1. Introduction
Skin cancer is a significant health concern worldwide and is among the most diagnosed cancers [
1,
42,
43,
44]. It is primarily divided into two types: melanoma and non-melanoma. Although melanoma is less prevalent, representing only 1% of all skin cancer cases, it is responsible for most skin cancer-related deaths due to its aggressive behavior. In the United States, melanoma caused approximately 7,800 deaths in 2022, with new cases expected to rise to 98,000 in 2023 [
2]. The likelihood of developing skin cancer in the US is significant, with one in five Americans predicted to be affected, emphasizing the critical need for effective diagnostic and treatment approaches [
3]. The economic impact of skin cancer is substantial, with treatment costs in the United States alone surpassing
$8.1 billion annually. These costs extend beyond direct medical expenses, encompassing lost productivity and the extensive care required for advanced stages of the disease. Early and accurate detection of skin cancer, particularly malignant melanoma, is essential for improving patient outcomes and mitigating these considerable financial burdens [
4]. Dermatoscopy and dermoscopy are essential tools in the clinical evaluation of skin lesions, aiding dermatologists in detecting malignant characteristics [
5]. However, the manual interpretation of these images can be labor-intensive and subject to human error, depending greatly on the clinician's skill and experience. Advances in machine learning have recently brought forth new techniques that enhance diagnostic accuracy and efficiency in clinical settings [
6,
7]. These technologies are especially beneficial in computationally constrained environments, such as mobile health applications, where they provide reliable diagnostic support with minimal computational requirements. Adopting these advanced technologies not only improves patient care by enabling earlier and more precise diagnosis but also can reduce healthcare costs by preventing the progression of the disease to more severe and expensive stages. Skin cancer, particularly malignant melanoma, is known for its swift progression and high mortality rate, making early and accurate diagnosis crucial for enhancing patient outcomes.
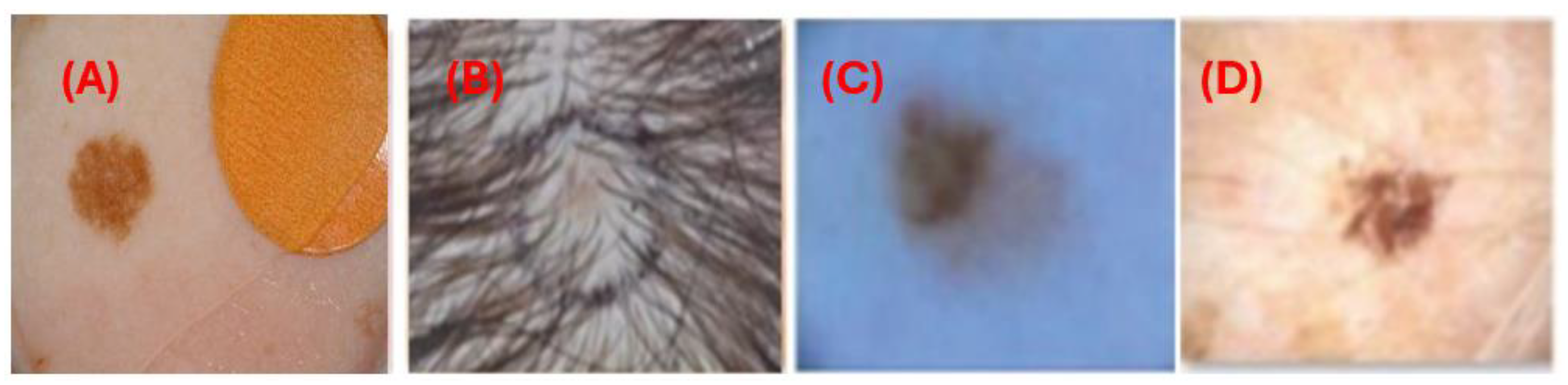
Manual interpretation of dermoscopic images can be labor-intensive, prone to errors, and highly dependent on the clinician's expertise. Furthermore, various medical samples present notable challenges. These include indistinct boundaries where lesions merge with the surrounding skin, variations in illumination that alter the appearance of lesions, artifacts such as hair and bubbles that obscure lesion borders, and differences in lesion size and shape. Additionally, variations in imaging conditions and resolutions, age-related skin texture changes, complex backgrounds that complicate segmentation, and differences in skin color due to racial and climatic factors also contribute to the difficulty of accurate interpretation.
Figure 1 illustrates challenges in skin lesion segmentation using dermoscopic images: (A) artifacts from image acquisition, (B) occlusion in lesions due to hair, (C) minor variation in the lesion and skin color, and (D) low contrast between wound and skin.
In recent years, integrating computer-aided tools and artificial intelligence (AI) into clinical practice has significantly enhanced the precision and efficiency of skin cancer diagnosis [
8,
9]. A fundamental technique in this field is skin cancer segmentation, which accurately delineates the boundaries of skin lesions in medical images. This process is crucial for evaluating lesion characteristics, tracking their development, and informing treatment strategies. The rapid progress in AI technology, combined with the widespread use of smart devices like point-of-care ultrasound (POCUS) systems and smartphones [
10,
11,
12], has made AI-driven methods for skin cancer detection increasingly popular. Patients now benefit from improved access to medical information, remote monitoring, and personalized care, leading to greater satisfaction with healthcare services. However, challenges persist, particularly in medical diagnostics. One significant hurdle is the precise and efficient segmentation of skin lesions, which is essential for accurate diagnosis but challenging to achieve on devices with limited computational power. Most AI-driven medical applications depend on deep learning techniques, as detailed in [
14], which generally demand substantial computational resources and extensive learning parameters, creating obstacles for integration into devices with constrained hardware capabilities [
15].
In this study, we extend our previous method, UCM-Net [
19], introducing UCM-NetV2, a new powerful, lightweight, and robust approach for skin lesion segmentation. UCM-NetV2 embeds a new hybrid module that combines Convolutional Neural Networks (CNN) and Multilayer Perceptions (MLP) to enhance feature learning. Utilizing new proposed group loss functions, our method surpasses existing lightweight techniques in skin lesion segmentation. Additionally, we explore applying XNOR-Net, a classical binary neural network (BNN), to UCM-NetV2 to reduce inference computation costs further. Key contributions of UCM-NetV2 include:
Hybrid Feature Learning: The UCM-NetV2 Block is a new proposed block that integrates CNN and MLP, enhancing the learning of complex and distinct lesion features.
Computational Efficiency: UCM-NetV2 contains an inference mode that reduces inference computation costs below 0.04 Glops with parameters similar to those of the previous UCM-Net.
Enhanced Loss Function: A new dynamic loss function integrates output and internal stage losses, ensuring efficient learning during the model's training process.
Superior Results: UCM-NetV2 achieves competitive prediction performance on the ISIC 2017 and 2018 datasets, compared to previous lightweight methods on metrics like Dice similarity, sensitivity, specificity, and accuracy.
BNN-UCM-NetV2 Network: We explore applying XNOR-Net to UCM-NetV2, reducing inference computation costs to below 0.02 GFLOPs without sacrificing the model's prediction performance.
The remainder of the paper is organized as follows:
Section 2 explores related works, such as TinyML for Healthcare and Supervised Methods of Segmentation. In
Section 3, we present the proposed UCM-NetV2 and BNN-UCM-NetV2 networks.
Section 4 presents the simulation results and analysis. Finally,
Section 5 presents the conclusion and future studies.
2. Related Works
TinyML for Healthcare: Segmentation in biomedical imaging involves precisely delineating anatomical structures and pathological regions from medical images, which is crucial for accurate diagnostics. Recent advancements in artificial intelligence (AI) have significantly improved segmentation techniques, enhancing both accuracy and efficiency. An exciting development in this field is the incorporation of TinyML, particularly for lesion segmentation, offering promising research opportunities and practical applications. TinyML involves deploying machine learning models on low-power, compact hardware, revolutionizing healthcare by enabling advanced analytics directly at the point of care. This technology supports real-time, on-device processing, making complex medical image analysis feasible even in resource-limited or mobile healthcare settings. Applying TinyML to lesion segmentation can provide immediate diagnostic insights during patient exams or in remote areas, reducing dependency on extensive infrastructure typically needed for detailed analysis. Integrating TinyML into medical devices is expected to enhance diagnostic procedures, improve patient outcomes, and extend advanced medical technologies to underserved areas. Researchers are exploring advanced techniques such as hyper-structure optimization [
20], Binary Neural Networks [
21], neural architecture search [
26], and Edge AI [
16,
17] to maximize the efficiency and feasibility of deploying TinyML in critical applications.
