Submitted:
11 July 2024
Posted:
15 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. The New Quantum-Chemistry Based LSER (QC-LSER) Molecular Descriptors
3. Linear Solvation Energy Relationships with the New QC-LSER Descriptors
4. Applications
4.1. Self-Solvation Calculations
4.2. Hydrogen-Bonding Calculations in Solute (1) – Solvent (2) Systems
5. Discussion and Conclusions
Supplementary Materials
References
- Moore, J.D., Mountain, R.D., Ross, R.B., Shen, V.K., Siderius, D.W., Smith, K.D., 2018. The 9th Industrial Fluid Properties Simulation Challenge. Fluid Phase Equilib., 476, 1–5.
- Tillotson, M.J., Diamantonis, N. I., Buda, C., Bolton, L.W., Mu¨ller, E.A., 2023, Molecular modelling of the thermophysical properties of fluids: expectations, limitations, gaps and opportunities. Phys. Chem. Chem. Phys., 25, 12607.
- Sapir, L., Harries, D., 2017. Revisiting Hydrogen Bond Thermodynamics in Molecular Simulations, J. Chem. Theory Comput. 13, 2851−2857 . [CrossRef]
- Duarte Ramos Matos, G., Kyu, D.Y., Loeffler, H.H., Chodera, J.D., Shirts, M.R., Mobley, D.L., 2017. Approaches for Calculating Solvation Free Energies and Enthalpies Demon strated with an Update of the FreeSolv Database. J. Chem. Eng. Data 62, 5, 1559–1569 . [CrossRef]
- Hirata, F. 2020. Exploring Life Phenomena with Statistical Mechanics of Molecular Liquids. Boca Raton, FL, CRC Press.
- L. Pereyaslavets, G. Kamath, O. Butin, A. Illarionov, M. Olevanov, I. Kurnikov,S. Sakipov, I. Leontyev, E. Voronina, T. Gannon, G. Nawrocki, M. Darkhovskiy, I. Ivahnenko, A. Kostikov, J. Scaranto, M. G. Kurnikova , S. Banik, H. Chan, M. G. Sternberg, S. K. R. S. Sankaranarayanan, B. Crawford, J. Potoff, M. Levitt, R. D. Kornberg, B Fain 2022. Accurate determination of solvation free energies of neutral organic compounds from first principles. Nature Communications . [CrossRef]
- Abraham, M. H., McGowan, J. C. 1987. The use of characteristic volumes to measure cavity terms in reversed phase liquid chromatography, Chromatographia, 23: 243-246 . [CrossRef]
- Abraham, M. H. 1993. Scales of solute hydrogen-bonding: their construction and application to physicochemical and biochemical processes, Chem. Soc. Rev., 22, 73-83. [CrossRef]
- Abraham, M. H., Ibrahim, A., Zissimos, A. M. 2004. Determination of sets of solute descriptors from chromatographic measurements, J. Chromatogr. A, 1037, 29-47. [CrossRef]
- Abraham, M. H., Smith, R. E., Luchtefeld, R., Boorem, A. J., Luo, R., Acree Jr., W. E.. 2010. Prediction of solubility of drugs and other compounds in organic solvents, J. Pharm. Sci., 99, 1500-1515.
- Goss, K.-U., 2005. Predicting the equilibrium partitioning of organic compounds using just one linear solvation energy relationship (LSER). Fluid Phase Equilibr., 233, 19–22. [CrossRef]
- Endo, S., Watanabe, N., Ulrich, N.., G. Bronner, K-U. Goss, UFZ-LSER database v 2.1 [Internet], Leipzig, Germany, Helmholtz Centre for Environmental Research-UFZ, 2015. [last accessed on 12.06.2024], available from https://www.ufz.de/index.php?en= 31698& contentonly=1&m=0&lserd_data[mvc]=Public/start.
