Submitted:
10 July 2024
Posted:
11 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Databases for BCE Epitopes
2.2. Dataset Used to Compare Performance
2.3. Computational Methods Evaluated for Epitope Prediction
2.3.1. ISPIPab
2.3.2. EPSVR
2.3.3. Epitope3D
2.3.4. BepiPred
2.3.5. SEPPA
2.3.6. DiscoTope
2.3.7. VORFFIP
2.3.8. Spatom
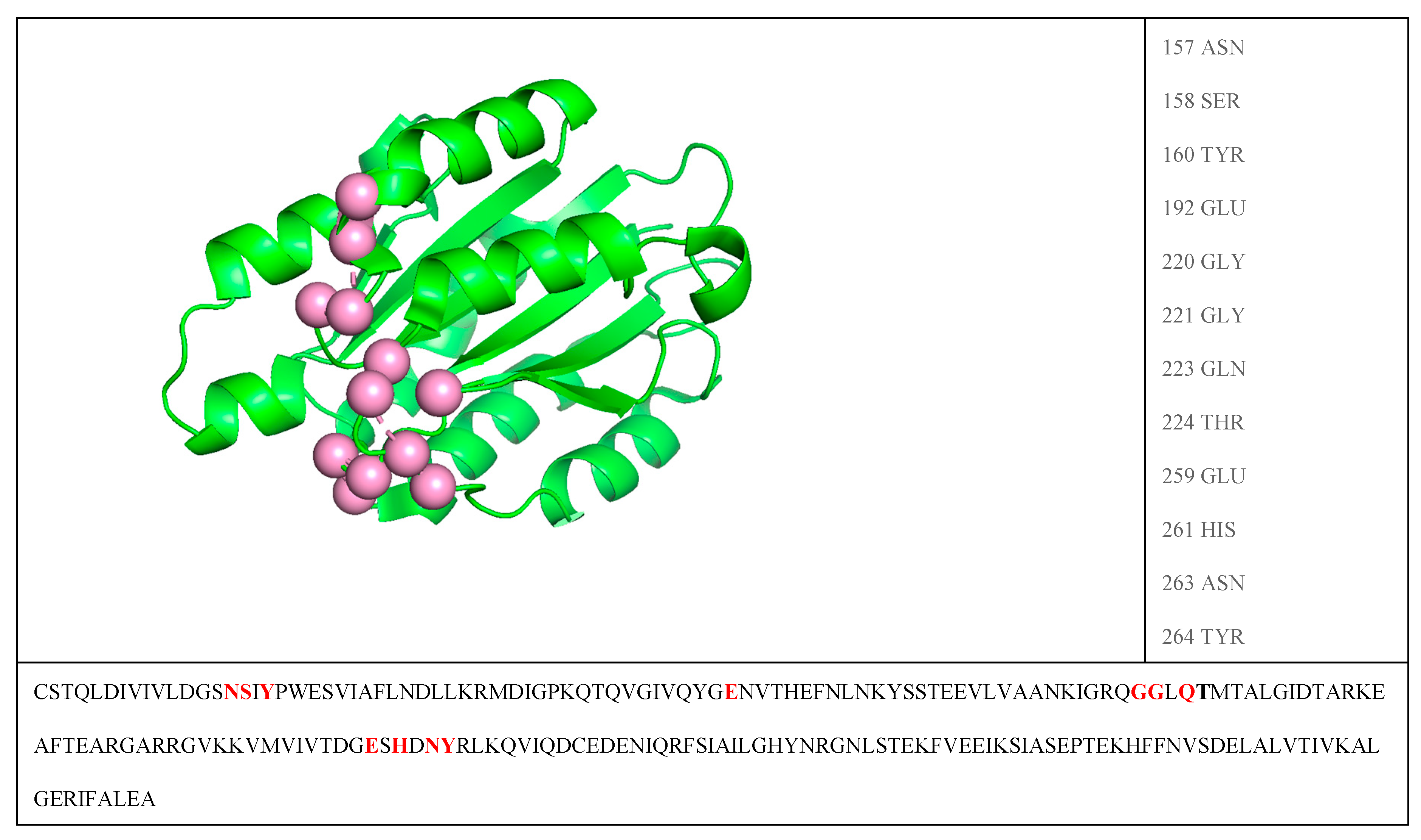
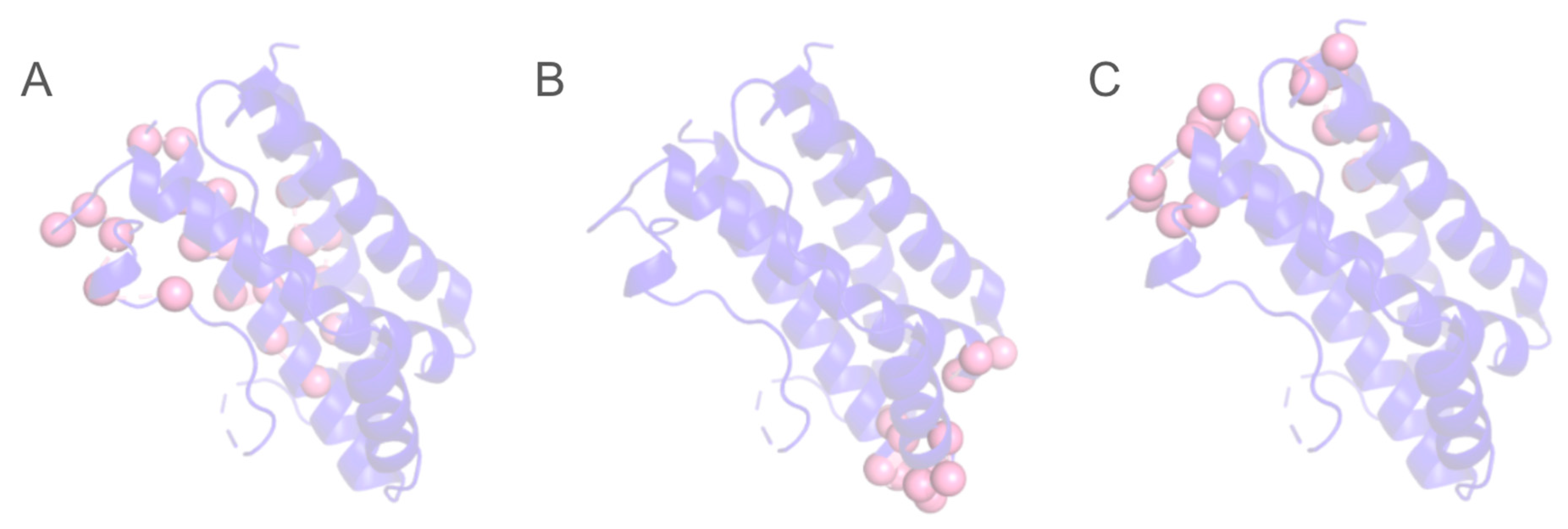
3. Results
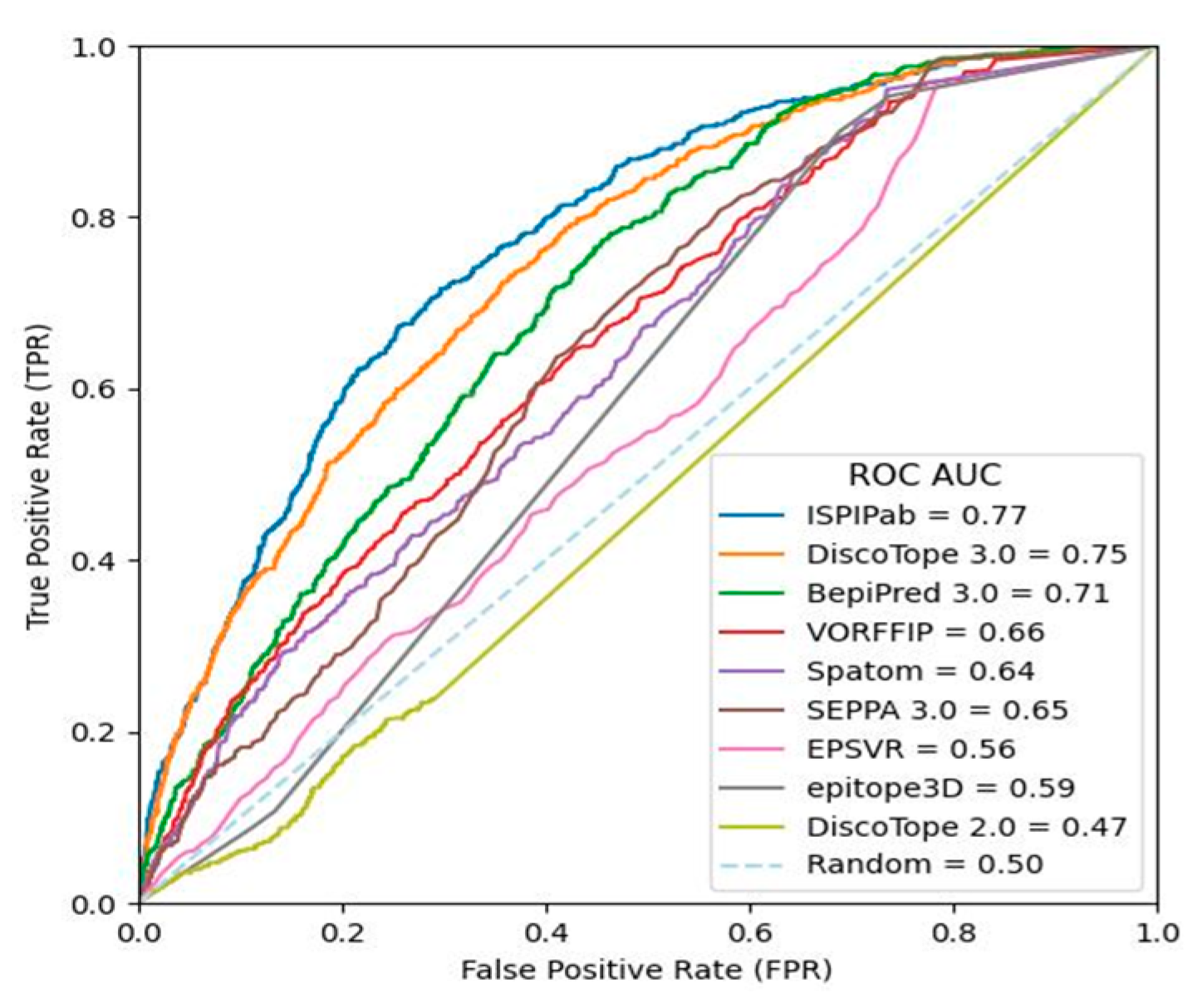
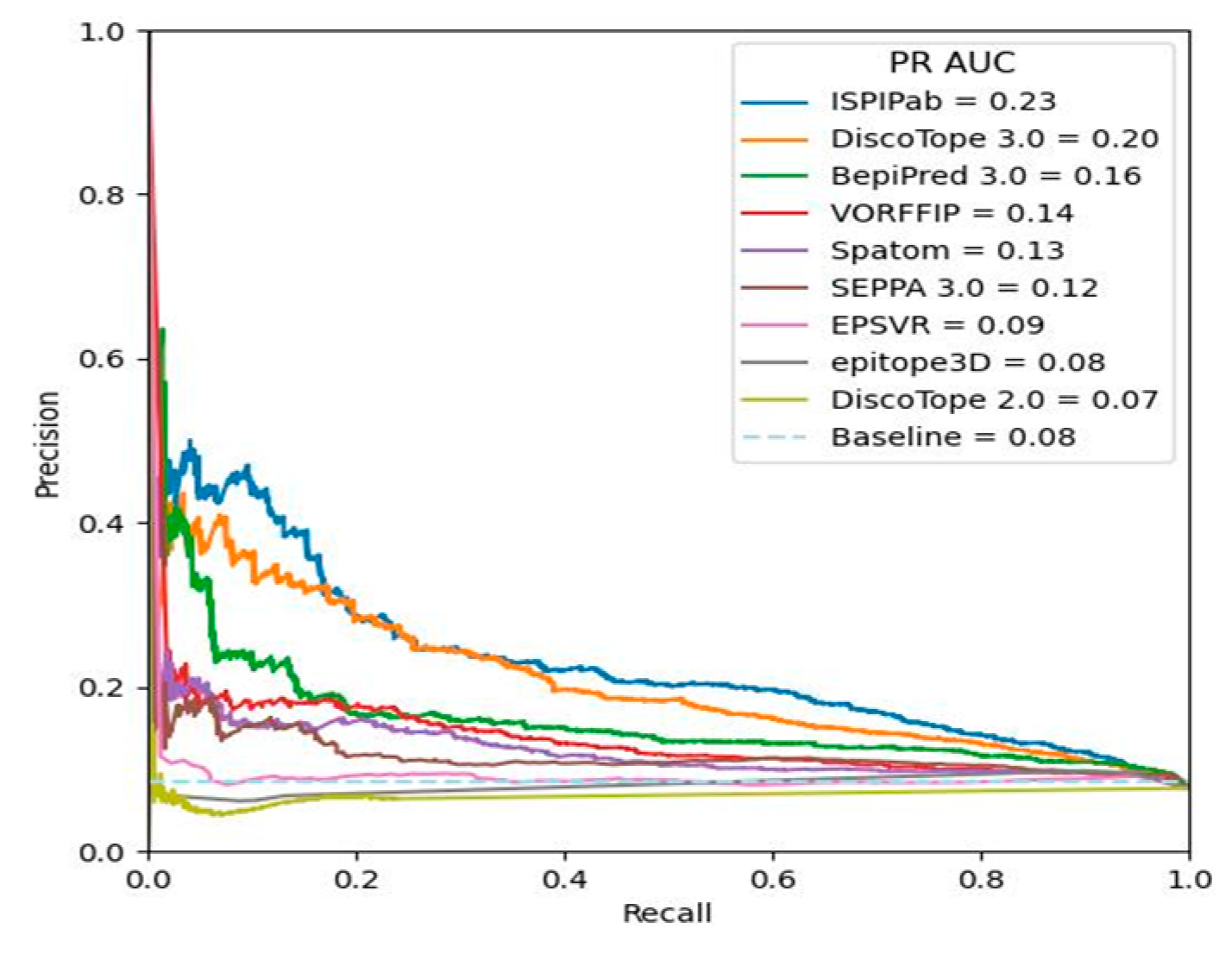
4. Conclusions
4.1. Inaccessible Web Servers
4.2. Limited Availability of Complexed Antibody-Antigen Structures
Supplementary Materials
Author Contributions
Conflicts of Interest
References
- Andersen, P.H.; Nielsen, M.; Lund, O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006, 15, 2558–2567. [Google Scholar] [CrossRef] [PubMed]
- Ferdous, S.; Kelm, S.; Baker, T.S.; Shi, J.; Martin, A.C. B-cell epitopes: Discontinuity and conformational analysis. Mol. Immunol. 2019, 114, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Van Regenmortel, M.H. Immunoinformatics may lead to a reappraisal of the nature of B cell epitopes and of the feasibility of synthetic peptide vaccines. J Mol Recognit 2006, 19, 183–7. [Google Scholar] [CrossRef]
- Dudek, N.L.; Perlmutter, P.; Aguilar, M.I.; Croft, N.P.; Purcell, A.W. Epitope Discovery and Their Use in Peptide Based Vaccines. Curr. Pharm. Des. 2010, 16, 3149–3157. [Google Scholar] [CrossRef] [PubMed]
- Leinikki, P.; et al. Synthetic peptides as diagnostic tools in virology. Adv Virus Res 1993, 42, 149–86. [Google Scholar] [PubMed]
