1. Introduction
The progressive digitalization of society impacts all conceivable domains, ranging from professional to mundane aspects, encompassing both the advantageous and detrimental. Notably, within these domains lies the field of healthcare in its diverse and extensive dimensions. Various medical facilities, including clinics, hospitals, offices, and research centers, are increasingly embracing the digitalization of medical information. This encompasses concepts such as electronic health records, bureaucratic records, and diagnostic imaging tests.
In recent years, especially following the globally imposed state of exceptionality due to the SARS-CoV-2 (commonly known as COVID-19) pandemic, a trend has emerged related to remote consultations (telemedicine). What was initially promoted to prevent contagion has proven beneficial in reducing time and travel for routine information inquiries or dosage consultations. It has also been instrumental in remote rural populations and contributed to the reduction of paper consumption. However, its applicability is limited when a physical examination is necessary.
This evolving landscape results in increasingly voluminous datasets generated at a higher velocity. This necessitates new approaches to facilitate sociological and clinical studies while supporting generative and predictive solutions. Leveraging Artificial Intelligence, these solutions aim to optimize clinical research and enhance diagnostic precision and early detection to the greatest extent possible.
Therefore, there is a need to explore the feasibility of generative solutions in extracting data from medical reports, categorized by specific criteria. The objective is to develop systems that enable healthcare professionals, through the use of generative chatbot prompts, to efficiently access key patient information in the shortest possible time. This approach eliminates the need for excessive manual searching for specific categories of information. The subsequent discussion delves into these considerations.
Large Language Models (LLMs) have the potential to revolutionize healthcare by improving diagnostic accuracy, supporting clinical decision-making, and bridging the gap between patients and healthcare providers [
1,
2]. LLMs can provide accurate basic knowledge, analyze specific situations, and offer patient-friendly information, making them valuable for patient education and consultation [
3]. They can also be integrated into medical practice responsibly and effectively, addressing the needs of various medical disciplines and diverse patient populations [
4].
LLMs are pre-trained models, meaning they possess the capacity to comprehend and generate text without the need for extensive additional training. This pre-training involves exposing the models to vast amounts of diverse textual data, allowing them to grasp language patterns and contextual nuances. LLMs, employing a transformer architecture, excel in a multitude of domains, demonstrating remarkable capabilities in natural language processing tasks and text comprehension. The essence of pre-training lies in enabling these models to predict the next word in a given text, a skill that underpins their ability to perform various other tasks, showcasing their inherent intelligence [
5]. Transformers are based on multi-layer neural networks which are trained with large datasets. Meanwhile, traditional LLMs, which were developed before the launch of ChatGPT, used programmed statistical techniques to predict the next word in a sentence [
6].
The utilization of recurrent neural networks (RNNs) is instrumental in eliminating the necessity for explicit word history modeling, allowing the incorporation of temporal dimensions while considering preceding words. This facilitates the retention of pertinent information from previous time steps. Notably, it is crucial to understand that RNNs operate by enhancing the feature vector encoding for each word or feature. The input vector is constructed using the word vector, with the output either copied or delayed from hidden neurons in the preceding time step. The activation function commonly employed is the softmax function. Additionally, it is essential to highlight that backpropagation through time is a key aspect of RNNs, and the output layer produces a probability distribution of a word based on the preceding word and contextual features. For clarity, it should be noted that RNNs, or recurrent neural networks, refer to a type of neural network architecture specifically designed for sequential data processing.
To enhance word prediction accuracy, we explored the utilization of socio-linguistic features, such as sequences of discourse-related tags that provide syntactic information. We also considered the delineation of conversation topics through clustering techniques, recognizing that word choices within a specific conversation are influenced by its topic, while incorporating log-scaled frequency considerations. Furthermore, we factored in the socio-situational context, which encompasses variables such as the conversational context (e.g., interview, spontaneous discussion, phone call, or academic seminar), the relationships between participants, and their quantity. These considerations collectively contribute to a more precise word prediction model [
7].
To equip Language Model Machines (LLMs) for tackling intricate challenges and transcending the constraints of generalized composition inherent in thought chain prompts, which are often based on limited examples, a novel “from more to less” prompting approach has been introduced. This innovative methodology aims to amalgamate the concept of natural language rationality with self-consistent decoding. The proposed approach unfolds in sequential phases, commencing with the decomposition and resolution of subproblems. This involves furnishing consistent examples showcasing the resolution of subproblems and compiling lists of previously answered subquestions along with their solutions. It is noteworthy to emphasize that consistent decoding, in this context, refers to the coherent and logical interpretation of information during the model’s generation process. This “from more to less” approach lays the foundation for leveraging bidirectional interactions, thereby amplifying the reasoning capabilities of LLMs [
8].