- Sinha, S., Yang, Ch., Wu, E., Acree, W.E. 2022. Abraham Solvation Parameter Model: Examination of Possible Intramolecular Hydrogen-Bonding using calculated solute descriptors, Liquids, 2, 131 – 146 . [CrossRef]
- C. Mintz, T. Ladlie, K. Burton, M. Clark, W.E. Acree Jr, M.H. Abraham, 2008, Enthalpy of solvation correlations for gaseous solutes dissolved in alcohol solvents based on the Abraham model, QSAR Comb. Sci. 27, 627–635. [CrossRef]
- Egert, T., Langowski, H. Ch., Linear Solvation Energy Relationships (LSERs) for Accurate Prediction of Partition Coefficients between Low Density Polyethylene and Water - Part II: Model Evaluation and Benchmarking, Eur. J. Pharm. Sci., 2022, 172, 1-10.
- Panayiotou, C., Zuburtikudis, I., Abu Khalifeh, H. 2023. Linear Free-Energy Relationships (LFER) and Solvation Thermodynamics: The Thermodynamic Basis of LFER Linearity. Ind. Eng. Chem. Res. 62 (6), 2989-3000. [CrossRef]
- Panayiotou, C., Zuburtikudis, I., Abu Khalifeh, H. 2023. Linear Solvation Energy Relationships (LSER) and Equation-of-State Thermodynamics: On the Extraction of Thermodynamic Information from LSER Database. Liquids 3, 66-89. [CrossRef]
- Panayiotou, C., Acree Jr, W.E/, Zuburtikudis, I., 2023. COSMO-RS and LSER Models of Solution Thermodynamics: Towards a COSMOLSER Equation of State Model of Fluids. J. Molec. Liq.
- Moine, E., Privat, R., Sirjean, B., Jaubert, J.N., 2017. Estimation of Solvation Quantities from Experimental Thermodynamic Data: Development of the Comprehensive CompSol Databank for Pure and Mixed Solutes. J. Phys. Chem. Ref. Data, 46 (3), 033102. https://hal.univ-lorraine.fr/hal-01703358.
- Joesten, M.D., Schaad, L.1974, Hydrogen bonding, Marcel Dekker, New York.
- Raevsky, O.A., Grigor'ev, V.Y., Kireev, D.B., Zefirov, N.S. 1992, Complete Thermodynamic Description of H-Bonding in the Framework of Multiplicative Approach. Quantitative Structure-Activity Relationships 11, 49-63. [CrossRef]
- Gilli G, Gilli P. 2009. The Nature of the Hydrogen Bond. Oxford, Oxford University Press.
- Baev, A.K. 2014, Specific Intermolecular Interactions of Nitrogenated and Bioorganic Compounds, Springer-Verlag Berlin Heidelberg.
- Katritzky, D., Fara, E., Yang, H., Tamm, K., Tamm, T., Karelson, M. 2004. Quantitative Measures of Solvent Polarity, Chem. Rev. 104: 175-198 . [CrossRef]
- Laurence, C., J.-F. Gal, J.-F. 2010. Lewis Basicity and Affinity Scales: Data and Measurements, Wiley, New York.
- Epley, T.D., Drago, R.S. 1967, Calorimetric Studies on Some Hydrogen-Bonded Adducts. Journal of the American Chemical Society 89, 5770-5773. [CrossRef]
- Murthy A.S., Rao, C. 1968, Spectroscopic Studies of the Hydrogen Bond. Applied Spectroscopy Reviews 2, 69-191.
- Arnett, E.M., Joris, L., Mitchell, E., Murty, T., Gorrie, T., Schleyer, P.V.R. 1970. Hydrogen-bonded complex formation. III. Thermodynamics of complexing by infrared spectroscopy and calorimetry Journal of the American Chemical Society 92, 2365-2377.