- Potocnakova, L.; Bhide, M.; Pulzova, L.B. An Introduction to B-Cell Epitope Mapping and In Silico Epitope Prediction. J. Immunol. Res. 2016, 2016, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Rux, J.J.; Burnett, R.M. Type-Specific Epitope Locations Revealed by X-Ray Crystallographic Study of Adenovirus Type 5 Hexon. Mol. Ther. 2000, 1, 18–30. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.; Meyer, B. Group Epitope Mapping by Saturation Transfer Difference NMR To Identify Segments of a Ligand in Direct Contact with a Protein Receptor. J. Am. Chem. Soc. 2001, 123, 6108–6117. [Google Scholar] [CrossRef]
- Sun, P.; Ju, H.; Liu, Z.; Ning, Q.; Zhang, J.; Zhao, X.; Huang, Y.; Ma, Z.; Li, Y. Bioinformatics Resources and Tools for Conformational B-Cell Epitope Prediction. Comput. Math. Methods Med. 2013, 2013, 1–11. [Google Scholar] [CrossRef]
- Huang, J.; Ru, B.; Dai, P. Bioinformatics Resources and Tools for Phage Display. Molecules 2011, 16, 694–709. [Google Scholar] [CrossRef]
- Moreau, V.; Granier, C.; Villard, S.; Laune, D.; Molina, F. Discontinuous epitope prediction based on mimotope analysis. Bioinformatics 2006, 22, 1088–1095. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.E. Epitope mapping. Methods Mol Biol 2005, 295, 255–68. [Google Scholar] [PubMed]
- Zheng, D.; Liang, S.; Zhang, C. B-Cell Epitope Predictions Using Computational Methods. Methods Mol Biol 2023, 2552, 239–254. [Google Scholar] [PubMed]
- Parker, J.M.R.; Guo, D.; Hodges, R.S. New hydrophilicity scale derived from high-performance liquid chromatography peptide retention data: correlation of predicted surface residues with antigenicity and x-ray-derived accessible sites. Biochemistry 1986, 25, 5425–5432. [Google Scholar] [CrossRef] [PubMed]
- Hopp, T.P.; Woods, K.R. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. 1981, 78, 3824–3828. [Google Scholar] [CrossRef] [PubMed]
- A Emini, E.; Hughes, J.V.; Perlow, D.S.; Boger, J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 1985, 55, 836–839. [Google Scholar] [CrossRef]
