1. Introduction
KP is an important gram-negative zoonotic conditional pathogen[
1]. It can infect humans and a wide range of farm animals, leading to infections such as pneumonia, meningitis and even sepsis[
2]. As a result, KP infections cause significant economic losses and pose a serious threat to public health.
Omp X acts as a virulence protein that can participate in bacterial pathogenesis, mediates bacterial adhesion to and invasion of epithelial cells. Additionally, it plays a role in bacterial adaptation to the osmotic pressure of the external environment[
3,
4]. A promising class of targeting molecules is represented by aptamers. Aptamers are short, single-stranded nucleic acid (ssDNA or RNA) molecular probes that bind the target molecule with high affinity and specificity, generated by the SELEX technology (Systematic Evolution of Exponentially Enriched Ligands)[
5,
6].
In this study, we used the proteins of Omp X and Omp A as targets for screening and counter-screening, respectively. We successfully screened the OmpX aptamer using MBs-SELEX and confirmed its high specificity and affinity by flow cytometry and qPCR. In addition, complexes of Klebsiella pneumoniae with specific FITC-labeled aptamer (FITC-Apt-3) and AuNCs were characterized using total internal reflection fluorescence microscopy (TIRFM).The blue fluorescent dots of AuNCs and the green fluorescent dots of FITC-labeled aptamer overlapped with the presence of bacteria. These results indicate that AuNCs and aptamers can specifically bind to target bacteria to form complexes. In addition, in vitro results showed that Apt-1, Apt-2 and Apt-3 inhibited bacterial growth, with Apt-3 exhibiting the strongest inhibitory effect. The results suggest that the aptamers can specifically recognize membrane proteins and thus inhibit the formation of bacterial cell membranes, thereby effectively inhibiting bacterial growth.
2. Material and Methods
The ssDNA library containing a 40 nt central random region flanked by two primer binding sites(
Table 1).Both the library and the primers were synthesized by Sangon Biotech (Shanghai, China). Carboxylated magnetic beads(MBs) were purchased from Thermo Fisher Scientific(USA). Omp X and Omp A proteins were purchased from MCE. KP strain(ATCC 21106) was purchased from China Industrial Culture Collection Center. Chloroauric acid (HAuCl
4) was purchased from MERCK. random DNA sequence were synthesized by Sangon Biological Science and Technology Company (Shanghai, China). Particle size and zeta-potential were used by the Malvern Nano Z Zetasizer (Malvern, UK). The fluorescence images were recorded using a confocal laser scanning microscope (IX2-DSU, Olympus, Japan).
2.1. Bacterial Culture
Bacteria were cultured in 20 mL of LB medium for 14 hours. The rough bacterial concentration was estimated by measuring the absorbance at 600 nm using a UV-Vis absorption spectrophotometer. The exact bacterial concentration was determined using the conventional agar plate counting method.
2.2. SELEX Screening for Omp X DNA Aptamers
The initial ssDNA library (10 nmol) was completely solubilized at 37
oC in 300 µL of binding buffer. Aptamers were prevented from intrastrand base pairing by heating at 95
oC for 5 min and then immediately cooling on ice for 10 min. to allow for better formation of specific spatial 3D structures. to provide rich binding spatial conformation for target binding[
7]. In addition, to eliminate non-specific aptamers, ssDNA was incubated with MBs-Omp A complexes and shaken at 37
oC for 1 h as a negative SELEX screen. The magnet separated unbound ssDNA. unbound ssDNA was incubated with MBs-Omp X complexes shaken at 37 °C for 1 h. After incubation, the supernatant containing unbound ssDNA was removed. Add 200 µL of ddH
2O to the MBs-Omp X-ssDNA complex and heat denaturation at 95
oCfor 5 min, and collect the supernatant as the secondary ssDNA library for the next round of screening.
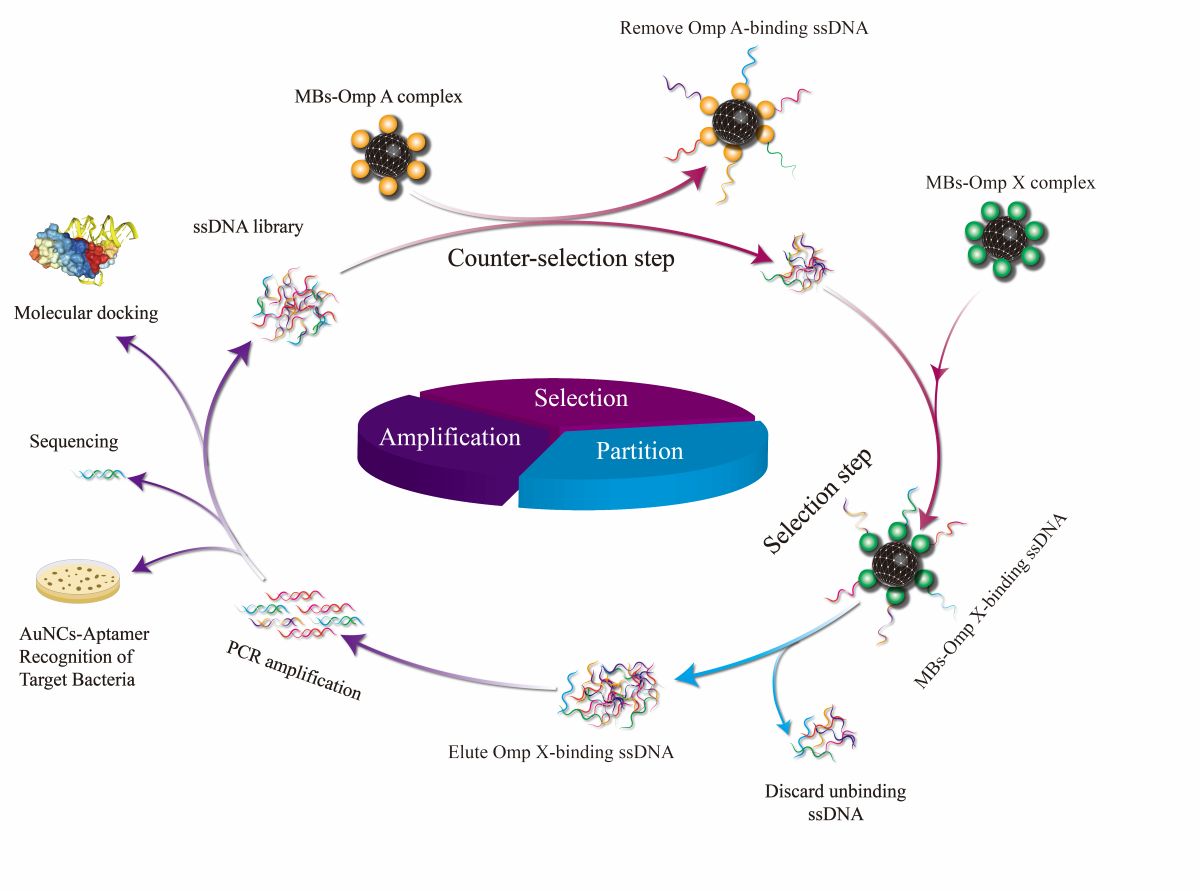
The MBs-SELEX screening was performed through 7 rounds of selection, including the steps of binding, elution, PCR amplification, and ssDNA regeneration, as shown in
Figure 1.