Supervised Methods of Segmentation: The evolution of AI technology has profoundly transformed medical image segmentation [
45]. Early advancements in this field primarily relied on convolutional neural networks (CNNs), such as the widely known U-Net and its attention-enhanced variant, Att-UNet [
22]. These models significantly improved segmentation accuracy by effectively focusing on relevant image features. As segmentation techniques advanced, hybrid architectures began to emerge. A notable category includes transformer-based models, which integrate the capabilities of transformers into segmentation tasks such as TransUNet [
23], TransFuse [
24], and Swin-UNet [
25]. Another noteworthy development is the integration of multilayer perceptron (MLP) architectures. UNeXt [
27] and MALUNet [
28], along with its advanced version, EGE-UNet [
29], utilize MLP-based designs to achieve superior segmentation results. More recently, Vision Mamba [
18]'s ability to deliver high-performance image processing with fewer parameters and lower computational requirements has led to the popularity of Mamba-based hybrid architectures. Examples include VM-UNet [
30] and its successor VM-UNetV2 [
31], LightM-UNet [
32], which emphasizes efficiency, and UltraLight VM-UNet [
33], designed for ultra-low computational environments. These innovations represent a significant shift towards more efficient and powerful medical image segmentation techniques. By integrating advanced AI methodologies, these new models provide improved accuracy, reduced computational demands, and greater applicability in diverse medical settings. In this paper, we extend our previous work, UCM-Net, to propose a new hybrid work, UCM-NetV2, which maintains fewer parameters and lower computations with high prediction performance. Additionally, we explore applying BNN to UCM-NetV2 to develop an Ultra-Fast version of a U-shaped Net.
3. Proposed Skin Lesion Segmentation Models: UCM-NetV2 and BNN-UCM-NetV2
This section introduces two innovative deep-learning models designed to address the critical challenge of high-precision skin lesion segmentation: UCM-NetV2 and BNN-UCM-NetV2. These models offer distinct advantages and are tailored for diverse computational environments, with a strong focus on real-world applicability in dermatology and healthcare. We will show that
UCM-NetV2 is a hybrid-based architecture that combines Convolutional Neural Networks (CNN) and Multilayer Perceptions (MLP) to enhance feature learning. Utilizing new proposed group loss functions, our method surpasses existing lightweight techniques in skin lesion segmentation. UCM-NetV2 is optimized on top of the previous generation, UCM-NetV2, for hardware-limited computing platforms. By focusing on maximizing the model's prediction performance, we proposed a new loss function for UCM-NetV2.
The BNN-UCM-NetV2 network extends the UCM-NetV2 model by incorporating binary neural networks (BNNs) for Binary Efficiency for tight resource-constrained devices: In BNNs, weights and activations are binarized into 1-bit values, drastically reducing the model size and computational demands. We especially proposed a new loss function for BNN-UCM-NetV2 to power the learning ability. We are the first to explore combining binary neural networks with U-shape networks for skin cancer segmentation.
Finally, we show that both UCM-NetV2 and BNN-UCM-NetV2 networks represent a significant step forward in developing practical and impactful solutions for skin lesion segmentation. By combining advanced CNN techniques with the efficiency of binary operations, these models offer a comprehensive approach to skin health monitoring and disease prevention.
3.1. UCM-NetV2 Network Structure
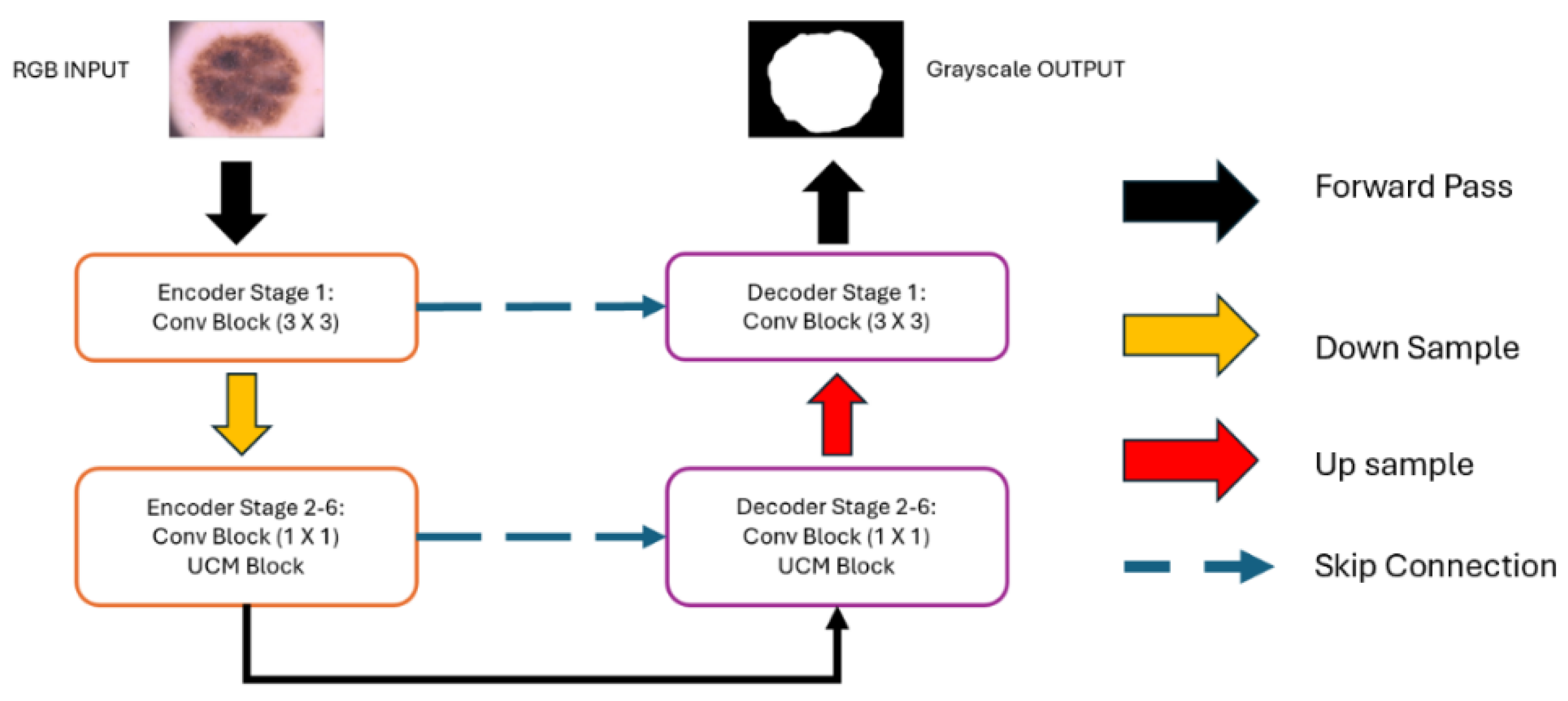
Network Structure Design:Figure 3 provides a detailed overview of the structural framework of UCM-NetV2, an advanced architecture with a distinctive U-shape design. This design builds upon the first generation of UCM-Net, incorporating both a down-sampling encoder and an up-sampling decoder to create a powerful network for skin lesion segmentation. The network is composed of six stages of encoder-decoder units, each with channel capacities of 8, 16, 24, 32, 48, and 64, respectively.
Convolution Block: In the initial encoder-decoder stage, our design uses a standard convolution layer with a 3x3 filter, effectively capturing spatial relationships within the input features. This kernel size is particularly beneficial for maintaining spatial integrity in the network's initial layers. For stages 2 through 6, we utilize a 1x1 convolution layer, significantly reducing the number of learnable parameters and the overall computational load, thus enhancing model efficiency. Each stage combines the 1x1 convolutional layer with UCM-Net blocks, contributing to advanced feature extraction and processing capabilities.