- Pellequer, J.-L.; Westhof, E.; Van Regenmortel, M.H. Correlation between the location of antigenic sites and the prediction of turns in proteins. Immunol. Lett. 1993, 36, 83–99. [Google Scholar] [CrossRef]
- Kolaskar, A.S.; Tongaonkar, P.C. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1990, 276, 172–174. [Google Scholar] [CrossRef]
- Karplus, P.A.; Schulz, G.E. Prediction of chain flexibility in proteins. Sci. Nat. 1985, 72, 212–213. [Google Scholar] [CrossRef]
- Sun, J.; Wu, D.; Xu, T.; Wang, X.; Xu, X.; Tao, L.; Li, Y.X.; Cao, Z.W. SEPPA: a computational server for spatial epitope prediction of protein antigens. Nucleic Acids Res. 2009, 37, W612–W616. [Google Scholar] [CrossRef]
- Ponomarenko, J.; Bui, H.-H.; Li, W.; Fusseder, N.; E Bourne, P.; Sette, A.; Peters, B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 2008, 9, 514–514. [Google Scholar] [CrossRef]
- Sweredoski, M.J.; Baldi, P. PEPITO: improved discontinuous B-cell epitope prediction using multiple distance thresholds and half sphere exposure. Bioinformatics 2008, 24, 1459–1460. [Google Scholar] [CrossRef] [PubMed]
- Kringelum, J.V.; Lundegaard, C.; Lund, O.; Nielsen, M. Reliable B Cell Epitope Predictions: Impacts of Method Development and Improved Benchmarking. PLOS Comput. Biol. 2012, 8, e1002829. [Google Scholar] [CrossRef] [PubMed]
- Bukhari, S.N.H.; et al. Machine Learning Techniques for the Prediction of B-Cell and T-Cell Epitopes as Potential Vaccine Targets with a Specific Focus on SARS-CoV-2 Pathogen: A Review. Pathogens 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Levitt, M. Nature of the protein universe. Proc. Natl. Acad. Sci. 2009, 106, 11079–11084. [Google Scholar] [CrossRef]
- Sela-Culang, I.; Ofran, Y.; Peters, B. Antibody specific epitope prediction—emergence of a new paradigm. Curr. Opin. Virol. 2015, 11, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wong, L.; Li, J. Antibody-Specified B-Cell Epitope Prediction in Line with the Principle of Context-Awareness. IEEE/ACM Trans. Comput. Biol. Bioinform. 2011, 8, 1483–1494. [Google Scholar] [CrossRef] [PubMed]