2.3. Preparation of Secondary ssDNA Library by Streptavidin Magnetic Beads(SA-MBs) Method
PCR amplification was conducted to generate double-stranded DNA (dsDNA). PCR reaction system: 5 µL ssDNA, 5 U TaqDNA polymerase, 1×PCR buffer, 1.5 mM MgCl2, 0.25 mM dNTP, 0.4 µM Bio-P7 primer, 0.4 µM P11 primer, and nuclease-free water in a total volume of 100 µL. PCR reaction conditions: 95 oC for 2 PCR reaction conditions: 95 oC, 2 min, (95 oC for 30 s, 55 oC for 30 s, 72 oC for 30 s, 15 cycles), 72 oC, 4 min, stored at 4 oC. Subsequently, 200 µL of Streptavidin-MBs were combined with 100 µL of PCR product and incubated at 37 oC for 30 min. 1 mL of 5% formamide was washed three times, each time for 3 min. 500 µL of BB were added to 95 oC heat denaturation for 10 min, and the supernatant was collected to obtain the secondary ssDNA. The secondary ssDNA library was prepared.
2.4. Real-Time Monitoring of the Screening Process by qPCR
For each round, 50 µL of secondary ssDNA was incubated with 100 µL of MBs-Omp A complex and 100 µL of MBs-Omp X complex at 37 ℃ for 1 h. The MBs were collected separately, the supernatant was discarded, and the MBs complexes were collected separately and washed three times with 1 mL of BB, each time for 3 min. Subsequently, 100 µL of ddH2O was added to each of the MBs complexes, and the supernatants were thermally denatured at 95 °C for 5 min. The supernatant was collected as qPCR template. qPCR reaction system: 5 µL ssDNA, 5 U TaqDNA polymerase, 1×PCR buffer, 1.5 mM MgCl2, 0.25 mM dNTP, 0.4 µM Bio-P7 primer, 0.4 µM P11 primer, and nuclease-free water in a total volume of 100 µL. PCR reaction conditions: 95 ℃, 2 min, (95 ℃ 30 min), (95 ℃ 30 min). 2 min, (95 ℃ for 30 s, 55°C for 30 s, 72°C for 30 s, 35 cycles), 72 oC for 4 min. ΔCt values were monitored in real time for ssDNA enrichment.
2.5. High Throughput Sequencing and Prediction of Secondary and Tertiary Structures and Molecular Docking Analysis
The secondary ssDNA library of the 7th round was used as a template, and PCR amplification was performed with P7 and P11 primers. The amplified products were identified by 1 % agarose gel electrophoresis, and the gel recovered and purified dsDNA of the target fragment was sent to Shanghai Biotech for high-throughput sequencing for comprehensive study. The aptamer sequences were analyzed for homology and sequence abundance using DNAMAN software. The most abundant ssDNA was selected as the candidate aptamer sequence(
Table 2). The secondary structure and Gibbs free energy (ΔG) of the aptamer sequences were predicted using RNA Structure software.
2.6. Evaluation of Aptamer Binding Dissociation Constants(Kd) and Specificity
To assess the affinity of candidate aptamers for Omp X. 100 μL of 500 ng/mL Omp X was incubated in 50 μL of MBs, and the reaction conditions were 37 °C for 2 h. After incubation, MBs-Omp X complex was washed three times with BB. Similarly, Omp A served as a negative control. Then 200 μL of FAM-labelled aptamers(FITC-Apt-1,FITC-Apt-2 and FITC-Apt-3) were mixed with the MBs-Omp X complex mixtures at concentrations of 0, 25, 50, 100, 200, and 400 nM respectively for 30 min. After two washes with 1 mL of BB, Samples were eluted by heat denaturation with 100 μL TE buffer at 100
oC for 10 min and the fluorescence intensity was analyzed by an MicroplateReader(λ
em=518 nm and λ
ex=492 nm). The
Kd value was derived by a non-linear regression equation Y =
Bmax×X / (
Kd + X) [
8,
9,
10] between the concentration of the aptamer(X) and the fluorescence intensity(Y) using Prism software.
The synthesized FITC-Apt-1, FITC-Apt-2 and FITC-Apt-3 were incubated with MBs-Omp A , MBs-Omp 37 and MBs-Omp 17, respectively, for 30 min at 37 ℃ in screening buffer (Cf = 200 nmol/L, Vf = 200 μL). The complexes were washed three times with screening buffer for 1 min each. The FITC-labelled aptamers were determined by flow cytometry.