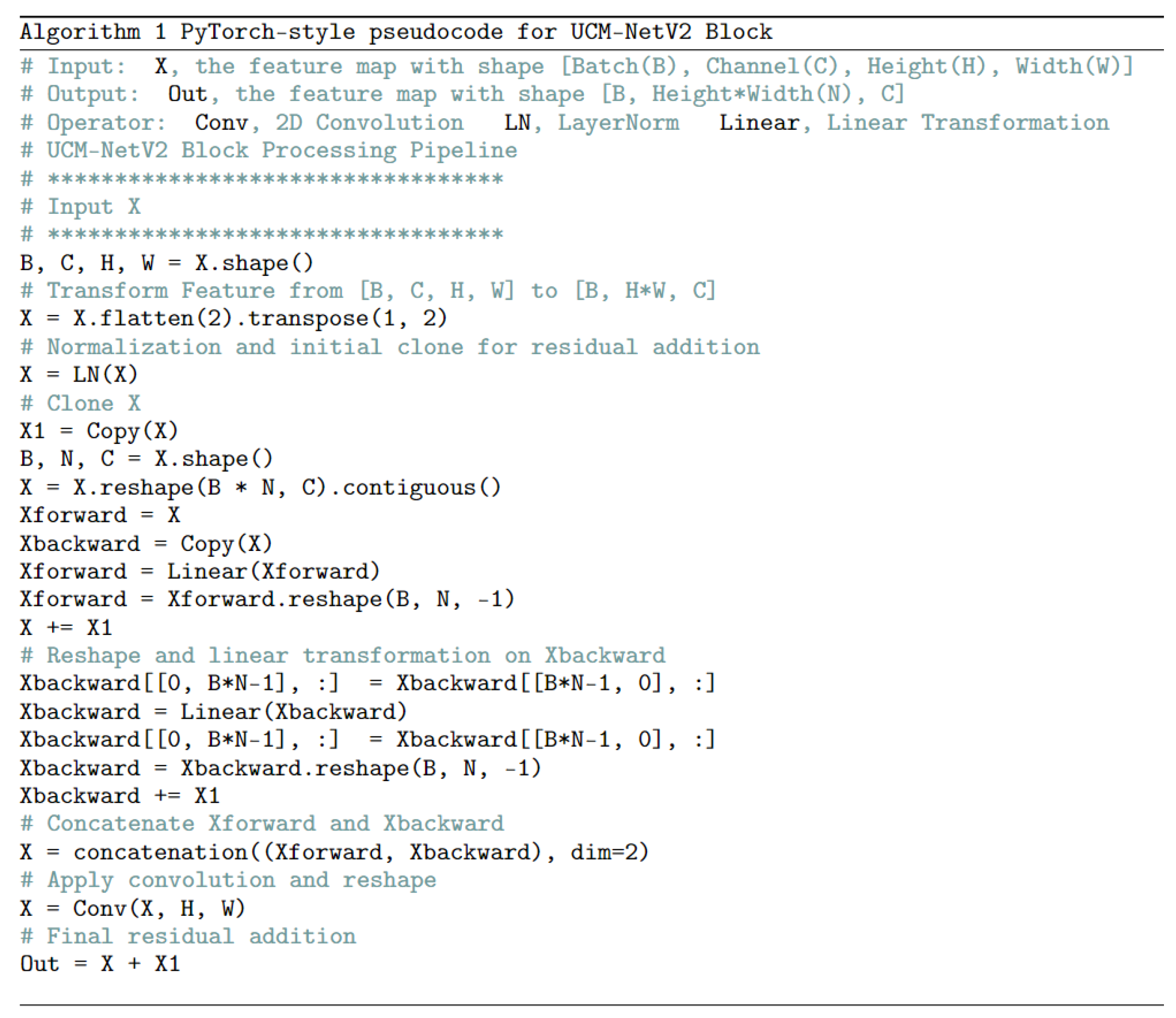
UCM-NetV2 Block: The 2nd-6th stages primarily utilize the UCM-NetV2 block for feature learning.
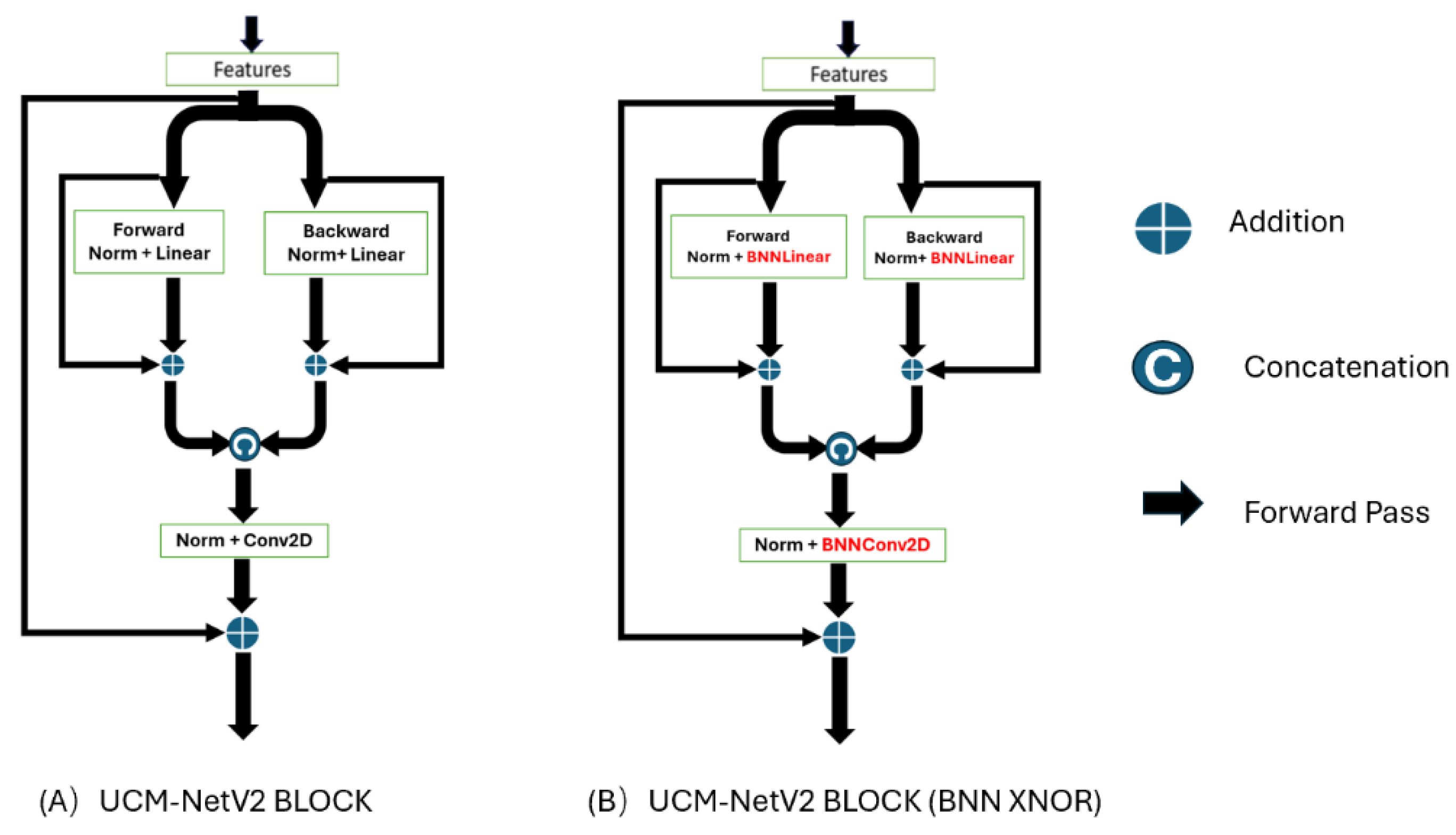
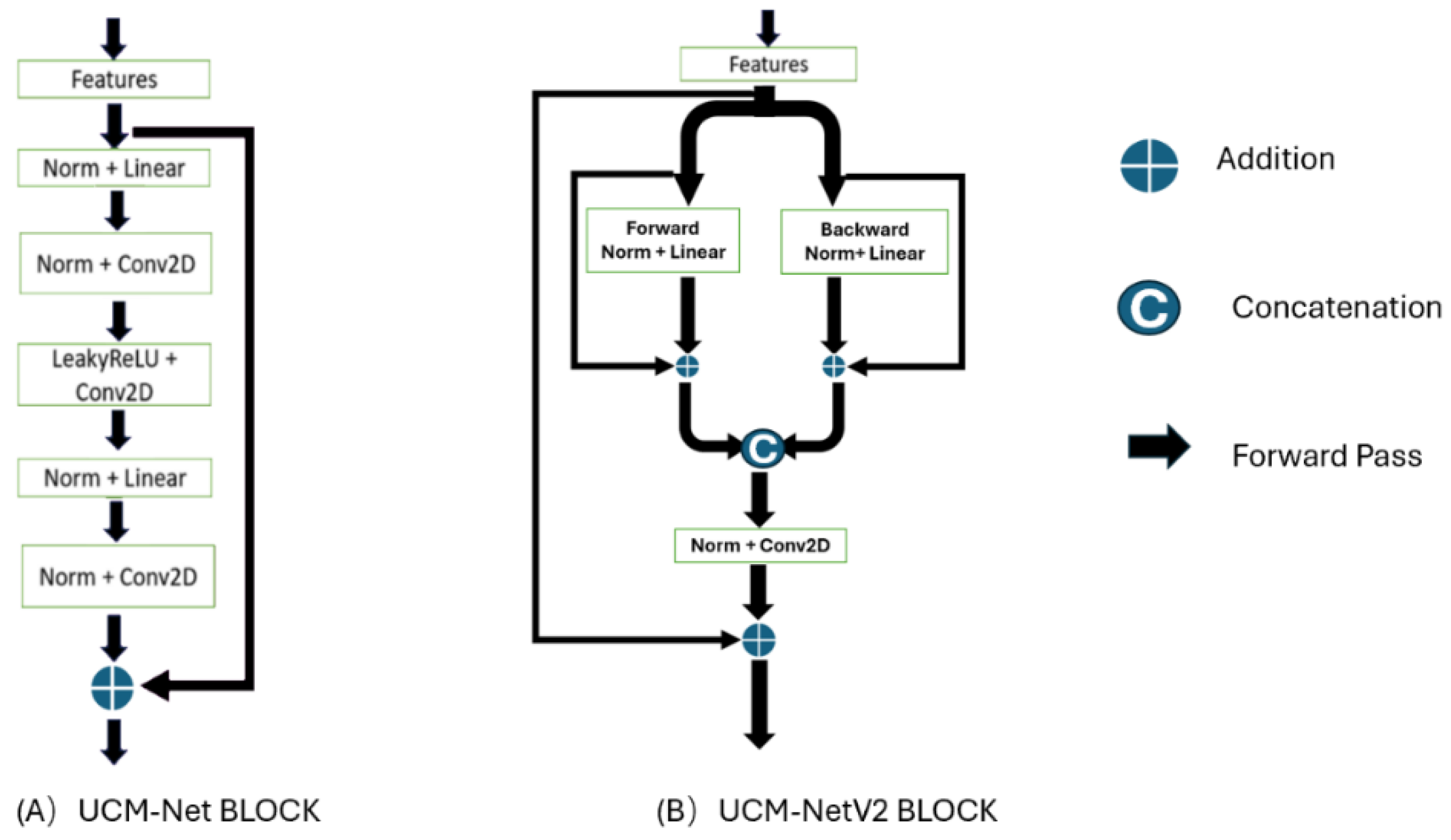
Figure 4 shows the differences between the UCM-Net and UCM-NetV2 blocks. Compared to its predecessor, the UCM-NetV2 block enhances feature learning by incorporating forward and backward paths with normalization, linear operations, concatenation, and additional convolutional layers. This block features twice the number of linear operations and an additional convolutional layer, reducing computational costs. With more residual operations, the UCM-NetV2 block enables more complex feature extraction and improved performance. The PyTorch-style pseudocode in
Figure 5 outlines the sequence of operations for feature learning.
