siRNA exposed to saline for 24 h at 37 °C) was quantified as 100%. The bands were quantified using Image Lab software version 6.0.1.
Figure 5.
Representative agarose gel images of gel retardation assay for evaluating the serum stability of the non-oleyl conjugated and oleyl-conjugated peptides/siRNA complexes at different N/P ratios using scrambled siRNA. Heparin sulfate dissociated siRNA from the peptide/siRNA complexes (FBS 25%, Incubation time 24 h). The band representing negative control (scrambled siRNA exposed to saline for 24 h at 37 °C) was quantified as 100%. The bands were quantified using Image Lab software version 6.0.1.
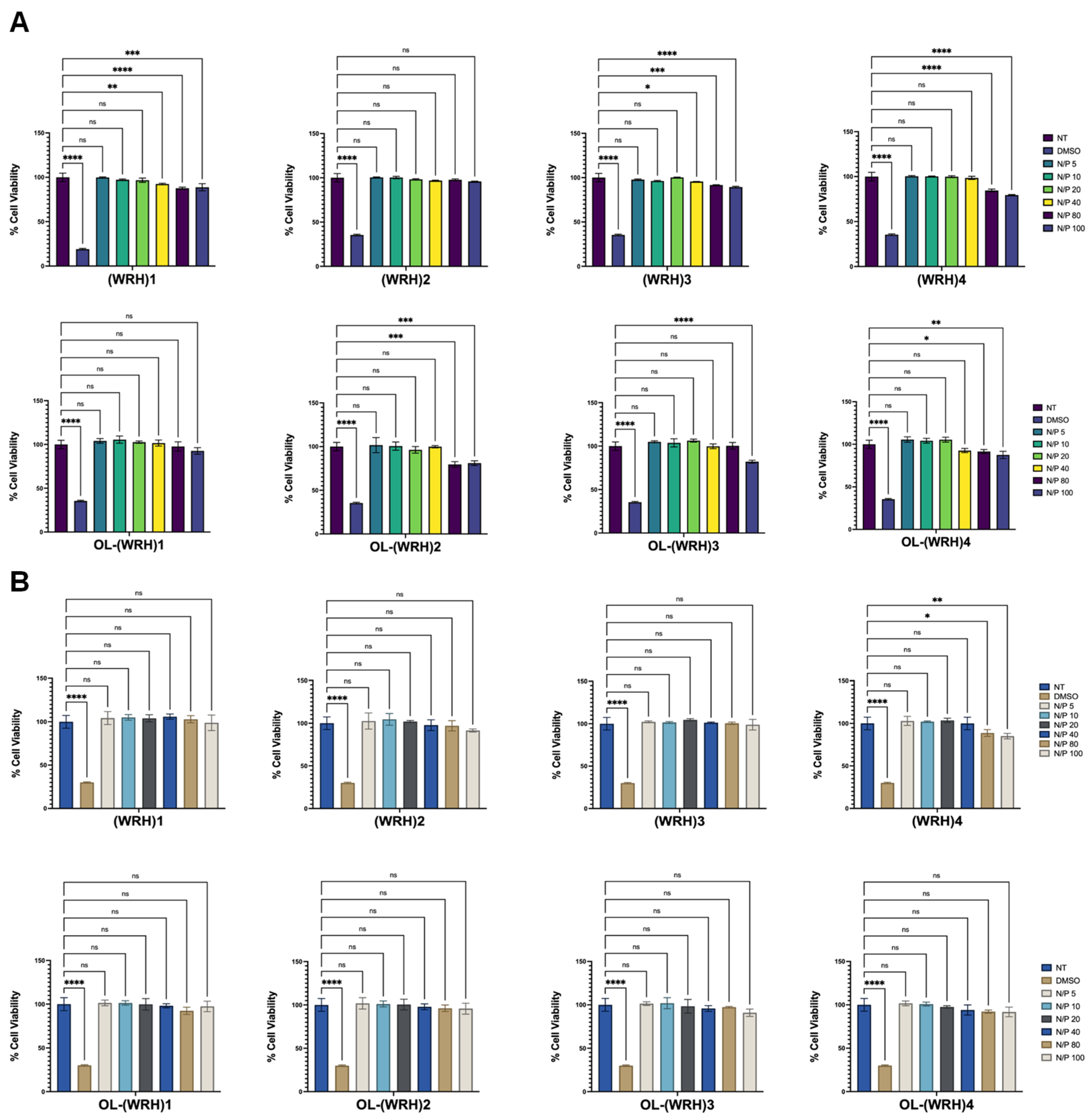
Figure 6.
Cytotoxicity of peptide-siRNA complexes in (A) MDA-MB-231 and (B) MCF-7 cells. Graphs represent mean normalized values (based on No Treatment or the NT group, n = 3). Error bars indicate standard deviation. ns = non-significant (p-value > 0.05), * (p-value < 0.05), ** p-value < 0.001, *** p-value < 0.0005, and **** p-value < 0.0001.
Figure 6.