2.7. Synthesis of AuNCs
Streptavidin was synthesized and modified with streptavidin according to a previously reported procedure with minor modifications[
11,
12]. Glutathione (31 mM, 8 mL) and HAuCl
4 solution (25 mM, 1 mL) were mixed and the pH was adjusted to 8.0. sodium borohydride (1 mg/mL, 2 mL) was added to the mixed solution and reacted at room temperature for 24 h. In the next step, 20 mg of AuNCs were washed three times with 10 mM PBS. The carboxyl terminus of glutathione-modified AuNCs was activated by EDC (10 mg/mL, 100 μL) and NHS (10 mg/mL, 100 μL) for 30 min[
13,
14,
15]. streptavidin (1 mg/mL, 50 μL, pH 9.0) was added to the activated AuNCs solution and reacted at room temperature for 24 h. The obtained streptavidin-modified AuNCs were washed three times and redispersed in PBS at a concentration of 0.1 mg/mL and stored at 4
oC for subsequent experiments [
16].
2.8. Characterization of Target Bacteria Recognition Based on AuNCs-Aptamers
KP bacterial fluids (100 μL, 108 CFU/mL) were mixed with their specific FITC-Apt-3 aptamer (10 μL, 20 μM) in the dark for 30 min. After centrifugation, the microspheres were washed three times with sterile water. Subsequently, the microspheres were resuspended in AuNCs solution in the dark for 30 min. Following three washes with PBS, the resulting FITC-labeled aptamers@bacteria@AuNCs complexes were resuspended in PBS and characterized using total internal reflection fluorescence microscopy. FITC-ssDNA library was used as a negative control.
2.9. Inhibition of Bacterial Growth by In Vitro Aptamers
200 μL of KP solution (108 CFU/mL) was divided into four parts in a 1.5 mL EP tube, and Apt-1, Apt-2, Apt-3 and ssDNA library (10 μM) were added respectively. After thorough mixing, the above liquids were spread evenly on LB culture plates respectively, and the number of colonies was counted after overnight incubation at 37 oC.
2.10. Statistical Analysis
All data were analyzed by calculating averages and standard deviations (SDs). No other statistical methods were used.
3. Results and Discussion
3.1. Screening of Omp X Aptamers
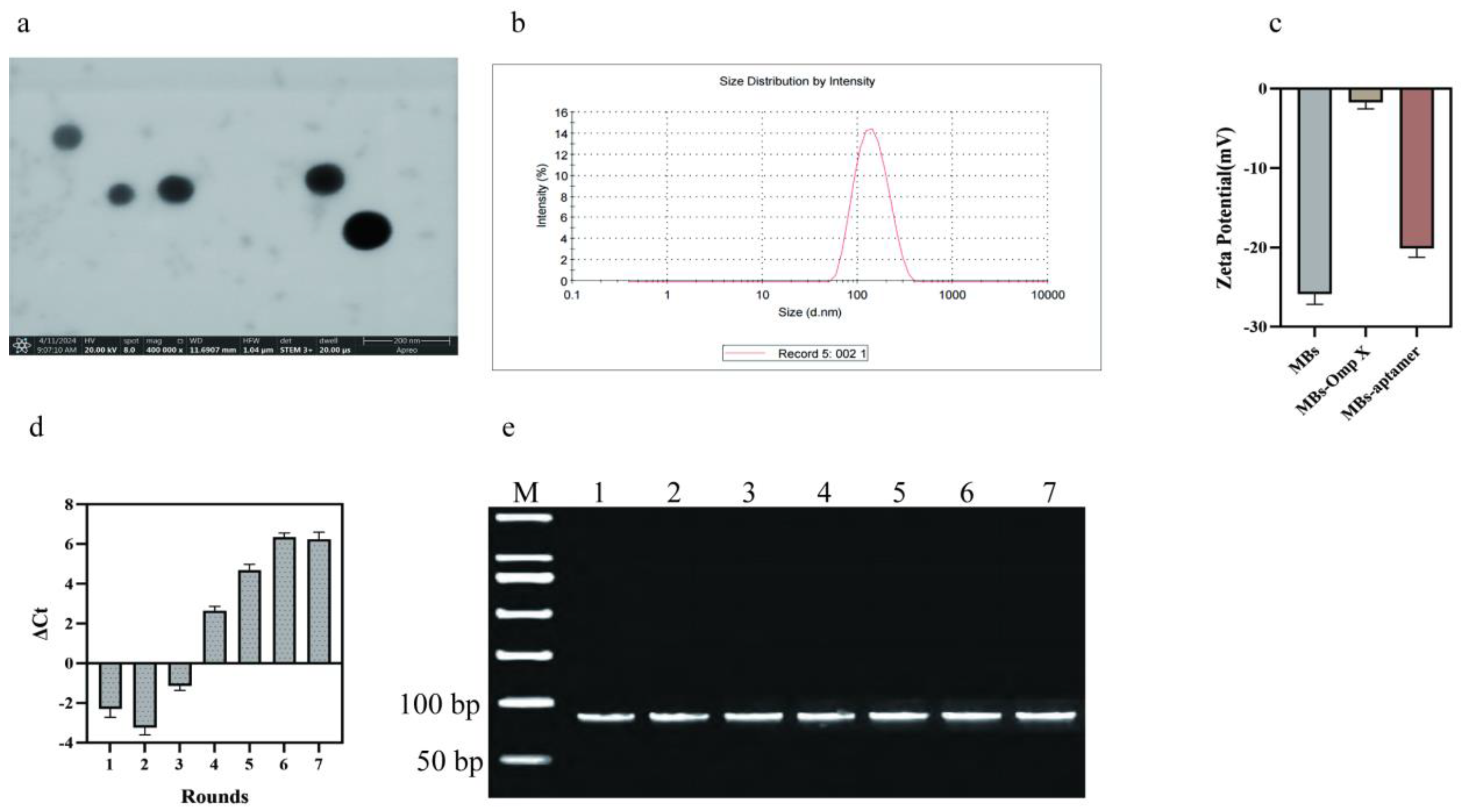
The MBs and MBs-coupled proteins were tested for potentials before and after binding to aptamers. As shown in
Figure 2. a-c, the particle size of the MBs is 122 nm, and the zeta potential value of the MBs is -26.8 mV, which indicates that the carboxyl group coupling rate is high. The zeta potential after binding to the protein is -1.6 mV, and the carboxyl group on the surface of the MBs binds to the amino group on the surface of the protein, which makes the zeta potential significantly smaller, and the zeta potential of the aptamer is -20 mV after binding to the protein, and the aptamer carries a negative charge, and therefore has a lot of phosphate groups on the surface.