Given the distinct characteristics of LLMs and specific operational considerations, our primary focus lies in addressing the challenges associated with healthcare digitalization. Our research places a significant emphasis on information extraction, with a notable shift towards document analysis as opposed to the conventional Extract, Transform, Load (ETL) or Extract, Load, Transform (ELT) processes commonly applied to structured datasets. This approach, broadly categorized, aims to unveil structured information from unstructured or semi-structured texts, representing a more expressive method that enhances communication.
It is essential to note that ETL typically refers to the process of extracting, transforming, and loading data into a structured format, while ELT reverses the sequence by loading data first and then transforming it. Our work underscores the significance of document analysis as a specialized area within the broader field of empirical Natural Language Processing (NLP), involving the extraction and encoding of information in the context of healthcare digitalization [
9].
To be more precise, in our experiment, we will collect various medical reports in PDF format. Using prompts, we will attempt to extract diverse clinical information, such as age, weight, family medical history, date of birth, or potential allergies. This information will be used to enhance our local data model, thus optimizing diagnostic monitoring by reducing the need for manual inquiries and the time spent on non-automated searches. The experiment will be conducted through the implementation of the Langchain framework in Python, with concurrent utilization of models from OpenAI (ChatGPT 3.5), Meta (LLaMa 2), and Vicuna.
The advancements in LLMs have significantly broadened their applications across various domains, with particular emphasis on their impactful role in healthcare. As we delve into the intricacies of LLMs, the imperative shift from foundational understanding to practical implementation becomes apparent. In the preceding chapter, an exploration of the fundamental architecture of LLMs underscores their training methods and transformative capacities across diverse disciplines.
Now, directing our attention to the practical implications of LLMs in healthcare digitalization, the pivotal integration of these models into medical practice commands attention. The ensuing discussion delves into the strategic application of LLMs to address intricate healthcare challenges, emphasizing their pivotal role in information extraction and meticulous document analysis. This discussion lays the groundwork for our empirical endeavors, where LLMs are utilized to extract critical clinical information from medical reports, contributing substantively to the optimization of diagnostic monitoring while streamlining manual efforts.
Prior to embarking on this empirical journey, due acknowledgment is accorded to related work that underscores the integration of language models in document analysis, offering an insightful understanding of the applied evaluation metrics and techniques. Subsequent chapters will progressively build upon these foundational aspects, casting light on network syntactic analysis, probabilistic experiments, and the incorporation of knowledge graph structures. This cohesive narrative aims to seamlessly bridge the theoretical underpinnings of LLMs with their pragmatic implementation within the dynamic landscape of healthcare digitalization, presenting valuable insights for scholarly discourse.
Multiple experiments have been conducted using LLMs to analyze documents, employing metrics that evaluate fluency (whether the generated text is coherent), correctness (if the prompt response is appropriate), and the quality of citations (if the cited passages are suitable). These experiments involve combining automated metrics with human evaluation, which is based on qualitative metrics that can cover aspects such as utility and the coherence of citations, providing scores on a scale from 1 to 5. The metrics have been tailored to each dataset, including the adoption of customized correctness metrics specific to the corresponding dataset [
10].
Network syntactic analysis, a method utilized for modeling knowledge about document components by delineating their geometric properties, lexical entities, and relationships, has emerged as a prominent technique. An example of its application is seen in the utilization of the FRESCO semantic network language. In this experiment, FRESCO was employed to analyze business letters, facilitating the extraction of structural elements such as the sender, recipient, date, and main body. This approach enables a comprehensive specification of knowledge concerning these structural components, ensuring both accuracy and completeness in the modeling process.
The accuracy of structural entity recognition is high when the visual organization of document elements (position, size, images, text formatting, etc.) can be used to identify the sender (as in the case of OneLineSender or BlockSender structures). However, this accuracy may decrease when the information is not concentrated in a specific location and is instead scattered across different sections of the letter. This situation can lead to document rejection, but the use of network analysis, combined with layer-specific knowledge, can optimize information extraction and automatic response generation [
11].