Cytotoxicity of peptide-siRNA complexes in (A) MDA-MB-231 and (B) MCF-7 cells. Graphs represent mean normalized values (based on No Treatment or the NT group, n = 3). Error bars indicate standard deviation. ns = non-significant (p-value > 0.05), * (p-value < 0.05), ** p-value < 0.001, *** p-value < 0.0005, and **** p-value < 0.0001.
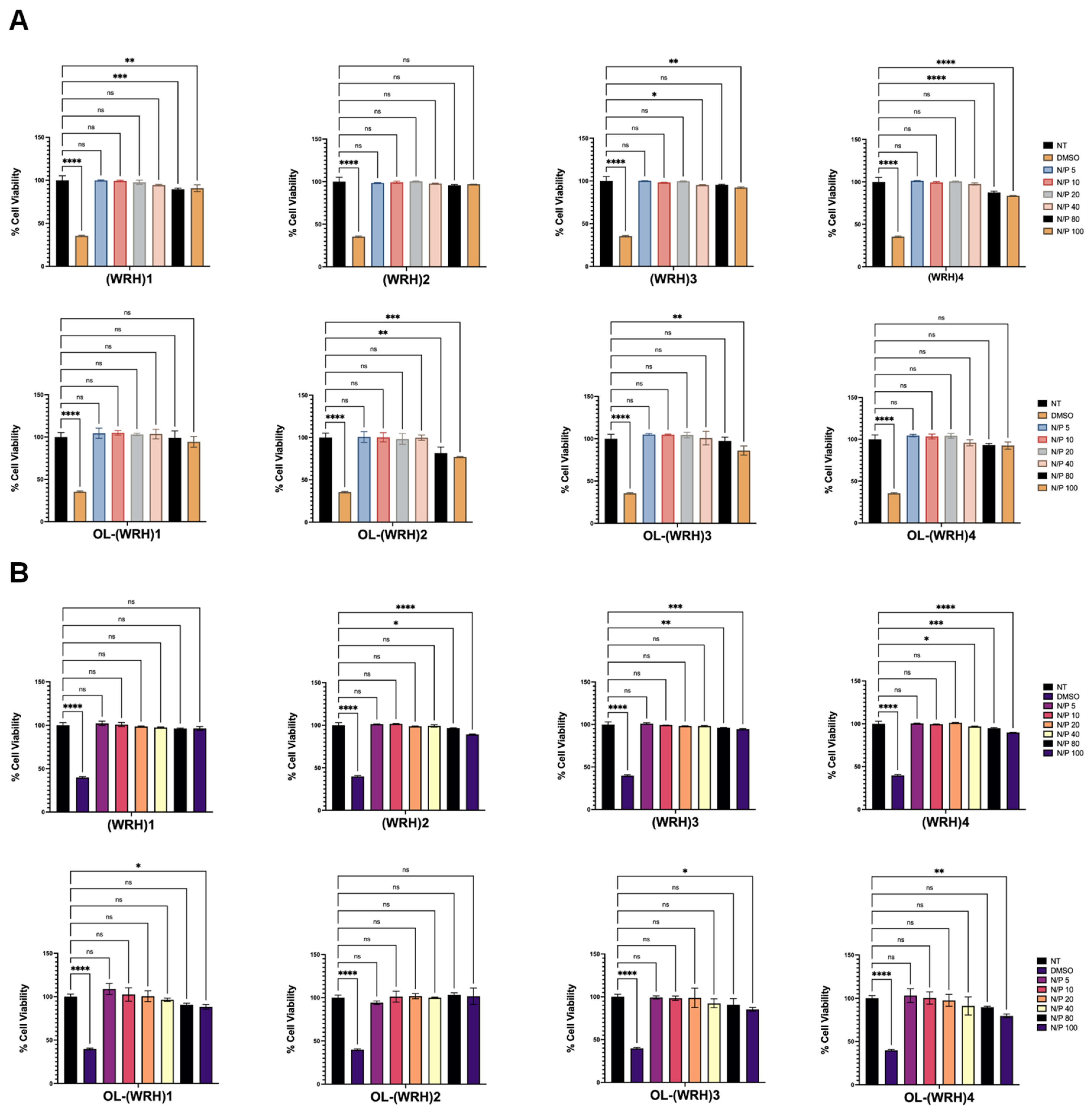
Figure 7.
Cytotoxicity of peptide-siRNA complexes in (A) SK-OV-3 and (B) HEK-293 cells. Graphs represent mean normalized values (based on No Treatment or the NT group, n = 3). Error bars indicate standard deviation. ns = non-significant (p-value > 0.05), * (p-value < 0.05), ** p-value < 0.001, *** p-value < 0.0005, and **** p-value < 0.0001.
Figure 7.
Cytotoxicity of peptide-siRNA complexes in (A) SK-OV-3 and (B) HEK-293 cells. Graphs represent mean normalized values (based on No Treatment or the NT group, n = 3). Error bars indicate standard deviation. ns = non-significant (p-value > 0.05), * (p-value < 0.05), ** p-value < 0.001, *** p-value < 0.0005, and **** p-value < 0.0001.