Loss Function for the UCM-NetV2 Network: In our solution, we designed a new group loss function similar to those used in TransFuse [
24], EGE-UNet [
29], and our previous work, UCM-Net [
19]. However, unlike these previous works, our proposed base loss function incorporates binary cross-entropy (BCE) (1), Dice Loss (2), and Squared Dice Loss (3) components. These components calculate the loss from the scaled layer masks at different stages than the ground truth masks. Equations (5) and (6) present the stage loss for different layers and the output loss in the output layer, respectively, using BCE and Dice loss components. Additionally, we introduce a dynamic coefficient for the output loss, equation (6), based on the training epoch, adding more weight to the loss during the earlier training stages. This approach ensures that the model focuses more on learning crucial features for the final output segmentation early in the training process. The advantages of using different loss functions are as follows:
Binary Cross-Entropy Losses (BCE): Widely used in classification tasks, including biomedical image segmentation, these losses are highly effective for pixel-level segmentation.
Dice Loss: Commonly used in biomedical image segmentation, Dice loss addresses class imbalance by focusing on the overlap between predicted and true regions.
Squared Dice Loss: Further enhances the Dice loss by emphasizing the contribution of well-predicted regions, thereby improving stability and performance.
where N is the total number of pixels (for image segmentation) or elements (for other tasks),
is the ground truth value, and
is the predicted probability for the i-th element.
where smooth is a small constant added to improve numerical stability.
Which represents an enhancement over the standard Dice loss by emphasizing the squared terms of intersections and union.
Equations (1), (2), and (3) define the base loss function (4) for our proposed model, incorporating the Dice loss and Squared Dice loss components. To fully understand how different combinations of loss functions affect the trained model's performance, we add an ablation experiment in the next section.
where
is the weight for different stages. In this paper, we set
to 0.1, 0.2, 0.3, 0.4, and 0.5 based on the i-th stage, as illustrated in
Figure 4. Equation (7) is our proposed group loss function that calculates the loss from the scaled layer masks in different stages compared to the ground truth masks. Equations (5) and (6) present the stage loss in different layers and the output loss in the output layer.
Figure 5.
UCM-NetV2 Block Pseudocode.
Figure 5.
UCM-NetV2 Block Pseudocode.
Figure 6.
BNN-UCM-NetV2 network.
Figure 6.
BNN-UCM-NetV2 network.
3.2. BNN-UCM-NetV2 Network Structure
Deep learning (DL) has revolutionized the development of intelligent systems with numerous real-world applications. However, deploying DL on resource-constrained devices with limited computational power and energy remains challenging. Binary Neural Networks (BNNs) offer a promising solution in this context. BNNs are a type of neural network where activations and weights are binarized to 1-bit values in all hidden layers, providing a highly compact version of a Convolutional Neural Network (CNN) [
21]. BNNs' ability to compress 32-bit activations and weights to 1-bit values significantly reduces storage, computation costs, and energy consumption, making them ideal for deployment on tiny, resource-limited devices. Key advantages of BNNs are
Compression: BNNs compress 32-bit floating-point values into 1-bit representations, leading to significant reductions in model size.
Computational Efficiency: Binary operations (e.g., XNOR) replace computationally expensive floating-point operations, resulting in faster inference.
Energy Savings: Lower computational demands translate to reduced energy consumption, vital for battery-powered devices.
To the best of our knowledge, no relevant research work has applied BNNs to skin lesion segmentation. We are the first to explore this topic, which we believe will be a potential direction to reduce computation costs for the segmentation model in the future. Our study focuses on the practical and efficient application of XNOR-Net to UCM-NetV2 to develop BNN-UCM-NetV2. This approach significantly reduces inference computation costs to under 0.02 GFLOPs while maintaining the model's prediction performance. BNNs achieve this by compressing 32-bit float weights and layer inputs into 1-bit numbers. Within each neural network layer's matrices, weights and inputs are represented using [-1, +1] based on the sign of the original value. This allows the substitution of original matrix multiplication with the XNOR operation, a highly efficient approach that drastically reduces computational overhead. XNOR-Net, a classic type of BNN, represents a significant milestone in efficient deep learning.
Figure 7 illustrates the BNN-UCM-NetV2 Block, a high-performing block that leverages the efficiency of binary operations to minimize computational complexity and memory usage. The BNN-UCM-NetV2 Block is a specific component of the BNN-UCM-NetV2 Network, and its efficient design is crucial for the network's overall performance. Full-precision operations are replaced with their binary counterparts, labeled as BNNLinear and BNNConv2D in the figure. Using binary weights and activations significantly reduces the memory and computational resources required. BNN layers employ the XNOR-Net approach, performing convolution operations with binary weights to enhance computational efficiency.
Loss Function for BNN-UCM-NetV2: The loss function in BNNs is a crucial component that balances the efficiency gains of binary operations with the need for effective learning. While BNNs offer significant advantages in computational and energy efficiency, they come with challenges related to gradient approximation and potential performance trade-offs. Careful design and tuning of the loss function are essential to unlock the full potential of BNNs for various applications. In traditional neural networks, loss functions like Binary Cross-Entropy Loss or Dice Loss for segmentation are commonplace. However, Binary Neural Networks (BNNs) require a more specialized approach to accommodate their binarized values and ensure smooth gradient flow during backpropagation. For the loss function for BNNs, we adapt UCM-NetV2's loss functions with additional terms or modifications to handle the difference between the binarization output and full-precision matrix output. A proposed approach incorporates the following components:
Primary Loss Function: This component focuses on the main task, calculating the difference between the ground truth and the model's prediction. We selected our proposed loss function (7).
Regularization Term: A regularization term, Manhattan norm (L1-norm), is added to each binary layer to learn the difference between the loss function's binarization matrix and the full-precision matrix. We record the difference between binary matrix output and full-precision matrix output in each layer. We calculate the L1-Norm value on those records and add the L1-Norm value to our loss function.