- Cabani, S., Gianni, P., Mollica, V., Lepori, L. 1981, Group Contributions to the Thermodynamic Properties of Non-Ionic Organic Solutes in Dilute Aqueous Solutions. J. Solution Chem. 10, 563- 595. [CrossRef]
- Abbott, S., Yamamoto, H., Hansen, C.M. 2010. Hansen Solubility Parameters in Practice, Complete with software, data and examples, third ed.-version 3.1.20. Book and Software published by Hansen-Solubility.com.
- Dortmund Data Bank [Last accessed on 28.06.2024]: https://www.ddbst.com/ddb.html.
- Sedov, L.A., Solomonov,, B.N., 2012. Hydrogen bonding in neat aliphatic alcohols: The Gibbs free energy of self-association and molar fraction of monomer. J. Mol. Liq., 167, 47–51.
- Solomonov, B.N., Yagofarov, M.I. ,2022, Compensation relationship in Thermodynamics of solvation and vaporization: features and applications. I. Non-hydrogen-bonded systems, J. Mol. Liq. 368. [CrossRef]
- Solomonov, B.N., Yagofarov, M.I., 2023. Compensation relationship in Thermodynamics of solvation and vaporization: features and applications. ΙΙ. Hydrogen-bonded systems, J. Mol. Liq. 372, 121205. [CrossRef]
- Dohnal, V. 2024, New QSPR molecular descriptors based on low-cost quantum chemistry computations using DFT/COSMO approach, J. Mol. Liq. 125256. [CrossRef]
- Goodsell, D.S. 2003, Bionanotechnology: Lessons from Nature. Wiley & Sons, New York.
- Qiu, X., Li, H., Ver Steeg, G., Godzik, A. 2024. Advances in AI for Protein Structure Prediction: Implications for Cancer Drug Discovery and Development. Biomolecules 14(3), 339; [CrossRef]
- Chapman, W.G., Gubbins, K.E., Jackson, G. and Radosz, M. 1990. Ind. Eng.Chem. Res. 29: 1709 – 1721.
- Vega LF, Llovall, F. 2016. Review and new insights into the application of molecular-based equations of state to water and aqueous solutions. Fluid Phase Equil. 416:150–173. [CrossRef]
- Torshizi, M. F, and Muller, E. A., 2021. Coarse-grained molecular dynamics study of the self-assembly of polyphilic bolaamphiphiles using the SAFT-γ Mie force field. Mol. Syst. Des. Eng. 6: 594 – 608.
- Wertheim, M. 1986. Fluids with highly directional attractive forces. III. Multiple attraction sites, J. Statist. Phys., 42, 459–476. [CrossRef]
- Kontogeorgis, G.M, Folas, G.K. 2010. Thermodynamic Models for Industrial Applications. From Classical and Advanced Mixing Rules to Association Theories. Chichester, U.K., John Wiley and Sons, Ltd.
- Panayiotou, C. and Sanchez, I.C. 1991. Hydrogen Bonding in Fluids: An Equation of-State Approach. J. Phys. Chem. 95, 10090 – 10097. [CrossRef]
- Panayiotou, C.; Stefanis, E.; Tsivintzelis, I.; Pantoula, M.; Economou, I. 2004. Nonrandom Hydrogen-Bonding Model of Fluids and Their Mixtures 1. Pure Fluids. Ind. Eng. Chem. Res. 43, 6592-6606 . [CrossRef]
- Panayiotou, C., Tsivintzelis, I., Economou., I.G. 2007. Nonrandom Hydrogen-Bonding Model of Fluids and their Mixtures. 2. Multicomponent Mixtures Ind. Eng. Chem. Res. 46, 2628 - 2636. [CrossRef]
- Veytsman, B. A.: 1990. Are lattice models valid for fluids with hydrogen bonds? J. Phys. Chem. 94, 8499 – 8500.