MBs-Omp X was used as the target for SELEX screening, and MBs-Omp A was used for counter-screening. A total of 7 rounds of screening were performed. During the screening process, each round of ssDNA secondary library was incubated with MBs-Omp X, and then unbound or weakly bound ssDNA was removed by magnetic separation and washing. the MBs-Omp X-ssDNA complexes were heat denatured at 95
oC, and bound ssDNAs were eluted to obtain bound ssDNAs. as the number of rounds of the screening increased, the ssDNA libraries showed a constant increase in the binding to Omp X, indicating that the specific aptamers that can bind to Omp X were continuously enriched. The ΔCt values stabilized in rounds 6 and 7. By the 7th round, the binding of ssDNA library to Omp X had reached saturation, so the screening was terminated. The whole screening process was dynamically detected by real-time quantitative PCR, and the enrichment results of the screening are shown in
Figure 2.d. The results of agarose gel electrophoresis of the secondary ssDNA libraries from the first to the seventh rounds show clear bands and band sizes that match the initial ssDNA libraries(
Figure 2.e).
3.2. High Throughput Sequencing Analysis of the Enriched Pools
We then performed high-throughput sequencing of the round 7 secondary library to identify aptamer candidate sequences. Fortunately, HTS has more sequences compared to Sanger sequencing, and the coverage allowed us to track changes in enrichment frequency across clusters during SELEX. Compared to copy number, enrichment rate is a more appropriate metric for selecting candidate aptamers with specific binding properties because it is a sensitive and direct reflection of target-induced enrichment rather than other factors such as PCR amplification bias. Aptamer candidates were categorised in groups based on their sequence repeatability, secondary structure and homology. We selected three representative sequences from different groups and performed synthetic DNA sequences for further characterisation.
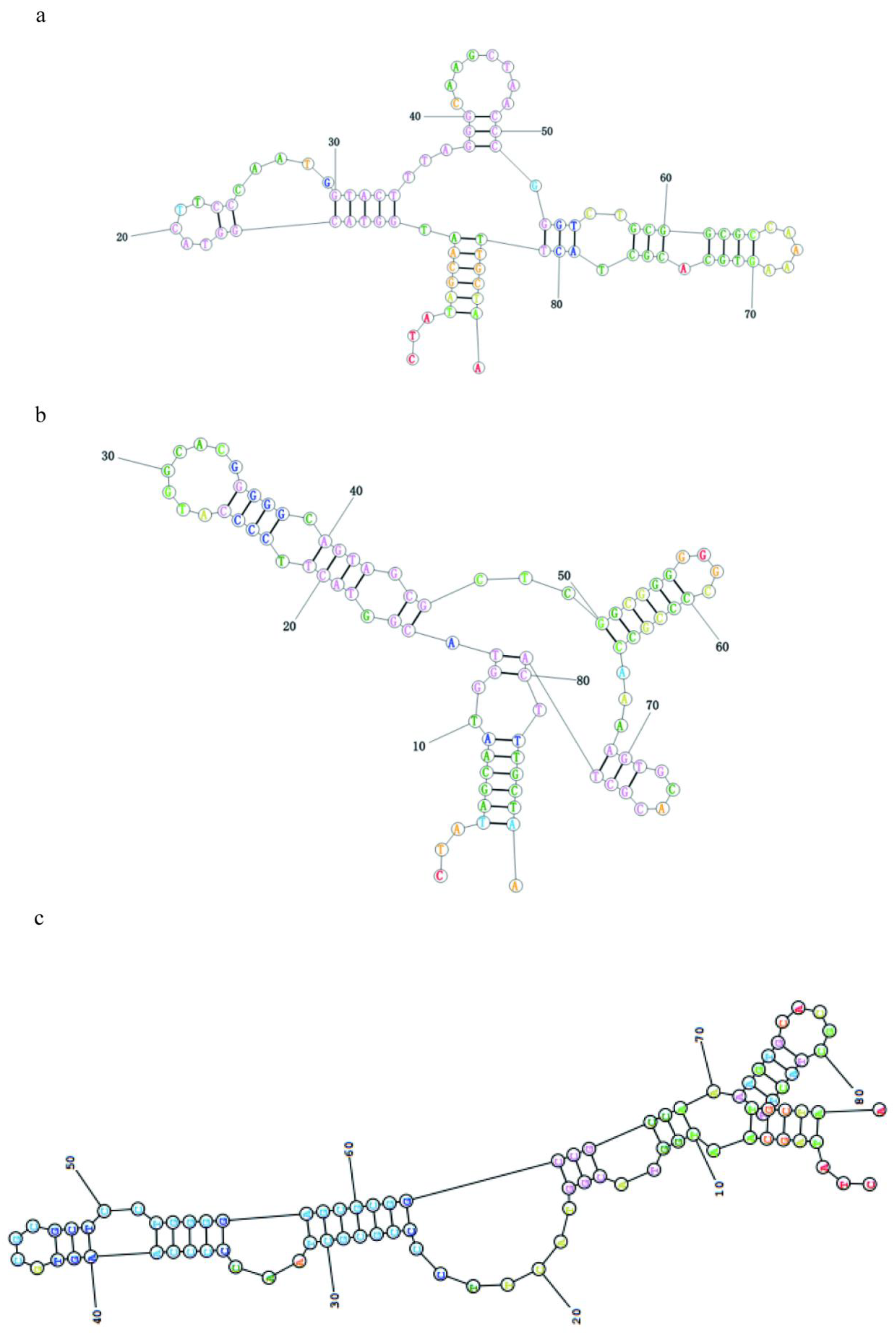
As shown in
Figure 3. a-c,the results of secondary structure simulations performed on the aptamer sequences of Apt-1, Apt-2 and Apt-3. The secondary structure of the aptamer was simulated using RNAstructure, and the results showed that the aptamer was dominated by stem-loop and pocket structures. This may be the structural basis of the aptamer binding to Omp X. In addition, docking simulations show that the binding of the Omp X to the candidate aptamer sequences is mainly dominated by hydrogen bonding. Thus, a stable complex structure is effectively formed.