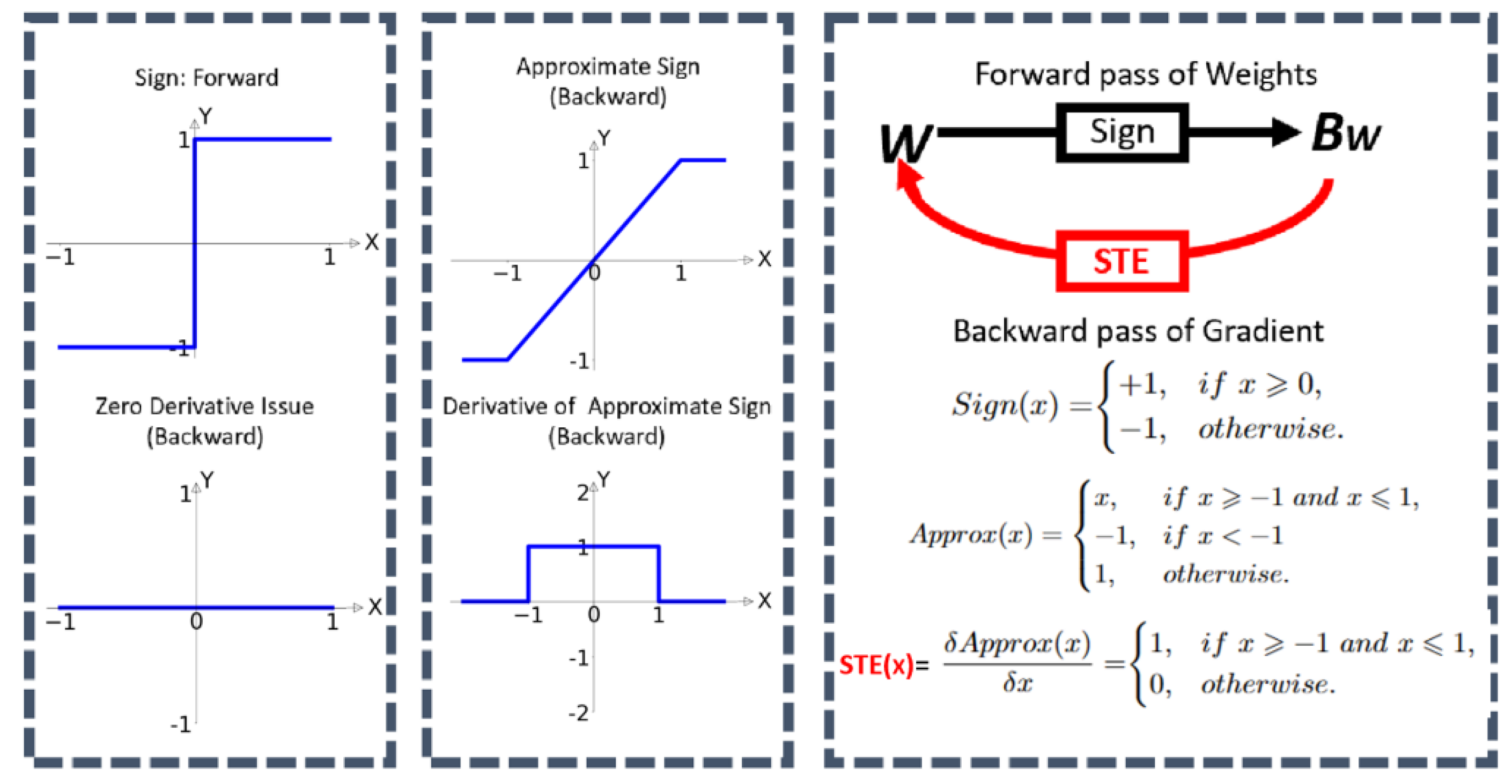
Gradient Approximation: Binarization from the sign function introduces discontinuities in the loss landscape, challenging direct gradient calculation. The reason is that the derivative value from the sign is always zero, which prevents the model from learning about loss during the training. A common method is gradually using approximation techniques like the straight-through estimator (STE).
The BNN-UCM-NetV2's loss function is:
where L1_Norm represents the Manhattan norm of the recorded difference binary matrix output with full-precision matrix output in each layer.
The loss function in BNNs is designed to balance the efficiency of binary operations with the need for effective learning. While BNNs offer significant advantages in terms of computational and energy efficiency, they come with challenges related to gradient approximation and performance trade-offs.
Figure 8.
BNNs STE process (cited from [
21]).
Figure 8.
BNNs STE process (cited from [
21]).
In conclusion, UCM-NetV2 and BNN-UCM-NetV2 represent a significant step forward in skin lesion segmentation. UCM-NetV2 delivers high-precision results on standard platforms, while BNN-UCM-NetV2 extends deep learning capabilities to resource-constrained environments. These models have the potential to revolutionize how dermatologists diagnose and monitor skin conditions, making accurate and efficient segmentation accessible to a broader range of practitioners and patients.
4. Results and Discussions
In this section, we describe different performance metrics and discuss their usage. We then provide details on the two test sets, ISIC2017 and ISIC2018, used for evaluation. Finally, we present the quantitative results of comparing our approach with state-of-the-art models on these test sets.
4.1. Experiment Setting and Preparation
Datasets: To evaluate the efficiency and performance of our proposed model against other published models, we utilized two public skin segmentation datasets from the International Skin Imaging Collaboration: ISIC2017 [
34,
35] and ISIC2018 [
36,
37]. The ISIC2017 dataset comprises 2000 dermoscopy images randomly divided into 1250 images for training, 150 for validation, and 600 for testing. The ISIC2018 dataset includes 2594 images, randomly divided into 1815 images for training, 259 for validation, and 520 for testing.
Evaluation Setting: Our UCM-NetV2 is implemented with the PyTorch [
38] framework. All experiments are conducted on the instance node at Lambda [
39] that has a single NVIDIA RTX A6000 GPU (24 GB), 14vCPUs, 46 GiB RAM, and 512 GiB SSD. The images are normalized and resized to 256×256. Simple data augmentations are applied, including horizontal flipping, vertical flipping, and random rotation. We select AdamW [
40] for the optimizer, initialized with a learning rate of 0.001 and a weight decay of 0.01. The CosineAnnealingLR [
41] is utilized as the scheduler with a maximum number of iterations of 50 and a minimum learning rate of 1e-5. Two hundred epochs are trained with a training batch size of 8 and a testing batch size of 1.
Evaluation Metrics: The model's performance is evaluated using the Dice Similarity Coefficient (DSC), sensitivity (SE), specificity (SP), and accuracy (ACC). Furthermore, the model's memory consumption is assessed based on the number of parameters and Gigaflops (GFLOPs). DSC measures the degree of similarity between the ground truth and the predicted segmentation map. SE measures the percentage of true positives with the sum of true positives and false negatives. SP measures the percentage of true negatives with the sum of true negatives and false positives. ACC measures the overall percentage of correct classifications. The formulas used are as follows:
Where TP denotes true positive, TN denotes true negative, FP denotes false positive, and FN denotes false negative.
4.2. Experimental Results Analysis
We compared our new model with published classical and efficient methods such as U-Net [
48], SCR-Net [
49], ATTENTION SWIN U-NET [
47], C2SDG [
50], VM-UNet [
30], VM-UNet v2 [
31], MALUNet [
28], UNeXt-S [
27], LightM-UNet [
32], EGE-UNet [
29] and UltraLight VM-Net [
33].