- Cia, G.; Pucci, F.; Rooman, M. Critical review of conformational B-cell epitope prediction methods. Briefings Bioinform. 2023, 24. [Google Scholar] [CrossRef]
- Briney, B.; Inderbitzin, A.; Joyce, C.; Burton, D.R. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature 2019, 566, 393–397. [Google Scholar] [CrossRef]
- Brown, M.C.; Joaquim, T.R.; Chambers, R.; Onisk, D.V.; Yin, F.; Moriango, J.M.; Xu, Y.; Fancy, D.A.; Crowgey, E.L.; He, Y.; et al. Impact of Immunization Technology and Assay Application on Antibody Performance – A Systematic Comparative Evaluation. PLOS ONE 2011, 6, e28718. [Google Scholar] [CrossRef]
- Kaur, M.; Chug, H.; Singh, H.; Chandra, S.; Mishra, M.; Sharma, M.; Bhatnagar, R. Identification and characterization of immunodominant B-cell epitope of the C-terminus of protective antigen of Bacillus anthracis. Mol. Immunol. 2009, 46, 2107–2115. [Google Scholar] [CrossRef]
- Ito, H.-O.; Nakashima, T.; So, T.; Hirata, M.; Inoue, M. Immunodominance of conformation-dependent B-cell epitopes of protein antigens. Biochem. Biophys. Res. Commun. 2003, 308, 770–776. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, X.; Sun, P.; Gao, B.; Ma, Z. Conformational B-Cell Epitopes Prediction from Sequences Using Cost-Sensitive Ensemble Classifiers and Spatial Clustering. BioMed Res. Int. 2014, 2014, 1–12. [Google Scholar] [CrossRef]
- Ren, J.; Song, J.; Ellis, J.; Li, J. Staged heterogeneity learning to identify conformational B-cell epitopes from antigen sequences. BMC Genom. 2017, 18, 113. [Google Scholar] [CrossRef] [PubMed]
- R.Viswanathan, M.C. R.Viswanathan, M.C., A. Roffe, J.E. Fajardo, A. Fiser, Computational Prediction of Multiple Antigen Epitopes. Bioinformatics, Under Review, 2024.