HDOCK SERVER simulates the binding of Omp X and the aptamer Apt-3. As shown in
Figure 3. d, Omp X is completely encapsulated by Apt-3.
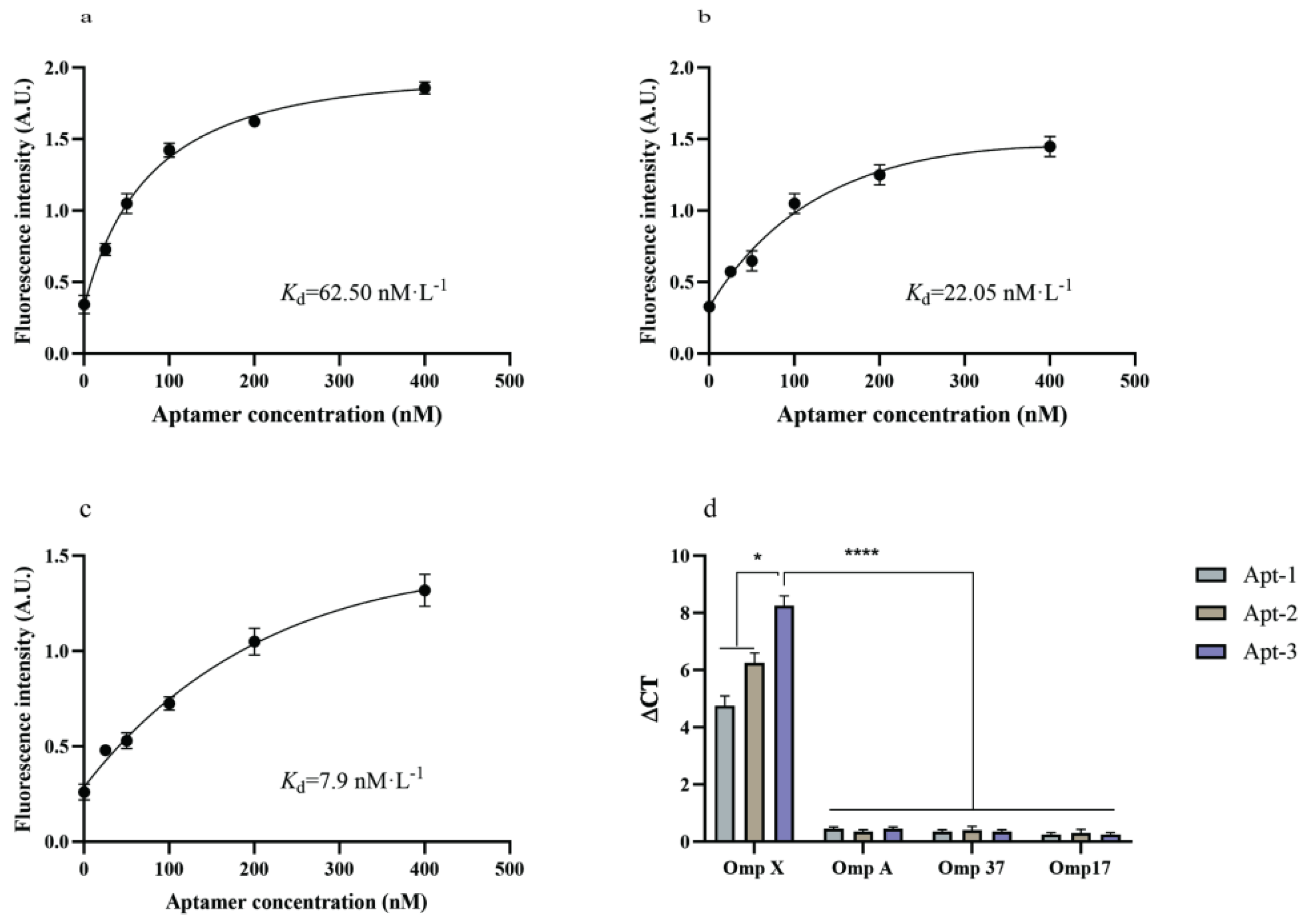
3.3. Binding Affinity and Specificity Studies
To determine the affinity of the candidate aptamer for Omp X, we used fluorescence intensity to determine the equilibrium dissociation constant (
Kd) between the candidate aptamer and the target, with the magnitude of the
Kd value calculated according to a specific formula. The value of
Kd of Apt-1 was 62.50 nM, Apt-2 was 22.05 nM and of Apt-3 was 7.9 nM (
Figure 4. a-c). The
Kd value is a reflection of the degree of affinity between the aptamer and the target; the higher the
Kd value, the lower the affinity, and vice versa[
17,
18].
The selected aptamer needs not only good affinity but also high specificity for Omp X. The specificity of the four candidate aptamers was determined by qPCR. Three different protein of Omp A, Omp 37 and Omp 17 were selected for testing. All three aptamers, Apt-1, Apt-2 and Apt-3, have high specificity to Omp X (
Figure 4. d).