Given that current transformer-based neural networks employ probabilistic techniques, it is interesting to note that decades ago, probabilistic experiments were conducted based on research into the use of logistic regression for obtaining ad hoc data, where a regression equation is fitted to learn data. The variables used in the equation are often statistical averages. The use of linear regression aims to find simple yet powerful paths, in the probabilistic sense, by combining search cues. The effectiveness of information retrieval has been enhanced through manual reformulations of topics [
12].
The integration of knowledge graph structures has emerged as a pivotal resource in the realm of text document analysis. Through the application of advanced natural language processing techniques, this approach facilitates the extraction of critical entities, such as geographical locations, temporal references, and personal names, followed by the utilization of specialized tools to address ambiguities and spelling variations. This approach, known as ‘occurrence data’, emphasizes the preservation of terms, phrases, and entities throughout the analytical process.
By amalgamating natural language processing and knowledge graph structures, this method enhances the comprehension of textual content, emphasizing contextual relations for more comprehensive information retrieval. Leveraging knowledge graph structures in text document analysis promises to provide deeper insights and a nuanced understanding of data, transcending the limitations of traditional keyword-based search and contributing to scientific exploration and data analysis.
Once the various entities have been extracted, the knowledge graph construction process commences. Each extracted entity represents a labeled node, and for each source of the various entities, a corresponding node is added. In the graph, a weight-1 edge is introduced between entities that co-occur within a document, signifying their simultaneous presence. However, when adding new nodes to the graph, care must be taken to ensure that no pre-existing node with the label of the entity already exists, as in such cases, the existing node is repurposed.
In order to account for the diverse nature of entities, each node is equipped with a set of nature properties, allowing us to record the type of entities (e.g., distinguishing between individuals and geolocations). If a vertex to be inserted already exists, as is the case with locations and dates, the vertex’s weight is increased incrementally. The resulting graph is both weighted and undirected, offering a wide range of query capabilities that can be tailored as needed. The structure of the links between nodes also provides flexibility in terms of the types of data that can be retrieved [
13].
In the medical field, a substantial portion of data remains unstructured today, encompassing concepts such as emails, data streams, voice and video recordings, as well as digital documents. Structured data’s growth tends to be more gradual. Automated text mining encompasses a range of methods designed to facilitate access to relevant information. Recent attention has been focused on natural language processing, as techniques from other domains, such as information retrieval and extraction (automated extraction of structured data from unstructured sources), are adapted and integrated into this context [
14].
Data extraction often leads to the discovery of tabular data, which is frequently embedded within text, particularly in medical diagnoses. Traditional machine learning models struggle to efficiently process information in this format, while large language models (LLMs) also face limitations in this regard. In response, methodologies like TEMED-LLM are proposed, featuring three crucial components: reasoning-extraction, result validation and correction, and training (preferably, of an interpretable model based on the extracted tabular data) [
15].
With the aforementioned goal in mind, efforts have been directed towards tasks such as SCHEMA-TO-JSON, a task focused on the extraction of structured records from tables and other semi-structured data sources, such as a web page. This task takes as input a table that can optionally be supplemented with context from the same document, along with an extraction schema that specifies the attributes to be extracted for different records that may contain varying numbers of attributes. As a result, it generates a sequence of JSON objects represented by an array of key-value pairs, each paired with a record type, condensing the information into a more accessible format.
An approach for table extraction called InstructTE is applied, which demonstrates competitive performance in both accuracy and precision, with an emphasis on balancing the two. It only requires a human-constructed extraction schema, incorporating an error recovery strategy. The schema approach ensures that the extraction process adheres to a predefined structure, enhancing the accuracy and consistency of the extracted information. Primarily, human-driven prompting is used to guide language models in the extraction of data from complex tables [
16].
Additionally, other data extraction experiments have been conducted, focusing on radiological results that may not necessarily be textual reports. In the case of textual data, a state-of-the-art question-answering system was employed, contrasting with radiologist annotations [
17]. On the other hand, for non-textual data, a manual extraction of various tomographies was performed, where the reports were randomly partitioned into training and validation sets based on a natural language rule to extract report attributes (resulting in high precision in identifying occlusion, distal, or basilar, of several large blood vessels) [
18].
2. Materials and Methods
2.1. Models
We will harness the power of the Langchain framework in Python to streamline the development of applications centered around the implementation of Large Language Models (LLMs). This strategic approach provides us with the agility to seamlessly integrate an existing data model and customize it to craft more personalized applications. A pivotal element of our methodology involves the utilization of FAISS vector stores, where each vector store is designed to accommodate chunks of 1000 characters. These vector stores play a crucial role in our workflow, serving as repositories for encoded representations of textual data.