- Yao, B.; Zheng, D.; Liang, S.; Zhang, C. Conformational B-Cell Epitope Prediction on Antigen Protein Structures: A Review of Current Algorithms and Comparison with Common Binding Site Prediction Methods. PLOS ONE 2013, 8, e62249. [Google Scholar] [CrossRef] [PubMed]
- da Silva, B.M.; et al. epitope3D: a machine learning method for conformational B-cell epitope prediction. Brief Bioinform 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.; Zheng, D.; Zhang, C.; Zacharias, M. Prediction of antigenic epitopes on protein surfaces by consensus scoring. BMC Bioinform. 2009, 10, 302–302. [Google Scholar] [CrossRef] [PubMed]
- Ponomarenko, J.V.; E Bourne, P. Antibody-protein interactions: benchmark datasets and prediction tools evaluation. BMC Struct. Biol. 2007, 7, 64–64. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, W. Databases for B-cell epitopes. Methods Mol Biol. 2014, 1184, 135–48. [Google Scholar] [PubMed]
- Berman, H.M.; et al. The Protein Data Bank. Nucleic Acids Res 2000, 28, 235–42. [Google Scholar] [CrossRef]
- Allcorn, L.C.; Martin, A.C.R. SACS—Self-maintaining database of antibody crystal structure information. Bioinformatics 2002, 18, 175–181. [Google Scholar] [CrossRef]
- Vita, R.; Zarebski, L.; Greenbaum, J.A.; Emami, H.; Hoof, I.; Salimi, N.; Damle, R.; Sette, A.; Peters, B. The Immune Epitope Database 2.0. Nucleic Acids Res. 2009, 38, D854–D862. [Google Scholar] [CrossRef]
- Ponomarenko, J.; Papangelopoulos, N.; Zajonc, D.M.; Peters, B.; Sette, A.; Bourne, P.E. IEDB-3D: structural data within the immune epitope database. Nucleic Acids Res. 2010, 39, D1164–D1170. [Google Scholar] [CrossRef]
- Saha, S.; Bhasin, M.; Raghava, G.P. Bcipep: A database of B-cell epitopes. BMC Genom. 2005, 6, 79–79. [Google Scholar] [CrossRef]
- Huang, J.; Honda, W. CED: a conformational epitope database. BMC Immunol. 2006, 7, 7–7. [Google Scholar] [CrossRef]
- Schlessinger, A. Epitome: database of structure-inferred antigenic epitopes. Nucleic Acids Res. 2006, 34, D777–D780. [Google Scholar] [CrossRef]
- Toseland, C.P.; Clayton, D.J.; McSparron, H.; Hemsley, S.L.; Blythe, M.J.; Paine, K.; A Doytchinova, I.; Guan, P.; Hattotuwagama, C.K.; Flower, D.R. AntiJen: a quantitative immunology database integrating functional, thermodynamic, kinetic, biophysical, and cellular data. Immunome Res. 2005, 1, 4–4. [Google Scholar] [CrossRef]
- Korber B, M.J. , Brander C, Koup R, Haynes B, Walker BD, HIV Molecular Immunology Database, T.B.a.B. Los Alamos National Laboratory, Editor. 1998: Los Alamos, New Mexico.