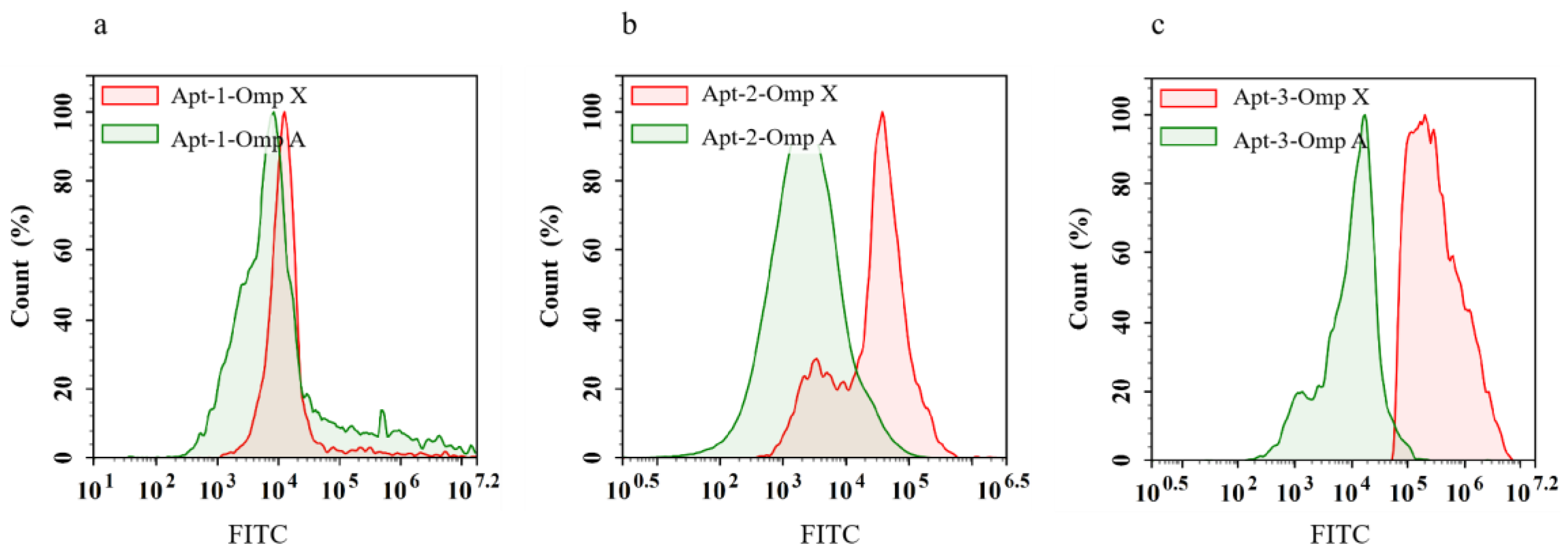
Flow cytometry results showed that FITC-Apt-1, FITC-Apt-2 and FITC-Apt-3 did not bind to Omp A and only specifically recognised Omp X(
Figure 5. a-c).
3.4. Results of the Recognition Characterization of Target Bacteria Based on AuNCs-Aptamers
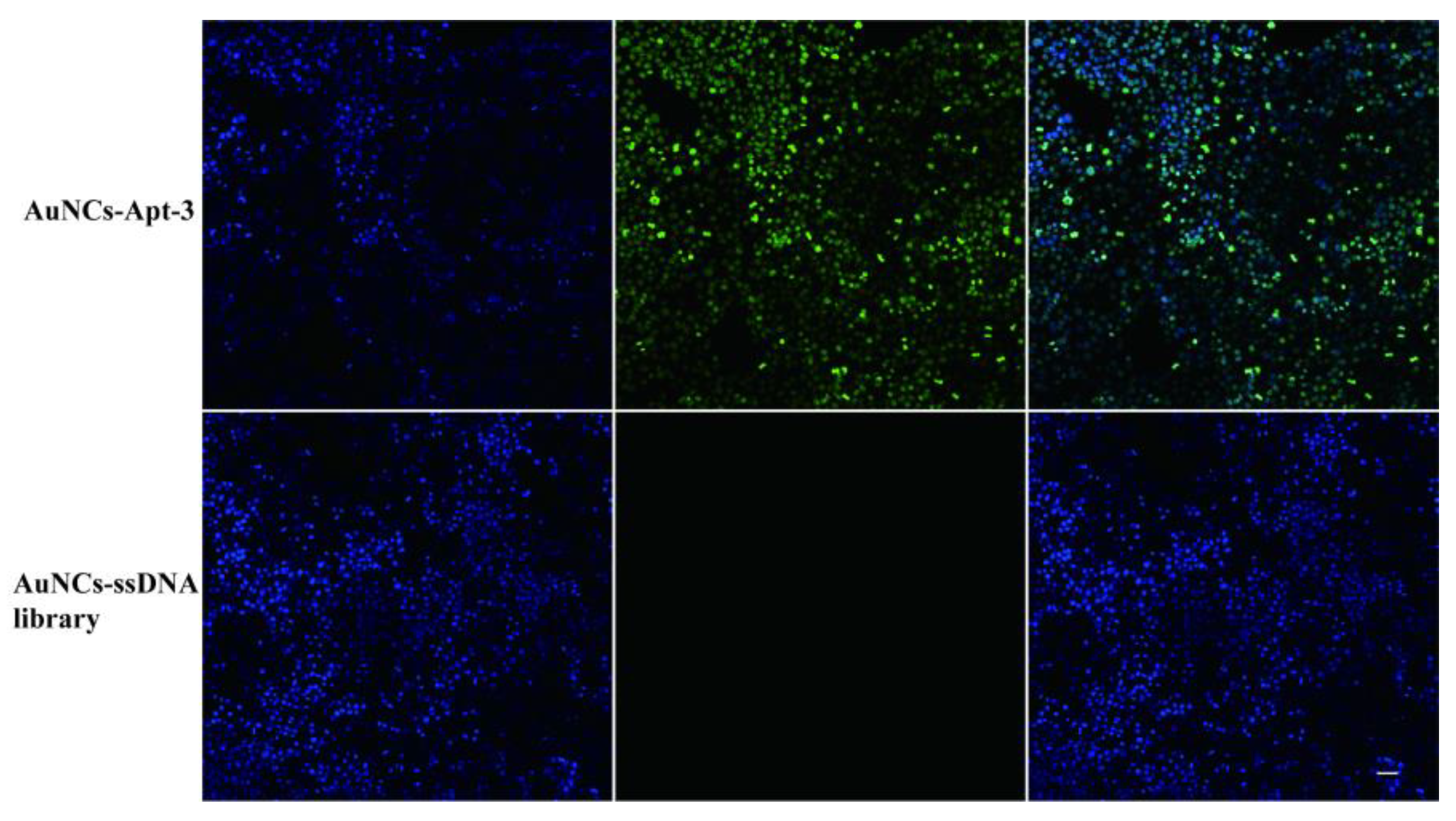
Prior to the bacterial assay, we demonstrated the specific recognition ability of the aptamer. The aptamer was labeled with fluorescein isothiocyanate (FITC) to characterize its recognition ability. Klebsiella pneumoniae was mixed with the specific FITC-labeled aptamer (FITC-Apt-3) and AuNCs, and the FITC-labeled ssDNA library served as a negative control. The complex was characterized using total internal reflection fluorescence microscopy (TIRFM). As shown in
Figure 6, the blue fluorescent dots of AuNCs and the green fluorescent dots of FITC overlapped with the presence of bacteria in the dark environment. These results indicate that AuNCs and aptamers can bind specifically to target bacteria to form complexes, providing good practical support for this strategy[
19,
20].