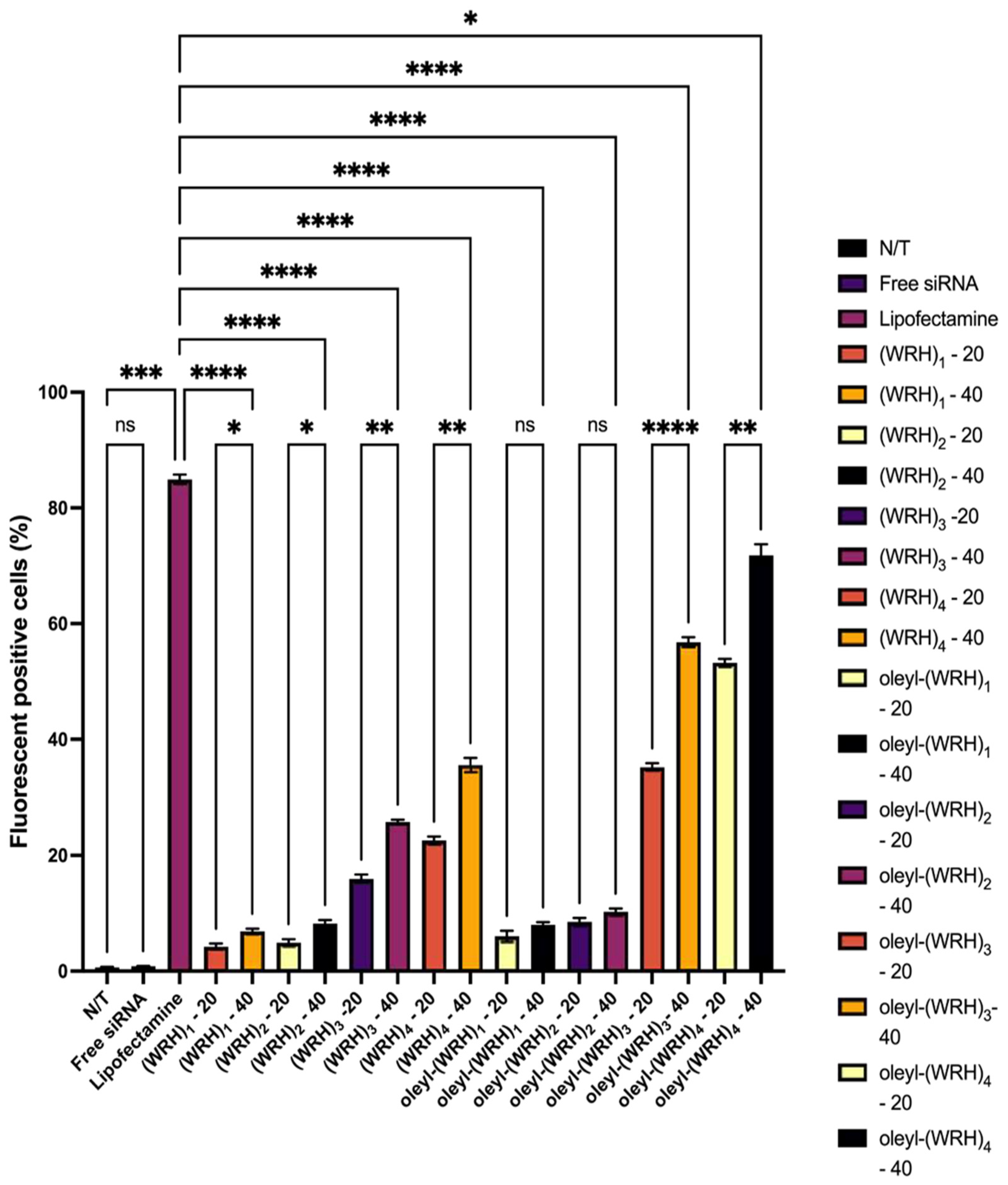
Figure 8.
Flow cytometry of non-oleyl and oleyl conjugated-siRNA complexes at N/P ratios of 20 and 40 in MDA-MB-231 cells using Alexa Fluor 488-labeled scrambled siRNA after 24 h incubation. Bars represent the mean value (n = 3), and the error bars represent standard deviations. ns = non-significant (p-value > 0.05), * (p-value < 0.05), ** p-value < 0.001, *** p-value < 0.0005, and **** p-value < 0.0001.
Figure 8.
Flow cytometry of non-oleyl and oleyl conjugated-siRNA complexes at N/P ratios of 20 and 40 in MDA-MB-231 cells using Alexa Fluor 488-labeled scrambled siRNA after 24 h incubation. Bars represent the mean value (n = 3), and the error bars represent standard deviations. ns = non-significant (p-value > 0.05), * (p-value < 0.05), ** p-value < 0.001, *** p-value < 0.0005, and **** p-value < 0.0001.