In the context of our workflow, a vector store refers to a storage mechanism that holds encoded vectors derived from textual information. Specifically, each vector store is configured to manage chunks of 1000 characters, ensuring a structured organization of the data. As we embark on similarity searches within our application, we will adhere to a specific search parameter for kwargs (keyword elements), denoted as the maximum number of documents involved in the search. This parameter, referred to with the nomenclature ‘n’, governs the retrieval of relevant documents and contributes to the precision and efficiency of our information retrieval process.
Our research will encompass tests with three distinct language models: GPT 3.5 (turbo mode), LLaMA 2, and Vicuna 7B (in the subsequent paragraphs there will be explanations about all those models). Each model will be evaluated separately by loading a story or medical report into the data model, progressively refining our personalized model. Questions designed for medical extraction, based on template prompts, will be posed, and the corresponding answers will be incorporated into the new model, which will be saved locally. Notably, it is crucial to recognize that each model may differ in features such as proprietary or open-source nature, parameters, and underlying architecture.
The GPT-3.5-turbo model, an advanced creation from OpenAI, is renowned for its cutting-edge natural language processing capabilities. Building upon the foundation of its predecessor, GPT-3, it boasts an extensive knowledge base and excels in comprehending and generating human-like text. Notably, its turbo mode is designed for rapid response times, catering to real-time applications. GPT-3.5-turbo is characterized by a vast number of parameters, showcasing its complexity and capacity to handle diverse language tasks. With a token limit that allows processing significant amounts of text in a single input, this model proves highly adept for our research in healthcare digitalization. Its robust parameters and token capabilities contribute to its effectiveness in comprehensively analyzing and generating insights from medical reports, aligning seamlessly with our information extraction goals. Its context window has 16385 tokens, and its training data is up to Sep 2021.
LLaMA 2, developed and publicly released by Meta, comprises a collection of pretrained and fine-tuned generative text models with varying scales, ranging from 7 billion to 70 billion parameters. Specifically, we are utilizing the 7B fine-tuned model within the Q4 quant mode, which emphasizes a range of parameters optimized for specific use cases. This model, designated as Llama-2-Chat, is tailored for dialogue scenarios and has been converted to the Hugging Face Transformers format for ease of integration. Notably, the range of parameters selected aligns with our research focus, striking a balance between computational efficiency and model performance. It is essential to acknowledge that the use of the Llama 2 model is governed by the Meta license, and access to the model weights and tokenizer requires acceptance of the license terms on the official website before requesting access. The Llama-2-Chat models, through fine-tuning, have demonstrated superior performance in dialogue-oriented benchmarks, outperforming several open-source chat models, and competing favorably with well-known closed-source models such as ChatGPT and PaLM in human evaluations for helpfulness and safety.
Vicuna 7B, an open-source chatbot refined through fine-tuning LLaMA on user-shared conversations from ShareGPT, is strategically augmented to enhance its Context of Thinking (CoT) capabilities using Single-Forward-Transfer (SFT). This enhancement is specifically tailored to leverage the distinctive attributes of the 7-billion parameter range within the Q5 quantization mode. In the Q5 quantization mode, the model parameters are meticulously tuned to achieve a harmonious balance between computational efficiency and model performance. Vicuna 7B, intricately tuned for nuanced language tasks, gains substantial benefits from the augmented CoT facilitated by SFT. This approach not only optimizes the model’s proficiency in comprehending and generating context-rich text but also seamlessly integrates the unique capabilities of the Q5 quantization mode. Our strategic use of Vicuna 7B, coupled with fine-tuning and quantization enhancements, underscores our dedication to maximizing efficiency and effectiveness in the domains of healthcare digitalization and information extraction.
2.2. Data
From an ethical and natural law perspective, privacy can be interpreted as intrinsic to the right to property, which is generally associated with bank savings as well as more tangible assets, including real estate. Individuals may be concerned about or wish to determine the extent to which different external entities should have knowledge of their personal data, including health, economic, social, and nutritional information.
However, in the realms of scientific and technological research, a significant dilemma arises. While one can understand and scrupulously defend the individual’s right to privacy, the trial-and-error phases inherent in advancements in fields such as computer science, medicine, and pharmacology necessitate experiments, in the broadest sense, with real samples.