3.5. Results of the Inhibitory Effect of In Vitro Aptamers on Bacterial Growth
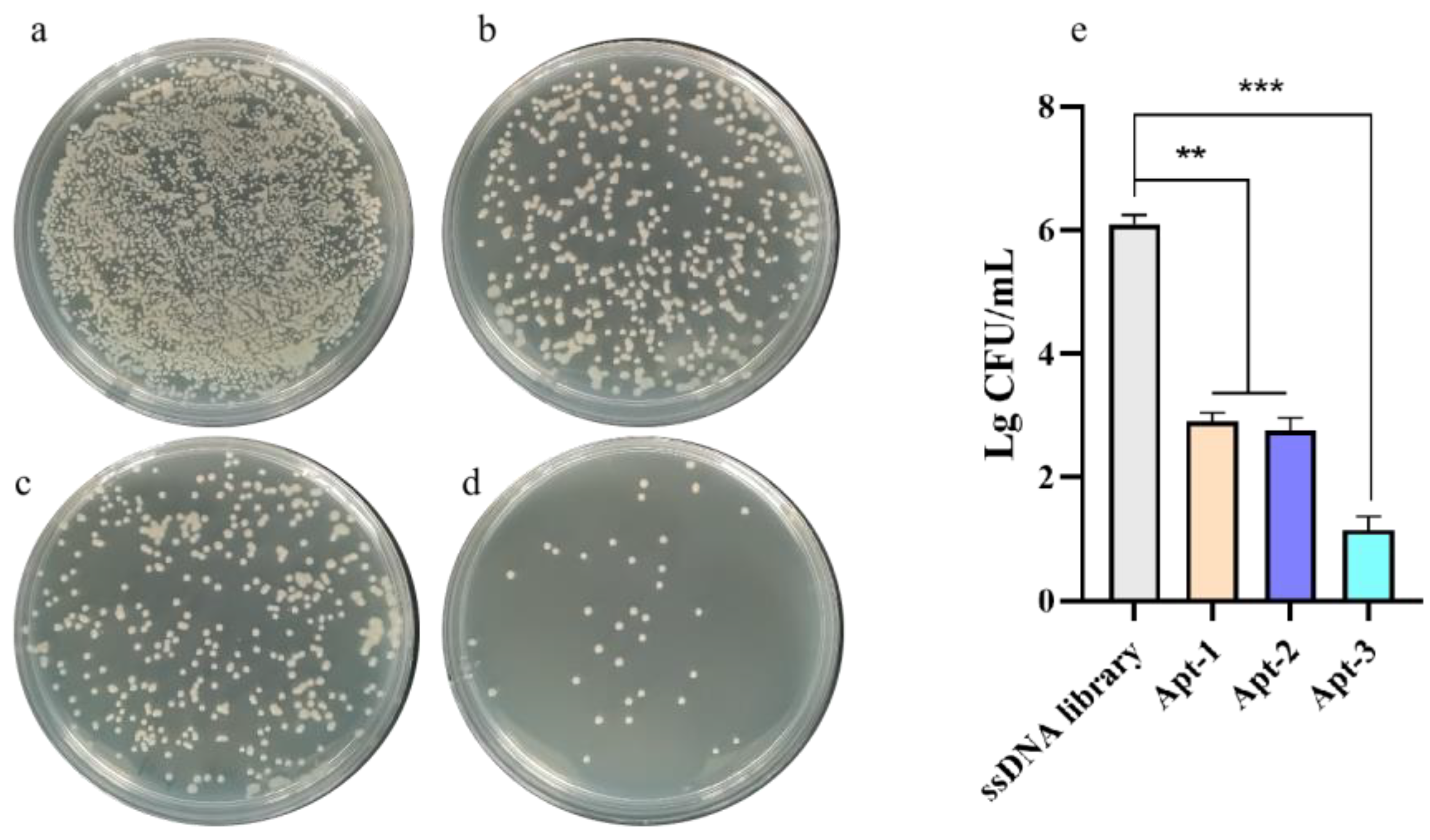
The outer membrane proteins OmpA and OmpX can synergistically participate in the invasion of Enterobacter sakazakii from the intestinal basal lamina into ileal tissues, and the absence of ompX significantly reduced the bacterial invasive adhesion to Caco-II cells. The aptamer can specifically bind Omp X can inhibit the growth of bacteria. As shown in
Figure 7, Apt-1,Apt-2 and Apt-3 could inhibit bacterial growth, among which Apt-3 inhibited the most obvious effect. The results suggest that the aptamer can specifically recognize membrane proteins, which may inhibit the formation of bacterial cell membrane and thus effectively inhibit bacterial growth. The aptamer has a potential mechanism of inhibiting bacterial growth. The mechanism of bacterial growth inhibition needs to be further verified by subsequent experiments.
4. Conclusions
In this work, we isolated for the first time a highly specific and high affinity 88-nucleotide DNA aptamer that was able to recognize Omp X by MBs-SELEX. qPCR was used to detect the enrichment of the ssDNA library. The affinity assay results showed that Apt-3 had the highest binding affinity and the lowest Kd value (7.9 nM). In addition, the results of flow cytometry experiments also indicated that Apt-3 had a high affinity for Omp X without binding to other targets. In addition, the secondary structures of these aptamers are able to fold into different secondary structures as predicted by the secondary structures. The stem-loop structure is common in the candidate aptamers. In addition, the primer regions of these sequences are also involved in the stem-loop structure. The binding mechanisms of the target molecules and aptamers will be further explored in subsequent work.