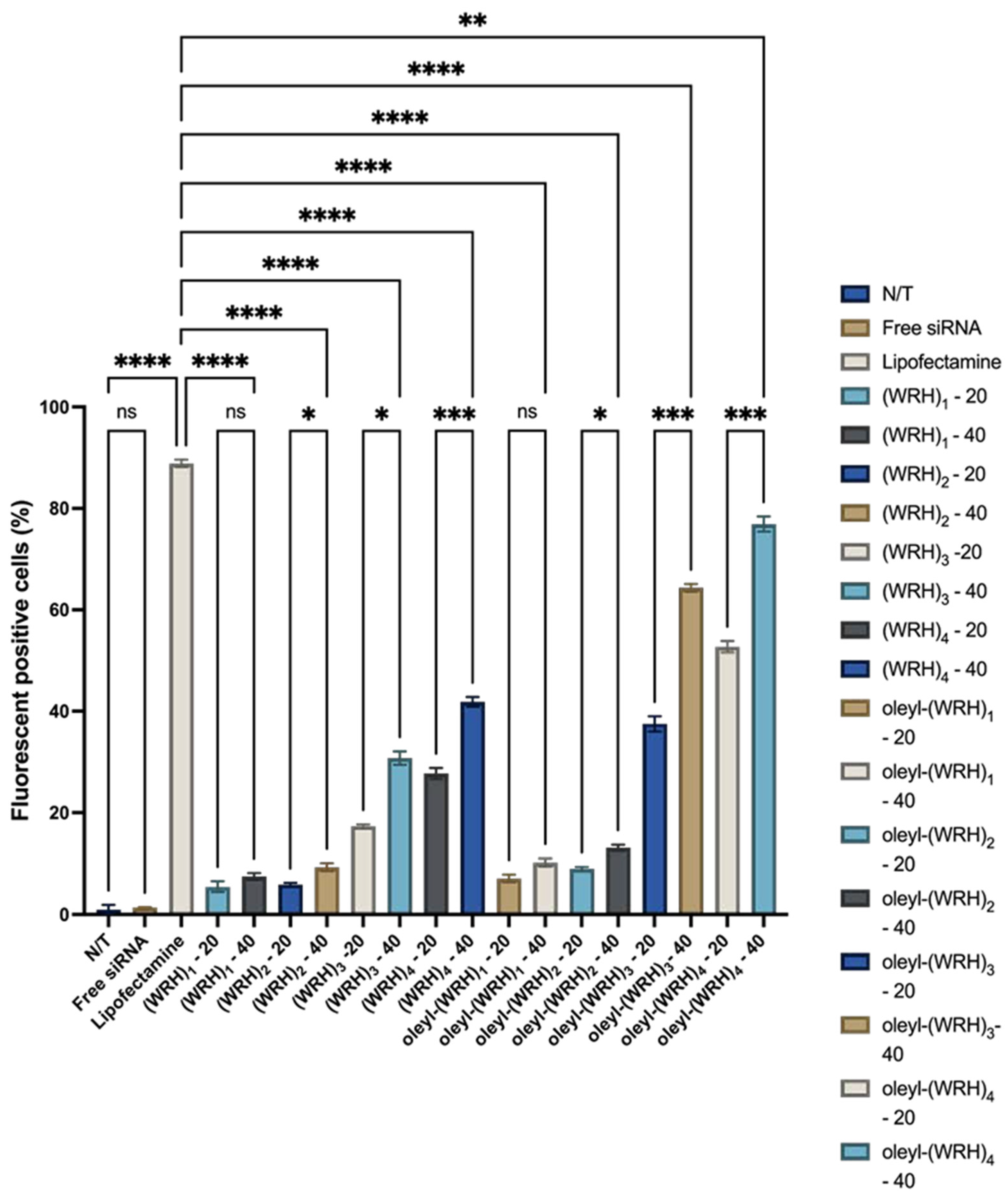
Figure 9.
Flow cytometry of non-oleyl and oleyl conjugated-siRNA complexes at N/P ratios of 20 and 40 in SK-OV-3 cells using Alexa Fluor 488-labeled scrambled siRNA after 24 h incubation. Bars represent the mean value (n = 3), and the error bars represent standard deviations. ns = non-significant (p-value > 0.05), * (p-value < 0.05), ** p-value < 0.001, *** p-value < 0.0005, and **** p-value < 0.0001.
Figure 9.
Flow cytometry of non-oleyl and oleyl conjugated-siRNA complexes at N/P ratios of 20 and 40 in SK-OV-3 cells using Alexa Fluor 488-labeled scrambled siRNA after 24 h incubation. Bars represent the mean value (n = 3), and the error bars represent standard deviations. ns = non-significant (p-value > 0.05), * (p-value < 0.05), ** p-value < 0.001, *** p-value < 0.0005, and **** p-value < 0.0001.