This situation underscores the importance of anonymization and pseudonymization. However, there is a lack of understanding about these strategies and the awareness of having, to a certain extent, what is known as open data, which can enhance research and technological innovation, in addition to serving commendable educational purposes. Not all hospitals have open records suitable for experimentation.
Therefore, the documents under examination consist of clinical histories with diverse origins and lack a standardized format. Originating from various hospitals and medical conventions with heterogeneous corporate structures, these documents present a unique challenge due to their non-conformity to a singular medical branch (e.g., cardiology, gynecology, psychiatry, etc.). This diversity results in a broad clinical and pharmacological spectrum, encompassing various clinical conditions and types of compounds found in medications.
To extract data from these documents, we employ a zero-shot prompting data extraction technique [
19], designed to enhance performance in tasks involving reasoning with linguistically untrained or previously unexposed information within a specific task or domain, utilizing Pydantic. Building on this approach, we create a predefined prompt based on a template querying specific categories: “nombre” (name and surname), “edad” (age), “diagnostico” (diagnosis), “medicamentos” (drugs), and “pruebas” (medical tests).
The output of each query instruction takes the form of a JSON schema containing the requested information. All resulting arrays are stored in a ‘vectorstore’ with model embeddings, progressively building a local model saved within the system using the pickle library, ensuring files are stored as local FAISS copies. This process allows the local model to be prepared for subsequent use after extracting data from each document.
It is important to note that our dataset comprises approximately 14 PDF documents written in the Spanish language. These documents, as mentioned earlier, lack any standardization and may include reports such as hospital discharge summaries or diagnostic components. The considered models have been OPENAI_3.5_TURBO (OpenAI 3.5), llama-2-7b-chat.Q4_K_M (LLaMA 2) and vicuna-7b-cot.Q5_K_M (Vicuna).