KP complexed with a specific FITC-labeled aptamer (FITC-Apt-3) and AuNCs was characterized using total internal reflection fluorescence microscopy (TIRFM).Blue fluorescent dots of AuNCs and green fluorescent dots of the FITC-labeled aptamer overlapped with the presence of bacteria. These results indicate that AuNCs and aptamers can bind specifically to target bacteria to form complexes, providing good practical support for this strategy. In addition, the results of in vitro experiments showed that Apt-1, Apt-2 and Apt-3 inhibited bacterial growth, with Apt-3 having the most obvious inhibitory effect. The results suggest that aptamers can specifically recognize membrane proteins and thus inhibit the formation of bacterial cell membranes, thus effectively inhibiting bacterial growth. Aptamers have a potential mechanism to inhibit bacterial growth. It provides a new potential molecular tool for inhibiting the growth of KP.
Declaration of competing interest
he authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
Data will be made available on request.
Acknowledgements
We thank the National Natural Science Foundation of China (81560346),the Heibei Key R&D Program (203777114D) and the Qinhuangdao Key R&D Program (201902A052), for their financial support.
References
- Hu G, Chen X, Chu W, et al. Immunogenic characteristics of the outer membrane phosphoporin as a vaccine candidate against Klebsiella pneumoniae. Vet Res. 2022, 21, 53(1):5. [CrossRef]
- Cerdeira L, Silva KC, Fernandes MR, et al.. Draft genome sequence of a CTX-M-15-producing Klebsiella pneumoniae sequence type 340 (clonal complex 258) isolate from a food-producing animal. J Glob Antimicrob Resist. 2016, 7:67-68. [CrossRef]
- Kim K, Choi J, Lim JA, et al. Outer membrane proteins A (OmpA)and X (OmpX)are essential for basolateral invasion of Cronobacter sakazakii. Appl Environ Microbiol, 2010a, 76(15): 5188-5198. [CrossRef]
- Iwanaga N, Chen K, Yang H, et al. Vaccine-driven lung TRM cells provide immunity against Klebsiella via fibroblast IL-17R signaling. Sci Immunol. 2021, 6(63):eabf1198. [CrossRef]
- Zhao Y, He L, Huang B, et al. Identification of a novel DNA aptamer that selectively targets lung cancer serum. RSC Adv. 2021, 11(53):33759-33769. [CrossRef]
- Zheng Y, Zhao Y, Di Y,et al. DNA aptamers from whole-serum SELEX as new diagnostic agents against gastric cancer. RSC Adv. 2019, 9(2):950-957.
- Santarpia G, Carnes E. Therapeutic Applications of Aptamers. International Journal of Molecular Sciences. 2024, 25(12):6742.
- Z. Hu, Z. Jiang, Z. Yang, L. et al. Development of a modularized aptamer targeting the nuclear T-cell suppressor PAC1, Analyst, 2023, 148(11): 2616-2625. [CrossRef]
- H.R. Kim, B.C. Kim, Development of multi-reactive aptamers for Cronobacter spp. using the sequential partitioning method to detect them in powdered infant formula, Anal Chim Acta, 2023, 1249:340935. [CrossRef]
- J. Li, M. Chen, S. Ke, J. Tian, H. Yu, X. Liu, B.Y. Yu, Generation of a high-affinity DNA aptamer for on-site screening of toxic aristolochic acid I in herbal medicines and botanical products, Anal Chim Acta, 2023, 1264:341302. [CrossRef]
- Huang X, Jain PK, El-Sayed IH, El-Sayed MA. Plasmonic photothermal therapy (PPTT) using gold nanoparticles. Lasers Med Sci. 2008, 23(3):217-28. [CrossRef]
- Jiaojiao Sun, Li Zhang, Yingchun Xu, et al. A platform for specific and sensitive detection of target bacteria by selective magnetic enrichment and a broad-spectrum fluorescent probe. Sensors and Actuators B: Chemical. 2021, (349):130762. [CrossRef]
- Kang JH, Super M, Yung CW, et al. An extracorporeal blood-cleansing device for sepsis therapy. Nat Med. 2014, 20(10):1211-6. [CrossRef]
- Q. Zhao, H. Yan, P. Liu,et al. An ultrasensitive and colorimetric sensor for copper and iron based on glutathionefunctionalized gold nanoclusters, Anal. Chim. Acta, 2016, (948):73-79.
- L. Shang, N. Azadfar, F. Stockmar, W. Send, et al. One-pot synthesis of near-infrared fluorescent gold clusters for cellular fluorescence lifetime imaging, Small. 2011, (7) :2614-2620. [CrossRef]
- J. Qiao, X. Meng, Y. Sun, et al. Aptamer-based fluorometric assay for direct identification of methicillin-resistant Staphylococcus aureus from clinical samples, J. Microbiol. Methods, 2018, (153): 92-98. [CrossRef]
- D. Chang, Z. Wang, C.D. Flynn, et al. A high-dimensional microfluidic approach for selection of aptamers with programmable binding affinities, Nat Chem., 2023, 15(6): 773-780. [CrossRef]
- H.B. Wu, C.H. Wang, Y.D. Chung, et al. Highly-specific aptamer targeting SARS-CoV-2 S1 protein screened on an automatic integrated microfluidic system for COVID-19 diagnosis, Anal Chim Acta. 2023, 1274: 341531.
- Fu Z, Huang J, Wei W, Wu Z, Shi X. A multimode biosensor based on prussian blue nanoparticles loaded with gold nanoclusters for the detection of aflatoxin B1. Anal Methods. 2024, 16(19):3088-3098.
- Wang C, Du X, Xie T, et al. Label- and modification-free-based in situ selection of bovine serum albumin specific aptamer. J Sep Sci., 2019, 42(23):3571-3578.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).