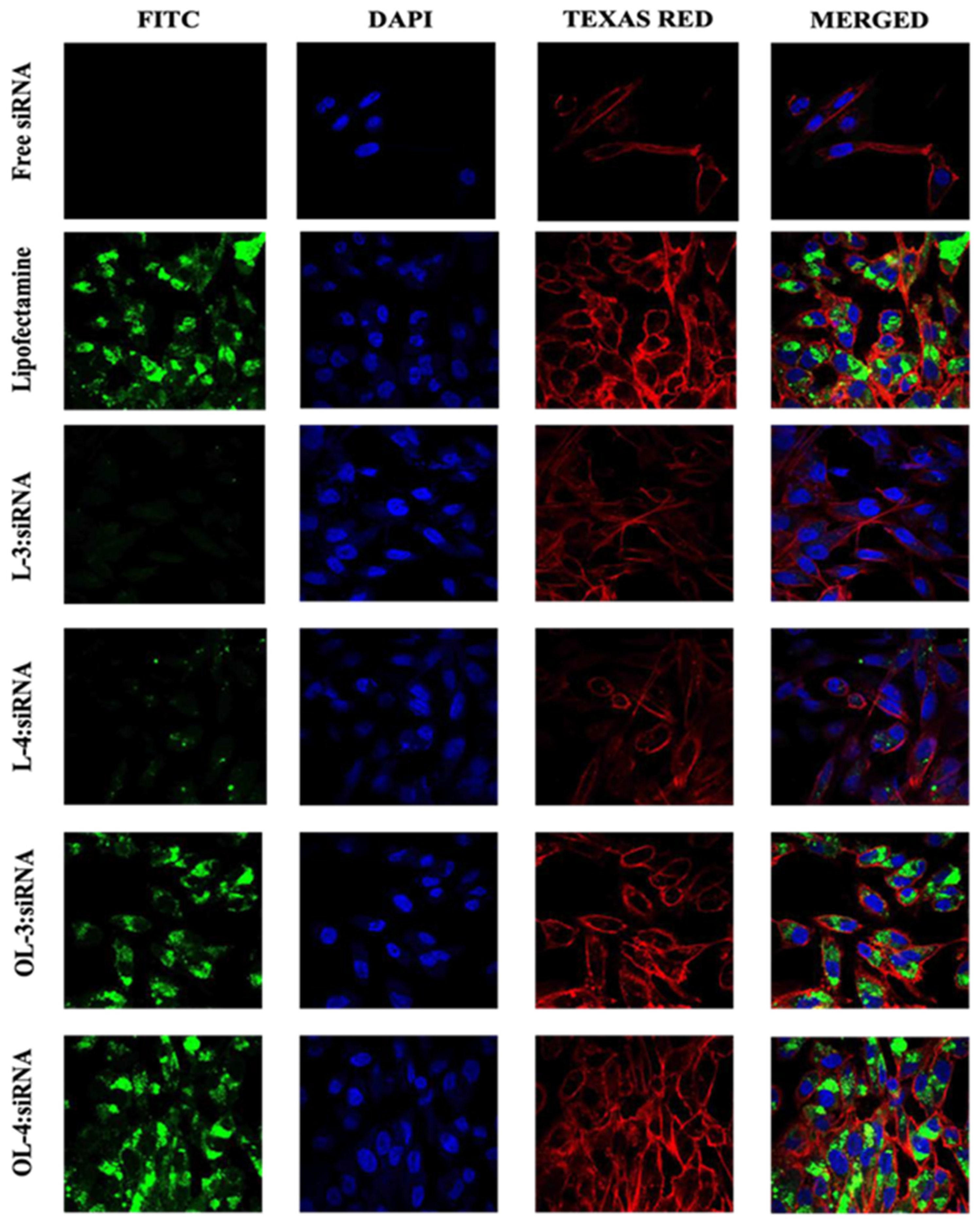
Figure 10.
Confocal microscopy images of MDA-MB-231 cells treated with (WRH)3, (WRH)4, oleyl-(WRH)3, and oleyl-(WRH)4 complexes with Alexa Fluor 488-labeled scrambled siRNA after 24 h incubation. The cell membrane cytoskeleton of the cells was stained using Texas Red (TR) Phalloidin while 4′,6-diamidino-2-phenylindole (DAPI) was used to stain the nuclei of the cells.
Figure 10.
Confocal microscopy images of MDA-MB-231 cells treated with (WRH)3, (WRH)4, oleyl-(WRH)3, and oleyl-(WRH)4 complexes with Alexa Fluor 488-labeled scrambled siRNA after 24 h incubation. The cell membrane cytoskeleton of the cells was stained using Texas Red (TR) Phalloidin while 4′,6-diamidino-2-phenylindole (DAPI) was used to stain the nuclei of the cells.