2.3. Retrieval Augmented Generation (RAG)
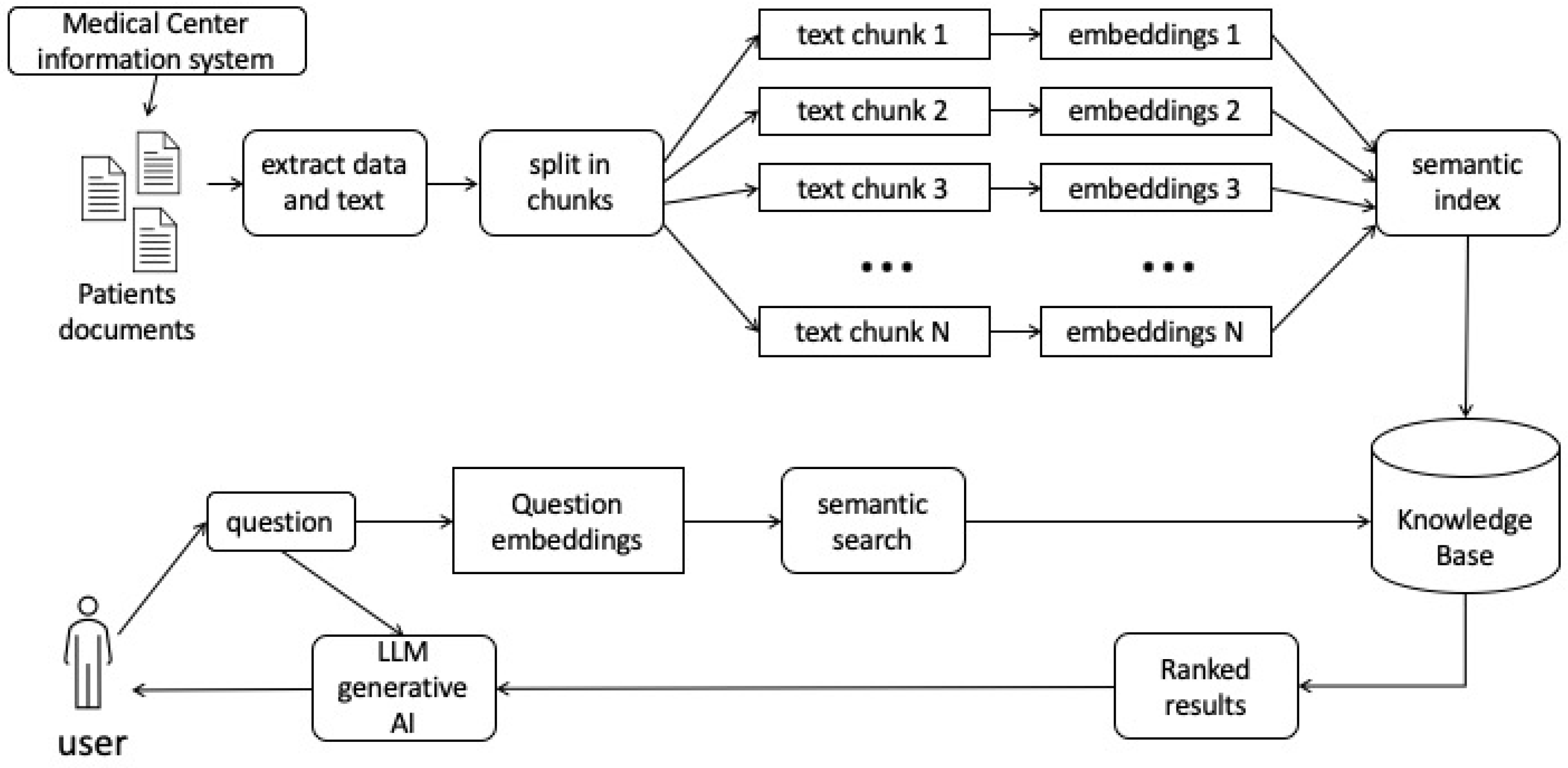
Retrieval Augmented Generation (RAG) is an approach that enhances language models by integrating information retrieval during the generation process, aiming to address issues like factual inaccuracies and hallucinations observed in the output of large language models [
20]. Traditional models, such as Naive RAG, follow a conventional methodology involving indexing, retrieval, and generation. In this paradigm, original data undergoes cleansing, conversion, and segmentation into manageable chunks represented as vectors through an embedding model. While Naive RAG has a structured approach, it grapples with challenges in retrieval precision, recall, dealing with outdated information, and issues in generation and augmentation. Addressing these challenges is crucial for implementing a robust retrieval-augmented generation framework [
21].
The Advanced RAG paradigm introduces optimization strategies in the pre-retrieval process, focusing on enhancing data indexing, fine-tuning embedding models, and post-retrieval processes such as re-ranking and prompt compression. Furthermore, the Modular RAG paradigm provides versatility and flexibility by integrating various methods to enhance functional modules, making it increasingly prevalent in the domain. Advanced RAG is considered a specialized form of Modular RAG, showcasing a relationship of inheritance and development among the three paradigms [
22]. A sort of schematic graphic abstraction is shown in
Figure 1.
Innovative approaches within the retrieval-augmented generation landscape leverage predictive modeling to iteratively anticipate and retrieve relevant documents during the generation process, showcasing the efficacy of integrating information retrieval with language models. In our upcoming experiment, we intend to delve into the RAG technique, with a specific focus on its fundamental aspects. While our exploration won’t navigate the intricacies of Advanced RAG, which optimizes the pre-retrieval process through strategies like enhancing data granularity and fine-tuning embedding models, our goal is to illuminate the basic yet impactful facets of RAG. Below is a specific flowchart outlining the iterative phases of our experiment (
Figure 2).