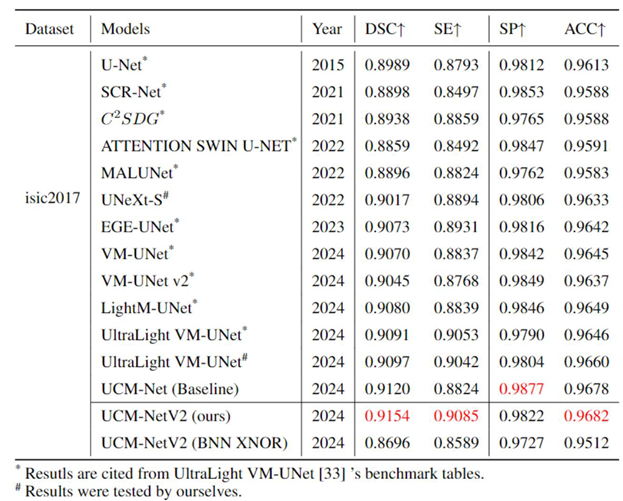
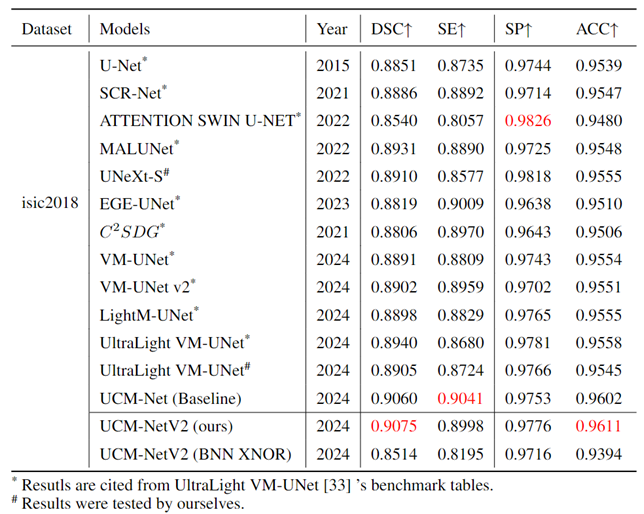
Table 1,
Table 2 and
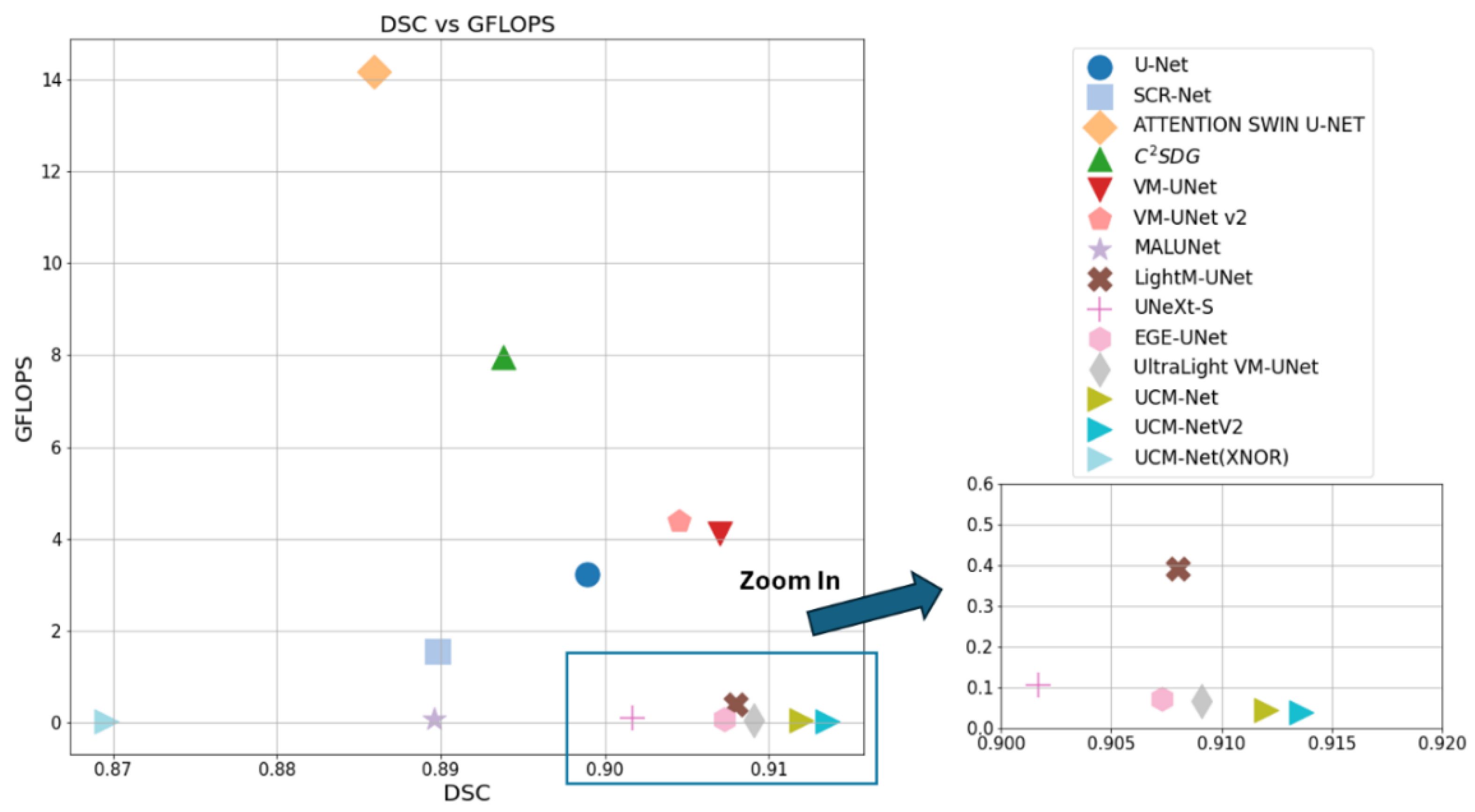
Table 3 comprehensively evaluate the performance of our UCM-NetV2, a novel and lightweight skin lesion segmentation model, against well-established models using the ISIC2017 and ISIC2018 datasets. UCM-NetV2 consistently outperforms the compared models across metrics such as DSC, SE, SP, and ACC, achieving the highest DSC of 0.9154 and SE of 0.9085 on ISIC2017, and DSC of 0.9075 and ACC of 0.9611 on ISIC2018.
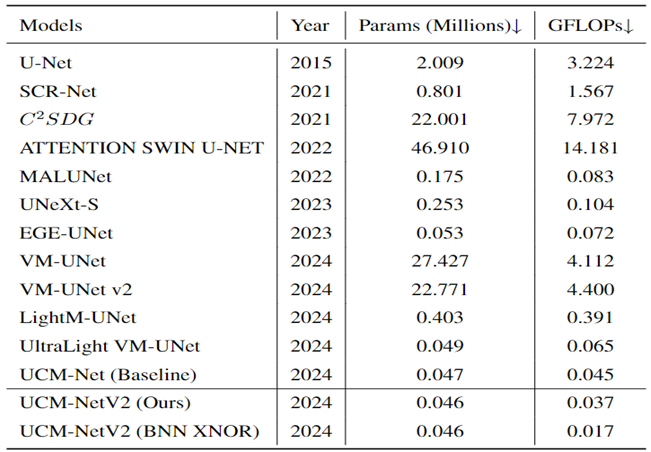
Table 3 highlights UCM-NetV2's computational efficiency, operating with significantly lower GFLOPs than other models, achieving this efficiency without compromising performance. We also explore applying BNN to our UCM-NetV2, which can further be speeded up without sacrificing the prediction performance. The key takeaway is that UCM-NetV2 establishes a new state-of-the-art for skin lesion segmentation, combining superior prediction performance with computational efficiency, thus redefining the standard for accurate and efficient skin lesion delineation in biomedical imaging.
4.3. Ablation Experiments with Different Loss Functions
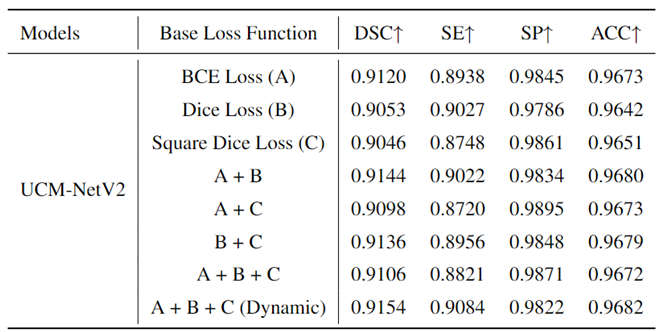
To prove the effects of our proposed loss function, we conducted an ablation experiment using different base loss functions to train our model, UCM-NetV2.
Table 4 presents the experimental results on the ISIC 2017 dataset, providing a comparative analysis of the performance metrics for each loss function. The base loss functions evaluated include Binary Cross-Entropy (BCE) Loss (A), Dice Loss (B), and Square Dice Loss (C). Additionally, combinations of these loss functions were tested: A + B, A + C, B + C, A + B + C, and a dynamic epoch-coefficient version of A + B + C. The performance metrics considered were the Dice Similarity Coefficient (DSC), Sensitivity (SE), Specificity (SP), and Accuracy (ACC). From the results, it is evident that the use of a dynamic combination of all three loss functions (A + B + C (Dynamic)) yields the highest DSC (0.9154) and SE (0.9084), indicating superior segmentation performance in terms of both overlap and true positive rate. The combination of A + B also shows a strong performance with a DSC of 0.9144 and an SE of 0.9022. While Dice Loss (B) and Square Dice Loss (C) individually achieve lower DSC values than their combinations, they still perform competitively. These findings highlight the importance of carefully selecting and combining loss functions to optimize the performance of segmentation models. The dynamic combination approach particularly stands out, suggesting that it effectively balances the strengths of each base loss function to achieve enhanced overall performance.