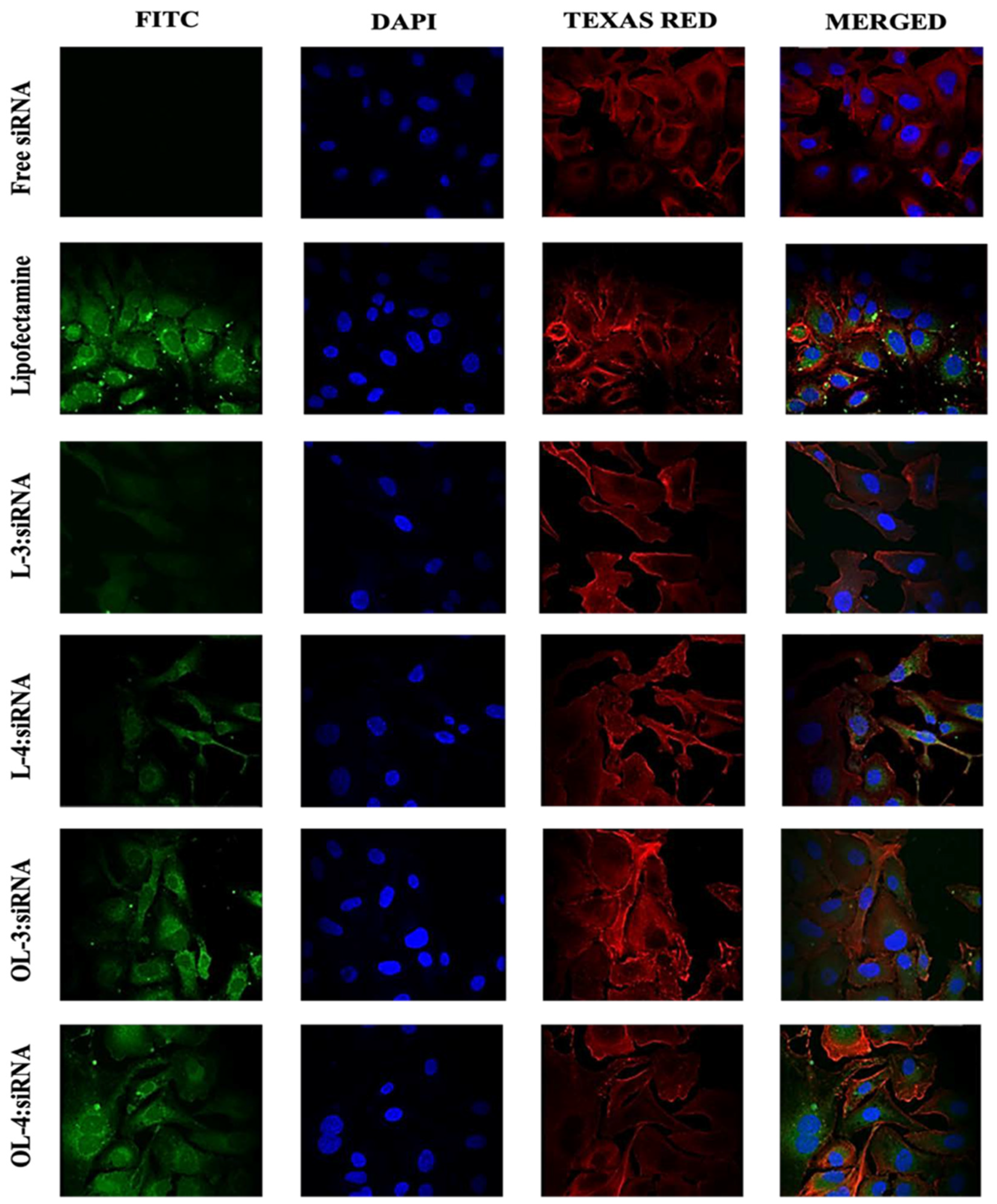
Figure 11.
Confocal microscopy images of SK-OV-3 cells treated with (WRH)3, (WRH)4, oleyl- (WRH)3, and oleyl-(WRH)4 complexes with Alexa Fluor 488-labeled scrambled siRNA after 24 h incubation. The cell membrane cytoskeleton of the cells was stained using Texas Red (TR) Phalloidin, while DAPI was used to stain the nuclei of the cells.
Figure 11.
Confocal microscopy images of SK-OV-3 cells treated with (WRH)3, (WRH)4, oleyl- (WRH)3, and oleyl-(WRH)4 complexes with Alexa Fluor 488-labeled scrambled siRNA after 24 h incubation. The cell membrane cytoskeleton of the cells was stained using Texas Red (TR) Phalloidin, while DAPI was used to stain the nuclei of the cells.