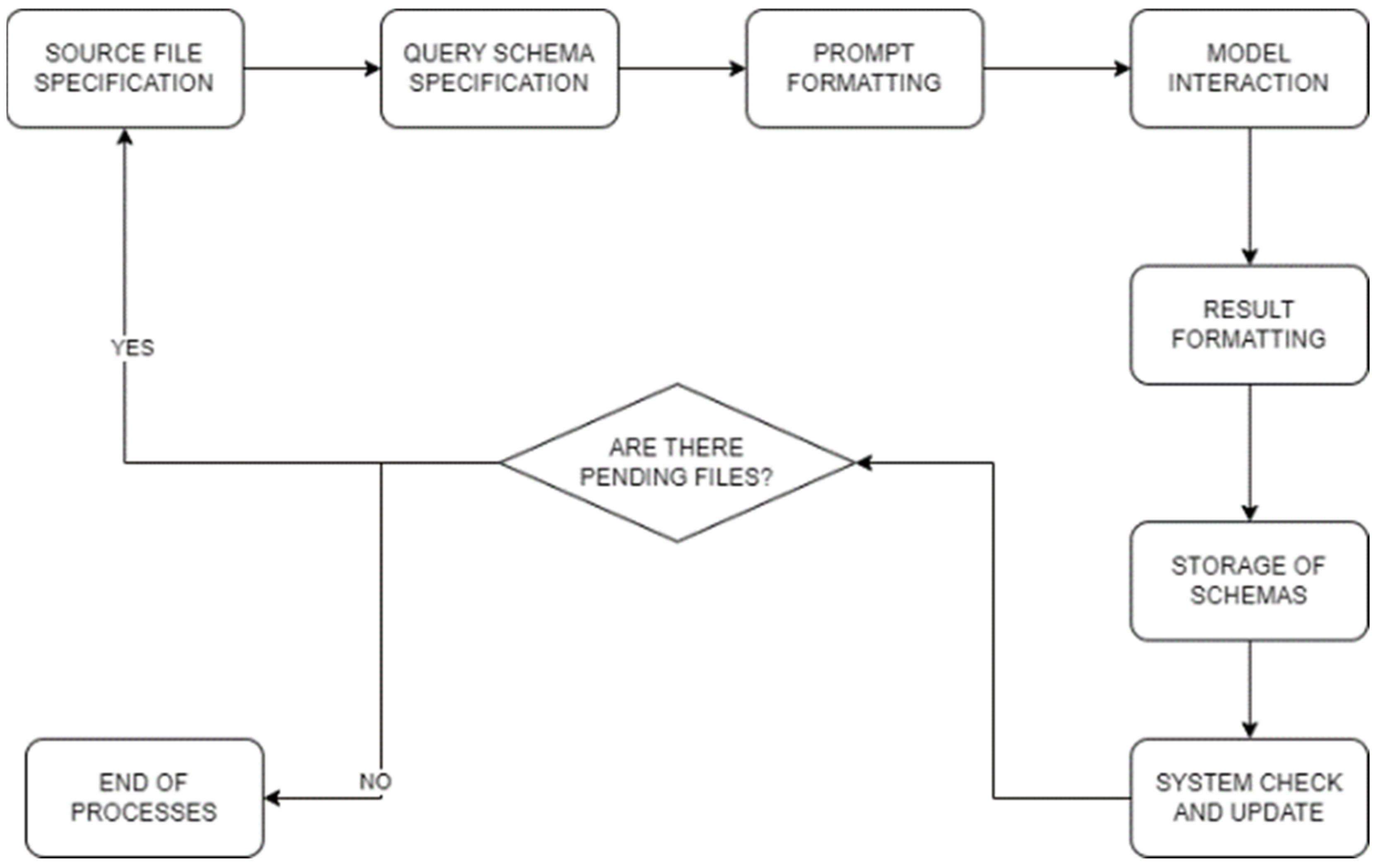
Upon mounting the file system that serves as the source for our data, the following steps are undertaken to structure and process the information systematically:
Source File (PDF) Specification: Leveraging the PyPDFLoader class from the pypdf package within the Python programming language, specifically executed in a Jupyter notebook environment, we systematically manage the content of PDF files. This includes the extraction of pertinent information from a predefined directory housing a curated selection of clinical documents across various categories. The subsequent utilization of PyPDFLoader facilitates the streamlined processing of content from each document with the corresponding Language Model (LLM).
Query Schema Creation: The formulation of a structured query is conducted to generate a JSON-style schema, meticulously aligning with key patient attributes such as name, age, diagnostic tests, diagnosis, and medication. Ensuring adherence to this specified schema is imperative. Python’s object-oriented programming paradigm, implemented within a Jupyter notebook, is instrumental in defining the class that underpins this schema, thereby ensuring seamless data extraction and subsequent processing.
Prompt Formatting: Prior to submitting the prompt for processing by the LLM, we meticulously format it to align precisely with the schema defined by Pydantic. This formatting process, executed in Python and complemented by the Pydantic library for data validation within the Jupyter notebook framework, ensures that the response from the LLM strictly adheres to the predefined schema.
Model Interaction: The transmission of the formatted prompt to the Language Model (LLM) is facilitated through serialization. This serialization process is executed using Python, either through the OpenAI API or the LlamaCpp library, contingent upon the specific case. The LLM, embedded within a Jupyter notebook, retrieves embeddings and pertinent data, applying predefined processing rules from various data models. The culmination of this interaction is directed towards a .bin file, serving as a repository for valuable embeddings.