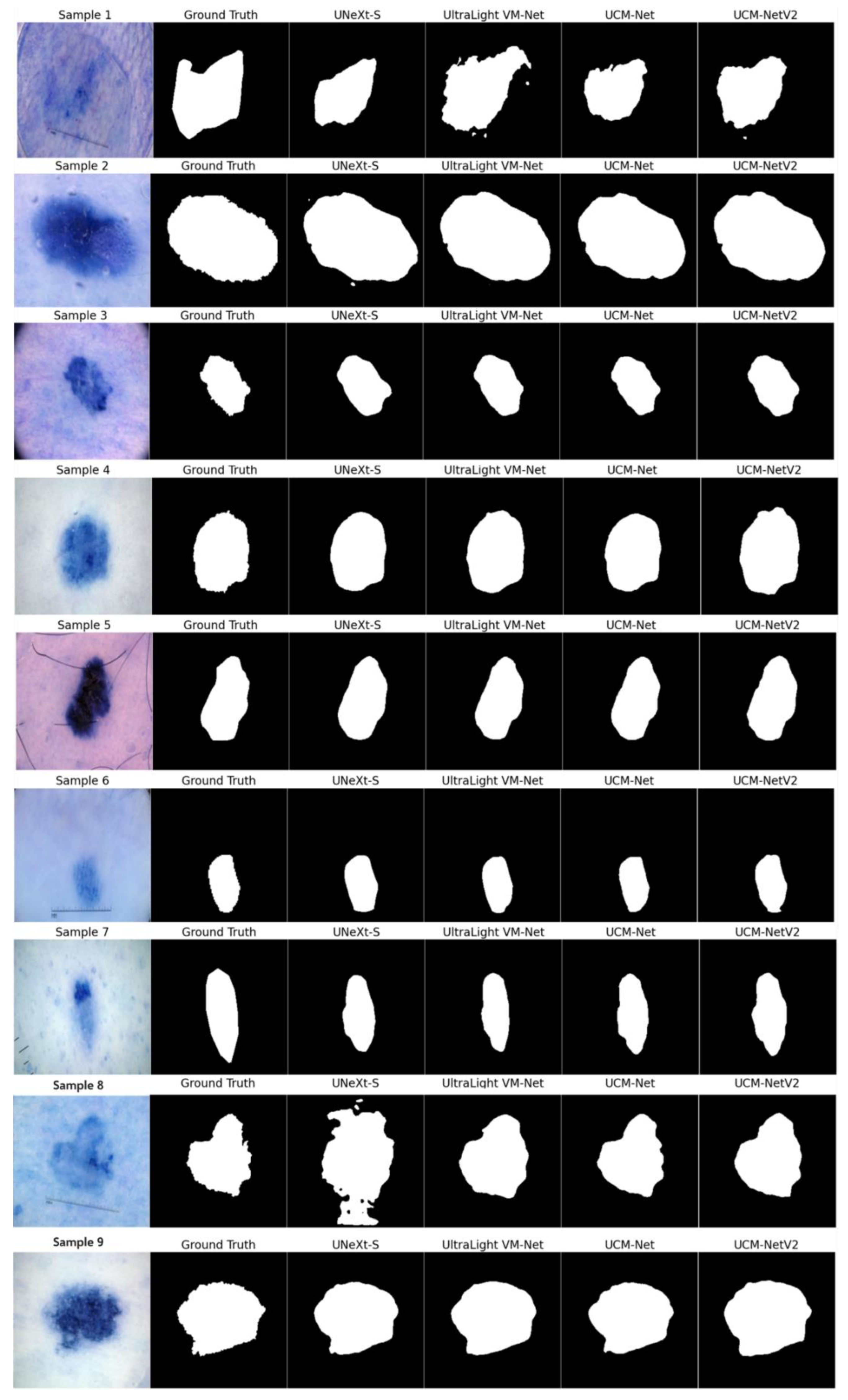
Figure 9 presents the segmentation results on nine sample inputs. Each row in the figure corresponds to a different sample input, where the columns display the original image, the ground truth, and the segmentation results from various models, including UNeXt-S, UltraLight VM-Net, UCM-Net, and UCM-NetV2. The segmentation masks are shown in grayscale for clear visibility, making it easy to compare the performance of each model against the ground truth. UCM-NetV2 captures the boundaries of the target regions with high precision, closely matching the ground truth annotations. This model demonstrates robustness across all samples, with minimal noise or artifacts, unlike other models that occasionally introduce errors, as seen with UltraLight VM-Net in samples 1 and 8. The clear and accurate segmentation provided by UCM-NetV2 highlights its effectiveness in medical image analysis tasks, where precision is critical. The model's ability to consistently produce high-quality segmentation masks makes it a reliable choice for practical applications. This visual comparison underscores UCM-NetV2's competitive performance compared to other published lightweight segmentation models, making it a standout among the tested models and suggesting its potential for further development and deployment in medical imaging workflows.
5. Conclusions
Lesion area segmentation is the first step to creating an artificial intelligence system to quantitatively compare lesion images captured at different moments. This paper introduces UCM-NetV2 and BNN-UCM-NetV2, which will be poised to make significant contributions to the field of dermatology by enabling high-precision skin lesion segmentation with lower computation costs. UCM-NetV2, the second generation of UCM-Net, boasts robust feature learning capabilities while maintaining a minimal parameter count and reduced computational demand. We applied this innovative approach to the challenging task of skin lesion segmentation, conducting comprehensive experiments using a range of evaluation metrics to demonstrate its effectiveness and efficiency. The results unequivocally show UCM-NetV2's superior performance compared to recent lightweight and Mamba-based models for skin lesion segmentation. Notably, UCM-NetV2 is the first model to consume less than 0.04 GFLOPs for this task. Additionally, we explore BNN-based segmentation on skin lesion area segmentation, which points to a new research topic for lightweight models.
Our future work will focus on expanding the applicability of UCM-NetV2 to a broader range of medical imaging tasks, particularly multi-class segmentation. We aim to maintain the model's efficiency while ensuring its performance remains competitive with state-of-the-art solutions. Additionally, we will investigate ways to enhance the prediction performance of the binary neural network (BNN) version in the next generation of UCM-Net. Also, we plan to create new image databases from various camera viewpoints under chosen lighting conditions since existing skin cancer-related datasets have significant limitations, such as a small number of image samples, limited disease conditions, insufficient annotations, and non-standardized image acquisitions. Furthermore, we encourage further research into translating suitable skin lesion analysis methodologies, including UCM-NetV2, into real-time clinical applications. This translation is essential for bridging the gap between research advancements and practical clinical use, ultimately improving patient care and diagnostic accuracy. the experimental results, their interpretation, as well as the experimental conclusions that can be drawn.
Author Contributions
Methodology, C.Y.; validation, C.Y.; formal analysis, C.Y.; investigation, C.Y.; writing—original draft preparation, C.Y.; writing—review and editing, D.Z. and S.A.; supervision, S.A.; All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data available within the article.
Conflicts of Interest
This study was internally funded and none of the authors have any conflicts of interest to disclose.
References
- Rundle, Chandler W., et al. "Epidemiologic burden of skin cancer in the US and worldwide." Current Dermatology Reports 9 (2020): 309-322. ACS, ACS. "Cancer Facts & Figures 2022." American Cancer Society, National Home Office, Atlanta (2022).
- Stephen L Brown, Peter Fisher, Laura Hope-Stone, Bertil Damato, Heinrich Heimann, Rumana Hussain, and M Gemma Cherry. Fear of cancer recurrence and adverse cancer treatment outcomes: predicting 2-to 5-year fear of recurrence from post-treatment symptoms and functional problems in uveal melanoma survivors. Journal of Cancer Survivorship, 17(1):187–196, 2023. [CrossRef]
- Rebecca L Siegel, Kimberly D Miller, and Nikita Sandeep Wagle. Cancer statistics, 2023.
- Robin Marks. An overview of skin cancers. Cancer, 75(S2):607–612, 1995.
- Thomas Martin Lehmann, Claudia Gonner, and Klaus Spitzer. Survey: Interpolation methods in medical image processing. IEEE transactions on medical imaging, 18(11):1049–1075, 1999. [CrossRef]
- Muhammad Nasir, Muhammad Attique Khan, Muhammad Sharif, Ikram Ullah Lali, Tanzila Saba, and Tassawar Iqbal. An improved strategy for skin lesion detection and classification using uniform segmentation and feature selection based approach. Microscopy research and technique, 81(6):528–543, 2018. [CrossRef]
- Catarina Barata, M Emre Celebi, and Jorge S Marques. Explainable skin lesion diagnosis using taxonomies. Pattern Recognition, 110:107413, 2021. [CrossRef]
- Isaac Sanchez and Sos Agaian. Computer aided diagnosis of lesions extracted from large skin surfaces. In 2012 IEEE International Conference on Systems, Man, and Cybernetics (SMC), pages 2879–2884. IEEE, 2012.