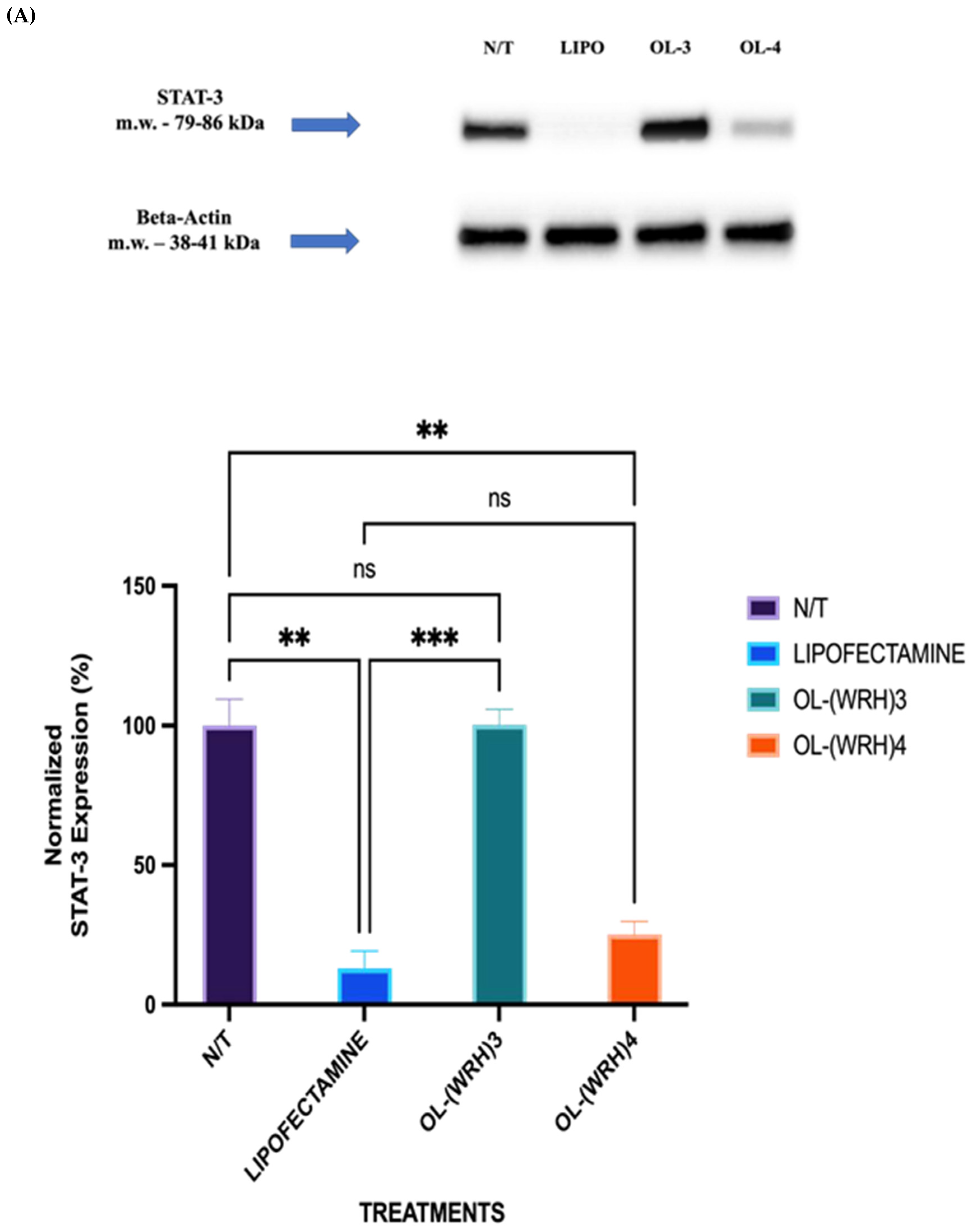
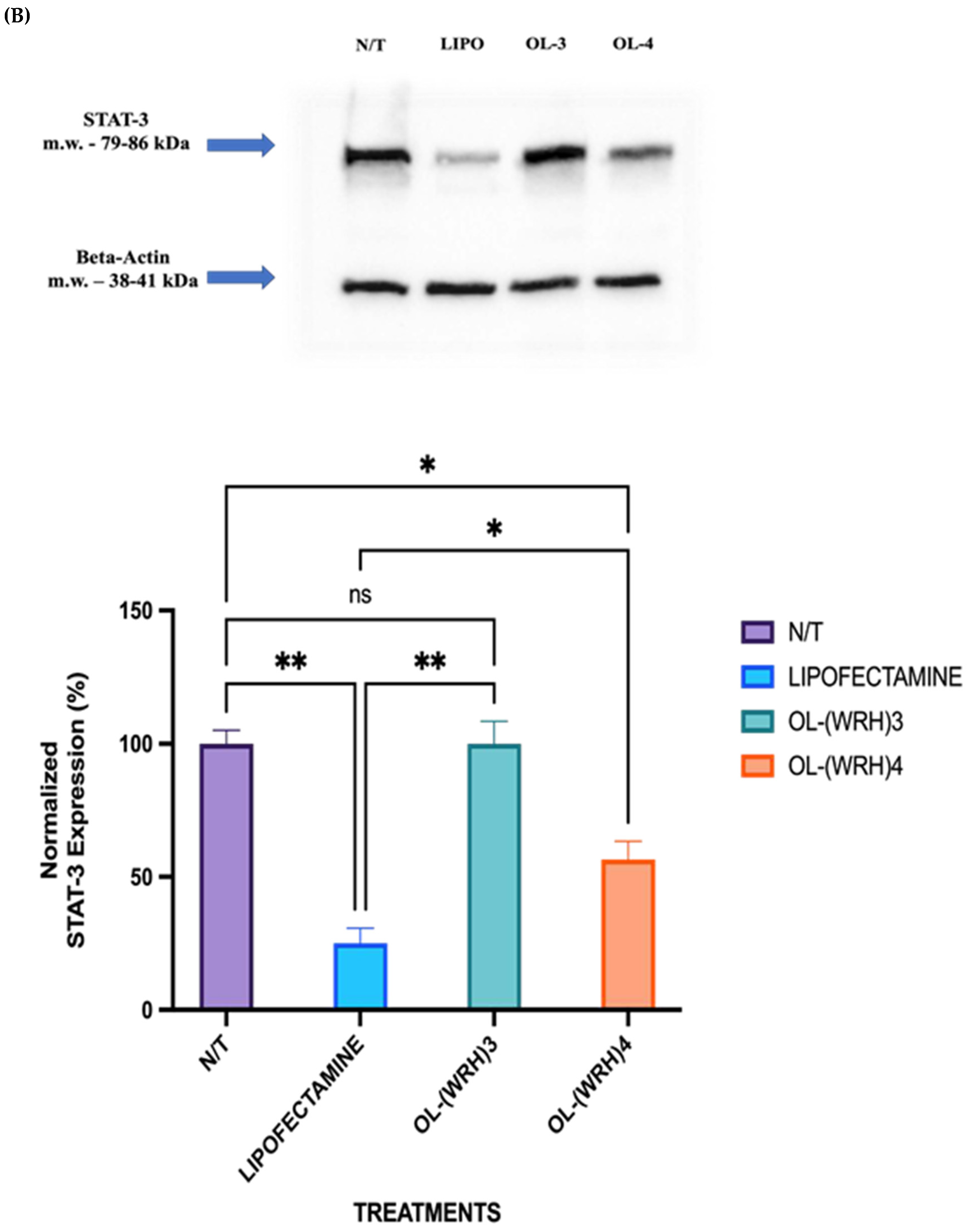
Figure 12.
Comparison of STAT protein downregulation efficiencies of oleyl-conjugated peptides oleyl-(WRH)3 and oleyl-(WRH)4 at N/P ratio of 40 in (A) MDA-MB-231 cells and (B) SK-OV3 cells. Error bars indicate standard deviation (n = 3). ns = non-significant (p-value > 0.05), * p-value < 0.05), ** p-value < 0.001, and *** p-value < 0.0005.
Figure 12.
Comparison of STAT protein downregulation efficiencies of oleyl-conjugated peptides oleyl-(WRH)3 and oleyl-(WRH)4 at N/P ratio of 40 in (A) MDA-MB-231 cells and (B) SK-OV3 cells. Error bars indicate standard deviation (n = 3). ns = non-significant (p-value > 0.05), * p-value < 0.05), ** p-value < 0.001, and *** p-value < 0.0005.
Table 1.
Optimization of manual peptide synthesis.
Table 1.
Optimization of manual peptide synthesis.
| STEP |
VOLUME (μL) |
TIME
(min) |
REAGENTS |
REACTION |
| 1 |
15,000 |
30 (10 x 3) |
DMF |
Resin Swelling |
| 2 |
25,000 |
120 |
Amino Acid 1 +
DMF + HCTU + DIPEA |
Coupling |
| 3 |
15,000 |
15 (5 x 3) |
DMF |
Washing |
| 4 |
10,000 |
30 (10 x 3) |
20% Piperidine in DMF (v/v) |
Deprotection |
| 5 |
15,000 |
15 (5 x 3) |
DMF |
Washing |