Result Formatting: The outcomes of the prompt, critically, are not processed as plain text but undergo transformation into JSON format. This strategic conversion enhances clarity and eases interpretation, ensuring a structured representation of the results. The Python-based implementation, within the Jupyter notebook environment, facilitates subsequent processing and detailed analysis.
Storage of Schemas: Post the JSON formatting, the structured results are systematically stored in an array. This array, acting as a repository, captures diverse schemas generated from different prompts. Within the Python-centric Jupyter notebook, this organized storage lays the foundation for subsequent processing stages and serves as a valuable resource for ongoing system development and meticulous data analysis.
System Check and Update: A comprehensive system check involves the verification of the presence of the local FAISS database file within the file system. This specific file system subset, resembling a decomposition into various files, is examined. If the database file is found, Python functionalities within the Jupyter notebook facilitate the seamless integration of new texts derived from the aforementioned JSON outcomes. The utilization of JSON format, even during system checks, enhances comprehension during future processing stages. In the event the database file is not found, the initiation of a new FAISS database involves the incorporation of embeddings obtained from the currently utilized Language Model (LLM) within the Python environment of the Jupyter notebook. Once this step is completed, the initial texts can be added to the database, establishing a foundational dataset for further utilization within the system.
The execution of this methodology has been meticulously carried out on high-performance hardware endowed with cutting-edge processors, ample memory, expansive storage, and any specialized hardware designed for the acceleration of Large Language Model (LLM) computations. The computational environment, seamlessly integrated with the efficiency of a Jupyter notebook, constitutes a critical component of our execution framework.
Our comprehensive workflow unfolds within the structured formalism of Python programming, harnessing the versatile capabilities of a Jupyter notebook. This robust combination not only facilitates the extraction and structuring of data from medical reports but also ensures dynamic and efficient handling throughout the entirety of the process. The utilization of advanced hardware and the integration of Python and Jupyter provide a solid foundation for the intricate tasks involved in our research, guaranteeing precision, speed, and adaptability at each stage of the workflow.
2.4. Evaluation
To define and quantify the metrics, a rubric based on the confusion matrix was established, applied across the five categories of the JSON schematic framework (name, age, diagnosis, tests, and medications). The confusion matrix (
Table 1) includes the following elements:
True Positives (TP): Instances where data is correctly extracted from the report.
False Positives (FP): Instances of erroneous data associations.
True Negatives (TN): Accurate identifications of the absence of data.
False Negatives (FN): Denials of data that actually exist but have not been correctly interpreted.
Table 1.
Confusion Matrix.
Table 1.
Confusion Matrix.
| |
|
PREDICTION |
| |
|
+ |
- |
| REAL CLASS |
+ |
TP |
FN |
| - |
FP |
TN |
Utilizing this matrix, we opted to consider accuracy (A), precision (P), recall (R), the Area Under Curve - Receiving Operating Characteristics (AUC-ROC), and the F1 score, five fundamental metrics that provide a comprehensive evaluation of the model’s performance. The formulas are as follows:
Accuracy (A): Accuracy, a fundamental metric in model evaluation, assesses the overall correctness of predictions by considering both true positives and true negatives. It is calculated as the ratio of the sum of true positives and true negatives to the total number of instances.
Precision (P): Precision, the ratio of true positives to the sum of true positives and false positives, provides insight into the accuracy of the extracted data, emphasizing the relevance of the identified information. It helps discern how many of the identified instances are indeed relevant to the task at hand.
Recall (R): It is the ratio of true positives to the sum of true positives and false negatives. It gauges the system’s ability to capture and retrieve all pertinent information, minimizing the likelihood of overlooking relevant data. In our context, recall is crucial to ensure that the system comprehensively identifies and retrieves all relevant data, reducing the chances of missing crucial information.
F1 Score (F1): The harmonic mean of precision and recall, in serving as a balanced measure that incorporates both false positives and false negatives. The F1 score is particularly valuable when there is an uneven distribution between positive and negative instances, preventing an undue influence on the evaluation due to class imbalance. The adoption of F1 score, along with precision and recall, aims to provide a comprehensive assessment that considers both the accuracy and completeness of the data extraction process across various categories within the JSON structure.
By incorporating these formulas and the confusion matrix, our evaluation aims to offer a detailed and nuanced assessment of the model’s performance. This approach ensures a thorough understanding not only of the correctness of extracted data but also the ability to avoid erroneous associations and capture all relevant information within the JSON structure.