- Isaac Sanchez and Sos Agaian. A new system of computer-aided diagnosis of skin lesions. In Image Processing: Algorithms and Systems X; and Parallel Processing for Imaging Applications II, volume 8295, pages 390–401. SPIE, 2012.
- Tiago M de Carvalho, Eline Noels, Marlies Wakkee, Andreea Udrea, and Tamar Nijsten. Development of smartphone apps for skin cancer risk assessment: progress and promise. JMIR Dermatology, 2(1):e13376, 2019.
- butterflynetwork.https://www.butterflynetwork.com/iq-ultrasound-individuals.
- phonemedical.https://blog.google/technology/health/ai-dermatology-preview-io-2021/.
- Andre Esteva, Alexandre Robicquet, Bharath Ramsundar, Volodymyr Kuleshov, Mark DePristo, Katherine Chou, Claire Cui, Greg Corrado, Sebastian Thrun, and Jeff Dean. A guide to deep learning in healthcare. Nature medicine, 25(1):24–29, 2019. [CrossRef]
- Chunlei Chen, Peng Zhang, Huixiang Zhang, Jiangyan Dai, Yugen Yi, Huihui Zhang, and Yonghui Zhang. Deep learning on computational-resource-limited platforms: a survey. Mobile Information Systems, 2020:1–19, 2020. [CrossRef]
- Neil C Thompson, Kristjan Greenewald, Keeheon Lee, and Gabriel F Manso. The computational limits of deep learning. arXiv preprint arXiv:2007.05558, 2020.
- Su, Weixing, et al. "AI on the edge: a comprehensive review." Artificial Intelligence Review 55.8 (2022): 6125-6183. [CrossRef]
- Singh, Raghubir, and Sukhpal Singh Gill. "Edge AI: a survey." Internet of Things and Cyber-Physical Systems 3 (2023): 71-92.
- Lianghui Zhu, Bencheng Liao, Qian Zhang, Xinlong Wang, Wenyu Liu, and Xinggang Wang. Vision mamba: Efficient visual representation learning with bidirectional state space model. arXiv preprint arXiv:2401.09417, 2024.
- Yuan, Chunyu, Dongfang Zhao, and Sos S. Agaian. "UCM-Net: A lightweight and efficient solution for skin lesion segmentation using MLP and CNN." Biomedical Signal Processing and Control 96 (2024): 106573. [CrossRef]
- Hyeji Kim, Muhammad Umar Karim Khan, and Chong-Min Kyung. Efficient neural network compression. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition, pages 12569–12577, 2019.
- Chunyu Yuan and Sos S Agaian. A comprehensive review of binary neural network. Artificial Intelligence Review, 56(11):12949–13013, 2023. [CrossRef]
- Ozan Oktay, Jo Schlemper, Loic Le Folgoc, Matthew Lee, Mattias Heinrich, Kazunari Misawa, Kensaku Mori, Steven McDonagh, Nils Y Hammerla, Bernhard Kainz, et al. Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999, 2018.
- Jieneng Chen, Yongyi Lu, Qihang Yu, Xiangde Luo, Ehsan Adeli, Yan Wang, Le Lu, Alan L Yuille, and Yuyin Zhou. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306, 2021.
- Yundong Zhang, Huiye Liu, and Qiang Hu. Transfuse: Fusing transformers and cnns for medical image segmen-tation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part I 24, pages 14–24. Springer, 2021.
- Hu Cao, Yueyue Wang, Joy Chen, Dongsheng Jiang, Xiaopeng Zhang, Qi Tian, and Manning Wang. Swin-unet: Unet-like pure transformer for medical image segmentation. In European conference on computer vision, pages 205–218. Springer, 2022.
- White, Colin, et al. "Neural architecture search: Insights from 1000 papers." arXiv preprint arXiv:2301.08727 (2023).
- Jeya Maria Jose Valanarasu and Vishal M Patel. Unext: Mlp-based rapid medical image segmentation network. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 23–33. Springer, 2022.
- Jiacheng Ruan, Suncheng Xiang, Mingye Xie, Ting Liu, and Yuzhuo Fu. Malunet: A multi-attention and lightweight unet for skin lesion segmentation. In 2022 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pages 1150–1156. IEEE, 2022.
- Jiacheng Ruan, Mingye Xie, Jingsheng Gao, Ting Liu, and Yuzhuo Fu. Ege-unet: an efficient group enhanced unet for skin lesion segmentation. arXiv preprint arXiv:2307.08473, 2023.
- Jiacheng Ruan and Suncheng Xiang. Vm-unet: Vision mamba unet for medical image segmentation. arXiv preprint arXiv:2402.02491, 2024.
- Mingya Zhang, Yue Yu, Limei Gu, Tingsheng Lin, and Xianping Tao. Vm-unet-v2 rethinking vision mamba unet for medical image segmentation. arXiv preprint arXiv:2403.09157, 2024.
- Weibin Liao, Yinghao Zhu, Xinyuan Wang, Cehngwei Pan, Yasha Wang, and Liantao Ma. Lightm-unet: Mamba assists in lightweight unet for medical image segmentation. arXiv preprint arXiv:2403.05246, 2024.
- Renkai Wu, Yinghao Liu, Pengchen Liang, and Qing Chang. Ultralight vm-unet: Parallel vision mamba significantly reduces parameters for skin lesion segmentation. arXiv preprint arXiv:2403.20035, 2024.
- Isic 2017 challenge dataset. https://challenge.isic-archive.com/data/#2017.
- Matt Berseth. Isic 2017-skin lesion analysis towards melanoma detection. arXiv preprint arXiv:1703.00523, 2017.
- Isic 2018 challenge dataset. https://challenge.isic-archive.com/data/#2018.
- Noel Codella, Veronica Rotemberg, Philipp Tschandl, M Emre Celebi, Stephen Dusza, David Gutman, Brian Helba, Aadi Kalloo, Konstantinos Liopyris, Michael Marchetti, et al. Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). arXiv preprint arXiv:1902.03368, 2019.
- Adam Paszke, Sam Gross, Francisco Massa, Adam Lerer, James Bradbury, Gregory Chanan, Trevor Killeen, Zeming Lin, Natalia Gimelshein, Luca Antiga, et al. Pytorch: An imperative style, high-performance deep learning library. Advances in neural information processing systems, 32, 2019.
- Lambda cloud gpu. https://cloud.lambdalabs.com/instances.
- Ilya Loshchilov and Frank Hutter. Decoupled weight decay regularization. In International Conference on Learning Representations, 2018.
- Ilya Loshchilov and Frank Hutter. Sgdr: Stochastic gradient descent with warm restarts. arXiv preprint arXiv:1608.03983, 2016.
- Linares, Miguel A., Alan Zakaria, and Parminder Nizran. "Skin cancer." Prim Care 42.4 (2015): 645-59.
- Armstrong, Bruce K., and Anne Kricker. "Skin cancer." Dermatologic Clinics 13.3 (1995): 583-594.
- Marks, Robin. "An overview of skin cancers." Cancer 75.S2 (1995): 607-612.
- Khalid M Hosny, Doaa Elshora, Ehab R Mohamed, Eleni Vrochidou, and George A Papakostas. Deep learning and optimization-based methods for skin lesions segmentation: A review. IEEE Access, 2023.
- Rastegari, Mohammad, et al. "Xnor-net: Imagenet classification using binary convolutional neural networks." European conference on computer vision. Cham: Springer International Publishing, 2016.
- Aghdam, Ehsan Khodapanah, et al. "Attention swin u-net: Cross-contextual attention mechanism for skin lesion segmentation." 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI). IEEE, 2023.
- Ronneberger, Olaf, Philipp Fischer, and Thomas Brox. "U-net: Convolutional networks for biomedical image segmentation." Medical image computing and computer-assisted intervention–MICCAI 2015: 18th international conference, Munich, Germany, October 5-9, 201.
- Lei, Dajiang, et al. "SCRNet: an efficient spatial channel attention residual network for spatiotemporal fusion." Journal of Applied Remote Sensing 16.3 (2022): 036512-036512.
- Shishuai Hu, Zehui Liao, and Yong Xia. Devil is in channels: Contrastive single domain generalization for medical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 14–23. Springer, 2023.Bio-Implants—Global Market Outlook (2017–2026): Expected to Grow at a CAGR of 10.1% ResearchAndMarkets.com. 2018. Available online: https://www.businesswire.com/news/home/20181109005315/en/Bio-Implants---Global-Market-Outlook2017-2026-Expected-to-Grow-at-a-CAGR-of-10.1---ResearchAndMarkets.com (accessed on 6 April 2024).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).