| 6 |
25,000 |
120 |
Amino Acid 2 +
DMF + HCTU + DIPEA |
Coupling |
| 7 |
15,000 |
15 (5 x 3) |
DMF |
Washing |
| 8 |
10,000 |
30 (10 x 3) |
20% Piperidine in DMF (v/v) |
Deprotection |
| 9 |
15,000 |
15 (5 x 3) |
DMF |
Washing |
| Steps 3 to 9 were repeated until the desired sequence was obtained. |
Table 2.
Range of Zeta Potential values corresponding to the extent of stability.
Table 2.
Range of Zeta Potential values corresponding to the extent of stability.
| Zeta Potential |
Stability |
| ±0-10 mV |
Highly Unstable |
| ±10-20 mV |
Relatively Stable |
| ±20-30 mV |
Moderately Stable |
| ±30 mV |
Highly Stable |
Table 3.
Particle size analysis of Peptide:siRNA complexes at N/P ratio 40.
Table 3.
Particle size analysis of Peptide:siRNA complexes at N/P ratio 40.
| Peptide-siRNA complex at N/P 40 |
Z-Ave (d. nm) |
PDI |
| (WRH)1 |
78.94 ± 1.3 |
0.213 ± 0.03 |
| (WRH)2 |
71.43 ± 2.5 |
0.201 ± 0.06 |
| (WRH)3 |
67.38 ± 1.6 |
0.217 ± 0.05 |
| (WRH)4 |
61.84 ± 1.8 |
0.229 ± 0.03 |
| oleyl-(WRH)1 |
75.66 ± 2.3 |
0.237 ± 0.07 |
| oleyl-(WRH)2 |
69.76 ± 2.1 |
0.213 ± 0.04 |
| oleyl-(WRH)3 |
64.28 ± 2.1 |
0.213 ± 0.04 |
| oleyl-(WRH)4 |
58.74 ± 2.1 |
0.213 ± 0.04 |
| Average |
69.50 ± 1.10 |
|