- Mensitieri, G., Scherillo, G.. Panayiotou, C., Musto, P. 2020. Towards a predictive thermodynamic description of sorption processes in polymers: The synergy between theoretical EoS models and vibrational spectroscopy, Materials Sci. & Eng. R, 140, 100525 . [CrossRef]
- Klamt, A. 1995. Conductor-like Screening Model for Real Solvents: A New Approach to the Quantitative Calculation of Solvation Phenomena. J. Phys. Chem. 99 (7): 2224-2235. [CrossRef]
- Lin, S. T. and Sandler, S. I. 2002. A priori phase equilibrium prediction from a segment contribution solvation model. Ind. Eng. Chem. Res. 41: 899−913.
- Bell, I. A., Mickoleit, E., Hsieh, C-M., Lin, S-T., Vrabec, J., Breitkopf, C.,Jager, A., 2020. A Benchmark Open-Source Implementation of COSMO-SAC, J. Chem. Theory Comput. 16, 2635−2646 . [CrossRef]
- Klamt, A. 2005. COSMO-RS from Quantum Chemistry to Fluid Phase Thermodynamics and Drug Design; Amsterdam: Elsevier.
- Grensemann, H., Gmehling, J. 2005. Performance of a conductor-like screening model for real solvents model in comparison to classical group contribution methods. Ind. Eng. Chem. Res.44: 1610−1624. [CrossRef]
- Pye, C. C.; Ziegler, T.; van Lenthe, E., Louwen, J. N. 2009. An implementation of the conductor-like screening model of solvation within the Amsterdam density functional package, Part II. COSMO for real solvents. Can. J. Chem. 87, 790−797. [CrossRef]
- Klamt, A., Eckert, F., Arlt, W. 2010. COSMO-RS: An alternative to simulation for calculating thermodynamic properties of liquid mixtures, Annual Review of Chemical and Biomolecular Engineering, 1:101–122 . [CrossRef]
- Klamt, A., Eckert, F., Reinisch, J., Wichmann, K. 2016. Prediction of cyclohexane-water distribution coefficients with COSMO-RS on the SAMPL5 data set. J Comput Aided Mol Des. 30(11): 959–967. [CrossRef]
- Reinisch, J., Klamt, A. 2014. Prediction of free energies of hydration with COSMO-RS on the SAMPL4 data set. J Comput Aided Mol Des. 28(3): 169–173. [CrossRef]
- COSMObase, ver. 2019, COSMOlogic GmbH &CoKG (now, BIOVIA Dassault Systemes).
- M. Zissimos, M. H. Abraham, A. Klamt, F. Eckert, and J. Wood 2002. A Comparison between the Two General Sets of Linear Free Energy Descriptors of Abraham and Klamt. J. Chem. Inf. Comput. Sci., 42, 6, 1320–1331 . [CrossRef]
- https://www.turbomole.org.
- https://www.3ds.com/products/biovia/materials-studio.
- https://www.scm.com/product/cosmo-rs/.
- Sinha, S., Yang, Ch., Wu, E., Acree, W.E. 2022. Abraham Solvation Parameter Model: Examination of Possible Intramolecular Hydrogen-Bonding using calculated solute descriptors, Liquids, 2, 131 – 146 . [CrossRef]
- Mintz, C., Ladlie, T., Burton, K., Clark, M., Acree Jr, W.E., Abraham, M.H. 2008, Enthalpy of solvation correlations for gaseous solutes dissolved in alcohol solvents based on the Abraham model, QSAR Comb. Sci. 27, 627–635.