- Jespersen, M.C.; Mahajan, S.; Peters, B.; Nielsen, M.; Marcatili, P. Antibody Specific B-Cell Epitope Predictions: Leveraging Information From Antibody-Antigen Protein Complexes. Front. Immunol. 2019, 10, 298. [Google Scholar] [CrossRef]
- Sobolev, V.; Sorokine, A.; Prilusky, J.; E Abola, E.; Edelman, M. Automated analysis of interatomic contacts in proteins. Bioinformatics 1999, 15, 327–332. [Google Scholar] [CrossRef]
- Orengo, C.; Michie, A.; Jones, S.; Jones, D.; Swindells, M.; Thornton, J. CATH – a hierarchic classification of protein domain structures. Structure 1997, 5, 1093–1109. [Google Scholar] [CrossRef]
- Porollo, A.; Meller, J. Prediction-based fingerprints of protein-protein interactions. Proteins 2007, 66, 630–45. [Google Scholar] [CrossRef]
- Savojardo, C.; Fariselli, P.; Martelli, P.L.; Casadio, R. ISPRED4: interaction sites PREDiction in protein structures with a refining grammar model. Bioinformatics 2017, 33, 1656–1663. [Google Scholar] [CrossRef]
- Viswanathan, R.; et al. , Protein-protein binding supersites. PLoS Comput Biol 2019, 15, e1006704. [Google Scholar] [CrossRef]
- Walder, M.; Edelstein, E.; Carroll, M.; Lazarev, S.; Fajardo, J.E.; Fiser, A.; Viswanathan, R. Integrated structure-based protein interface prediction. BMC Bioinform. 2022, 23, 1–16. [Google Scholar] [CrossRef]
- Liang, S.; Zheng, D.; Standley, D.M.; Yao, B.; Zacharias, M.; Zhang, C. EPSVR and EPMeta: prediction of antigenic epitopes using support vector regression and multiple server results. BMC Bioinform. 2010, 11, 381–381. [Google Scholar] [CrossRef]
- Rubinstein, N.D.; Mayrose, I.; Martz, E.; Pupko, T. Epitopia: a web-server for predicting B-cell epitopes. BMC Bioinform. 2009, 10, 287–287. [Google Scholar] [CrossRef]
- Zhang, W.; Niu, Y.; Xiong, Y.; Zhao, M.; Yu, R.; Liu, J. Computational Prediction of Conformational B-Cell Epitopes from Antigen Primary Structures by Ensemble Learning. PLOS ONE 2012, 7, e43575. [Google Scholar] [CrossRef]
- Dunbar, J.; Krawczyk, K.; Leem, J.; Marks, C.; Nowak, J.; Regep, C.; Georges, G.; Kelm, S.; Popovic, B.; Deane, C.M. SAbPred: a structure-based antibody prediction server. Nucleic Acids Res. 2016, 44, W474–W478. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Zhou, C. , et al. SEPPA 3.0-enhanced spatial epitope prediction enabling glycoprotein antigens. Nucleic Acids Res 2019, 47, W388–W394. [Google Scholar] [CrossRef]
- Jespersen, M.C.; Peters, B.; Nielsen, M.; Marcatili, P. BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res. 2017, 45, W24–W29. [Google Scholar] [CrossRef]
- Steinegger, M.; Söding, J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat. Biotechnol. 2017, 35, 1026–1028. [Google Scholar] [CrossRef]
- Clifford, J.N.; et al. BepiPred-3.0: Improved B-cell epitope prediction using protein language models. Protein Sci 2022, 31, e4497. [Google Scholar] [CrossRef]
- Qi, T.; Qiu, T.; Zhang, Q.; Tang, K.; Fan, Y.; Qiu, J.; Wu, D.; Zhang, W.; Chen, Y.; Gao, J.; et al. SEPPA 2.0—more refined server to predict spatial epitope considering species of immune host and subcellular localization of protein antigen. Nucleic Acids Res. 2014, 42, W59–W63. [Google Scholar] [CrossRef]
- Ansari, H.R.; Raghava, G.P. Identification of conformational B-cell Epitopes in an antigen from its primary sequence. Immunome Res. 2010, 6, 6–6. [Google Scholar] [CrossRef]
- Høie, M.H.; Gade, F.S.; Johansen, J.M.; Würtzen, C.; Winther, O.; Nielsen, M.; Marcatili, P. DiscoTope-3.0: improved B-cell epitope prediction using inverse folding latent representations. Front. Immunol. 2024, 15, 1322712. [Google Scholar] [CrossRef]
- Tubiana, J.; Schneidman-Duhovny, D.; Wolfson, H.J. ScanNet: an interpretable geometric deep learning model for structure-based protein binding site prediction. Nat. Methods 2022, 19, 730–739. [Google Scholar] [CrossRef]
- Shashkova, T.I.; Umerenkov, D.; Salnikov, M.; Strashnov, P.V.; Konstantinova, A.V.; Lebed, I.; Shcherbinin, D.N.; Asatryan, M.N.; Kardymon, O.L.; Ivanisenko, N.V. SEMA: Antigen B-cell conformational epitope prediction using deep transfer learning. Front. Immunol. 2022, 13, 960985. [Google Scholar] [CrossRef]
- Segura, J.; Jones, P.F.; Fernandez-Fuentes, N. Improving the prediction of protein binding sites by combining heterogeneous data and Voronoi diagrams. BMC Bioinform. 2011, 12, 352–352. [Google Scholar] [CrossRef]
- Wu, H.; Han, J.; Zhang, S.; Xin, G.; Mou, C.; Liu, J. Spatom: a graph neural network for structure-based protein–protein interaction site prediction. Briefings Bioinform. 2023, 24. [Google Scholar] [CrossRef]
- Vreven, T.; Moal, I.H.; Vangone, A.; Pierce, B.G.; Kastritis, P.L.; Torchala, M.; Chaleil, R.; Jiménez-García, B.; Bates, P.A.; Fernandez-Recio, J.; et al. Updates to the Integrated Protein–Protein Interaction Benchmarks: Docking Benchmark Version 5 and Affinity Benchmark Version 2. J. Mol. Biol. 2015, 427, 3031–3041. [Google Scholar] [CrossRef]
- Zeng, M.; Zhang, F.; Wu, F.-X.; Li, Y.; Wang, J.; Li, M. Protein–protein interaction site prediction through combining local and global features with deep neural networks. Bioinformatics 2019, 36, 1114–1120. [Google Scholar] [CrossRef]
- Gainza, P.; Sverrisson, F.; Monti, F.; Rodolà, E.; Boscaini, D.; Bronstein, M.M.; Correia, B.E. Deciphering interaction fingerprints from protein molecular surfaces using geometric deep learning. Nat. Methods 2019, 17, 184–192. [Google Scholar] [CrossRef]
- Yuan, Q.; Chen, J.; Zhao, H.; Zhou, Y.; Yang, Y. Structure-aware protein–protein interaction site prediction using deep graph convolutional network. Bioinformatics 2021, 38, 125–132. [Google Scholar] [CrossRef]
- Zhang, Q.C.; Deng, L.; Fisher, M.; Guan, J.; Honig, B.; Petrey, D. PredUs: a web server for predicting protein interfaces using structural neighbors. Nucleic Acids Res. 2011, 39, W283–W287. [Google Scholar] [CrossRef]
- Hu, Y.-J.; Lin, S.-C.; Lin, Y.-L.; Lin, K.-H.; You, S.-N. A meta-learning approach for B-cell conformational epitope prediction. BMC Bioinform. 2014, 15, 378. [Google Scholar] [CrossRef]
- Kulkarni-Kale, U.; Bhosle, S.; Kolaskar, A.S. CEP: a conformational epitope prediction server. Nucleic Acids Res. 2005, 33, W168–W171. [Google Scholar] [CrossRef]
- Sela-Culang, I.; Ashkenazi, S.; Peters, B.; Ofran, Y. PEASE: predicting B-cell epitopes utilizing antibody sequence. Bioinformatics 2014, 31, 1313–1315. [Google Scholar] [CrossRef]
- Krawczyk, K.; Liu, X.; Baker, T.; Shi, J.; Deane, C.M. Improving B-cell epitope prediction and its application to global antibody-antigen docking. Bioinformatics 2014, 30, 2288–2294. [Google Scholar] [CrossRef]
- Demolombe, V.; de Brevern, A.G.; Felicori, L.; Nguyen, C.; de Avila, R.A.M.; Valera, L.; Jardin-Watelet, B.; Lavigne, G.; Lebreton, A.; Molina, F.; et al. PEPOP 2.0: new approaches to mimic non-continuous epitopes. BMC Bioinform. 2019, 20, 1–14. [Google Scholar] [CrossRef]
- Zeng, X.; Bai, G.; Sun, C.; Ma, B. Recent Progress in Antibody Epitope Prediction. Antibodies 2023, 12, 52. [Google Scholar] [CrossRef] [PubMed]

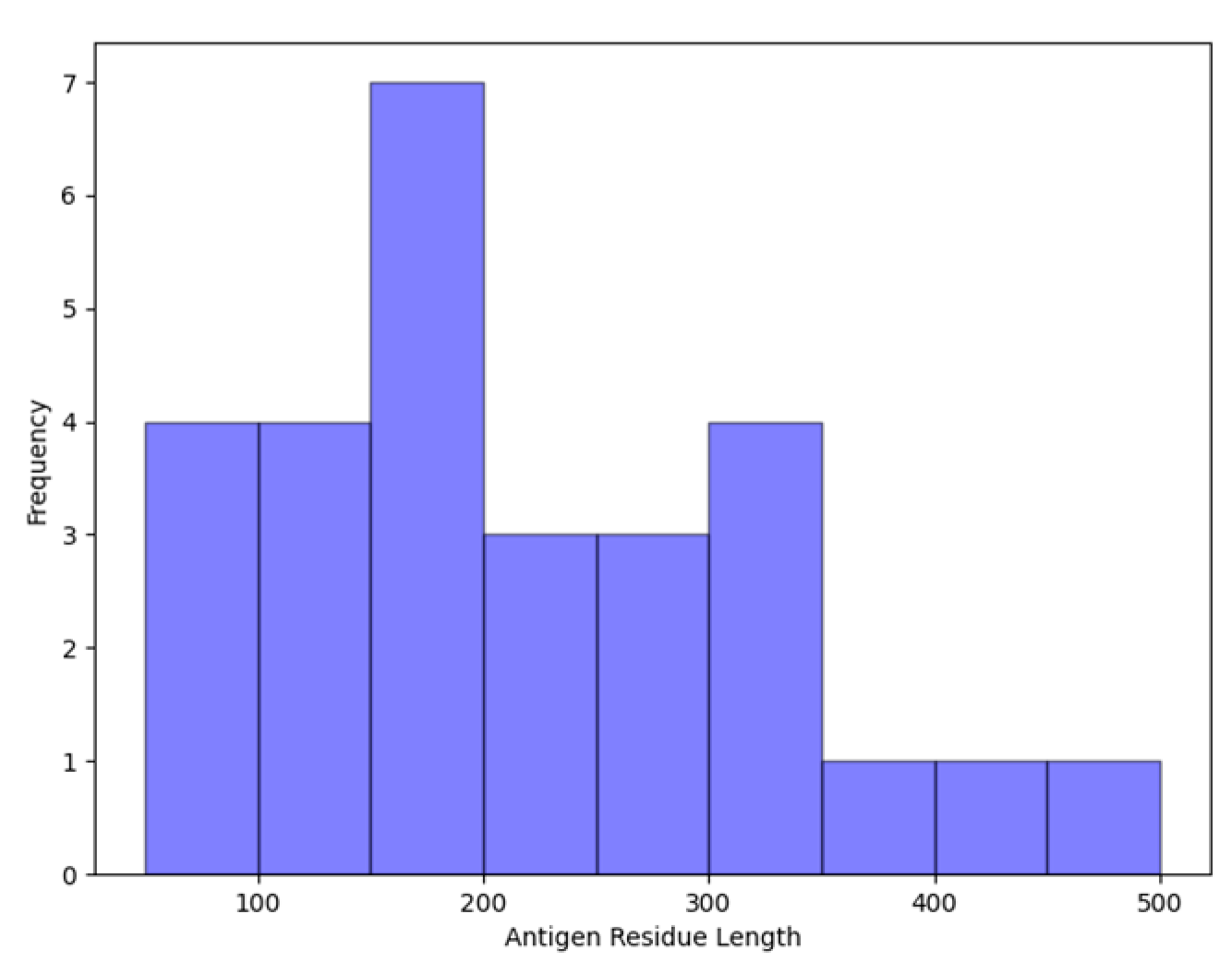
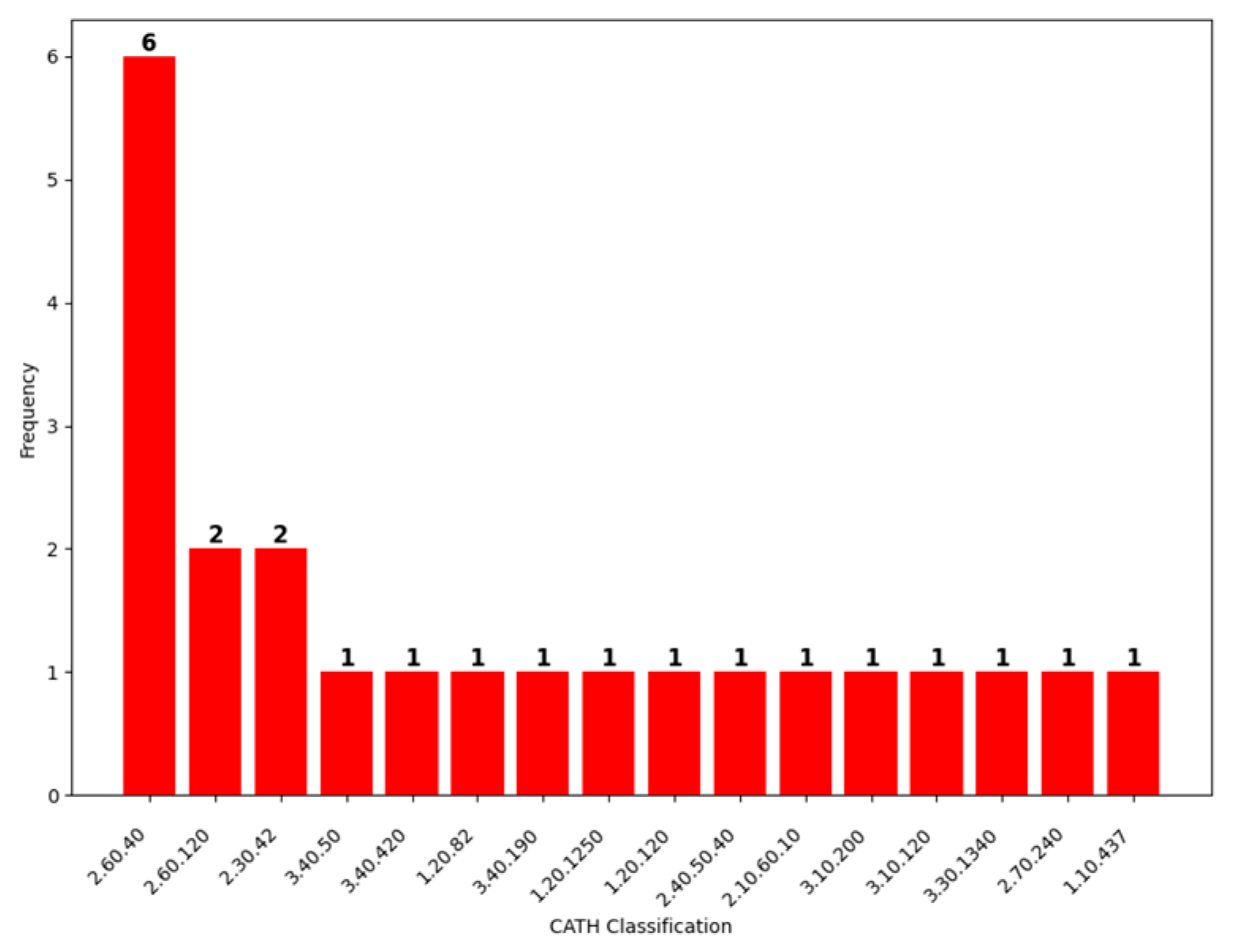
| ISPIPab | VORFFIP | DiscoTope 2.0 | DiscoTope 3.0 | EPSVR | Spatom | SEPPA 3.0 | BepiPred 3.0 | epitope3D | |
|---|---|---|---|---|---|---|---|---|---|
| <F1-score> | 0.312±0.16 | 0.192 ±0.18 | 0.133 ±0.17 | 0.241±0.16 | 0.162±0.21 | 0.161±0.17 | 0.179±0.14 | 0.241±0.16 | 0.109±0.10 |
| <MCC> | 0.230 ±0.17 | 0.090 ±0.18 | 0.017 ±0.19 | 0.143 ±0.17 | 0.054±0.23 | 0.048±0.19 | 0.067±0.18 | 0.145±0.17 | -0.015±0.09 |
| ROC-AUC | 0.77 | 0.66 | 0.47 | 0.75 | 0.56 | 0.64 | 0.65 | 0.71 | 0.59 |
| PR-AUC | 0.23 | 0.14 | 0.07 | 0.20 | 0.09 | 0.13 | 0.12 | 0.16 | 0.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