- Missopolinou, D., Ioannou, K., Prinos, I., and Panayiotou, C. 2002. Thermodynamics of alkoxyethanol + alkane mixtures. Z. Phys. Chem. 216 (7): 905 – 918. [CrossRef]
- Missopolinou, D., Tsivintzelis, I., and Panayiotou, C. 2006. Excess enthalpies of binary mixtures of 2-ethoxyethanol with four hydrocarbons at 298.15, 308.15, and 318.15K: An experimental and theoretical study. Fluid Phase Equil. 245 (2): 89–101. [CrossRef]
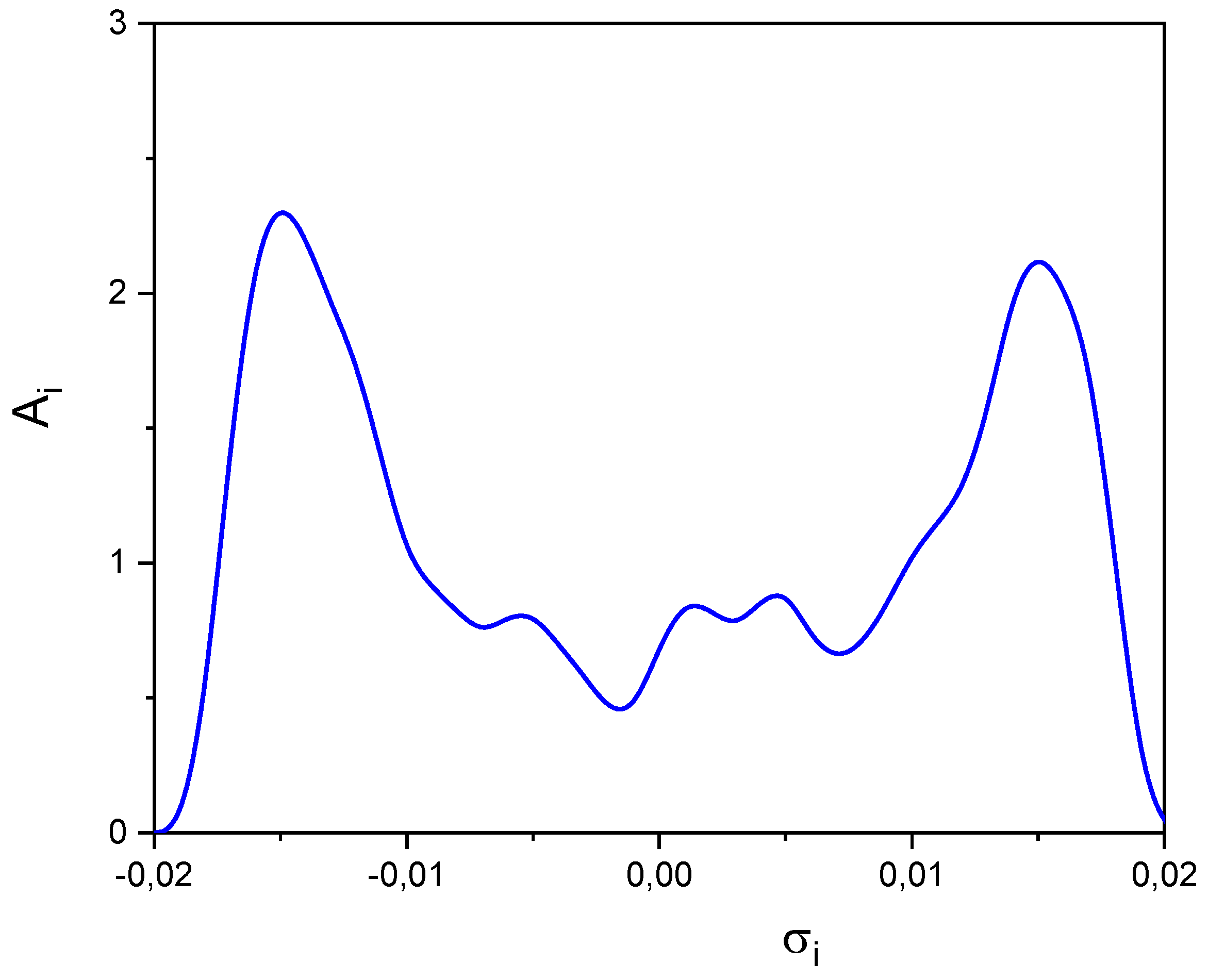
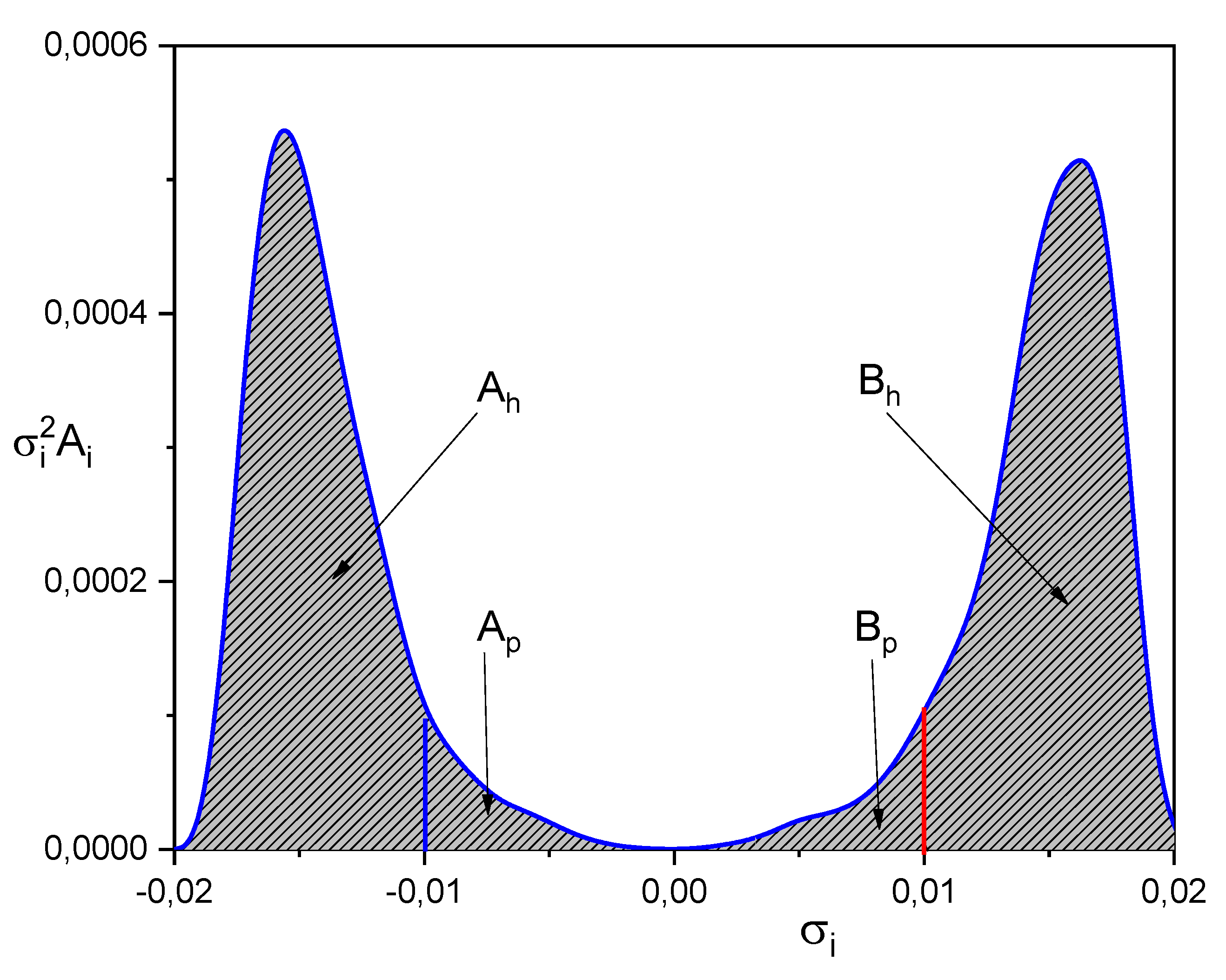
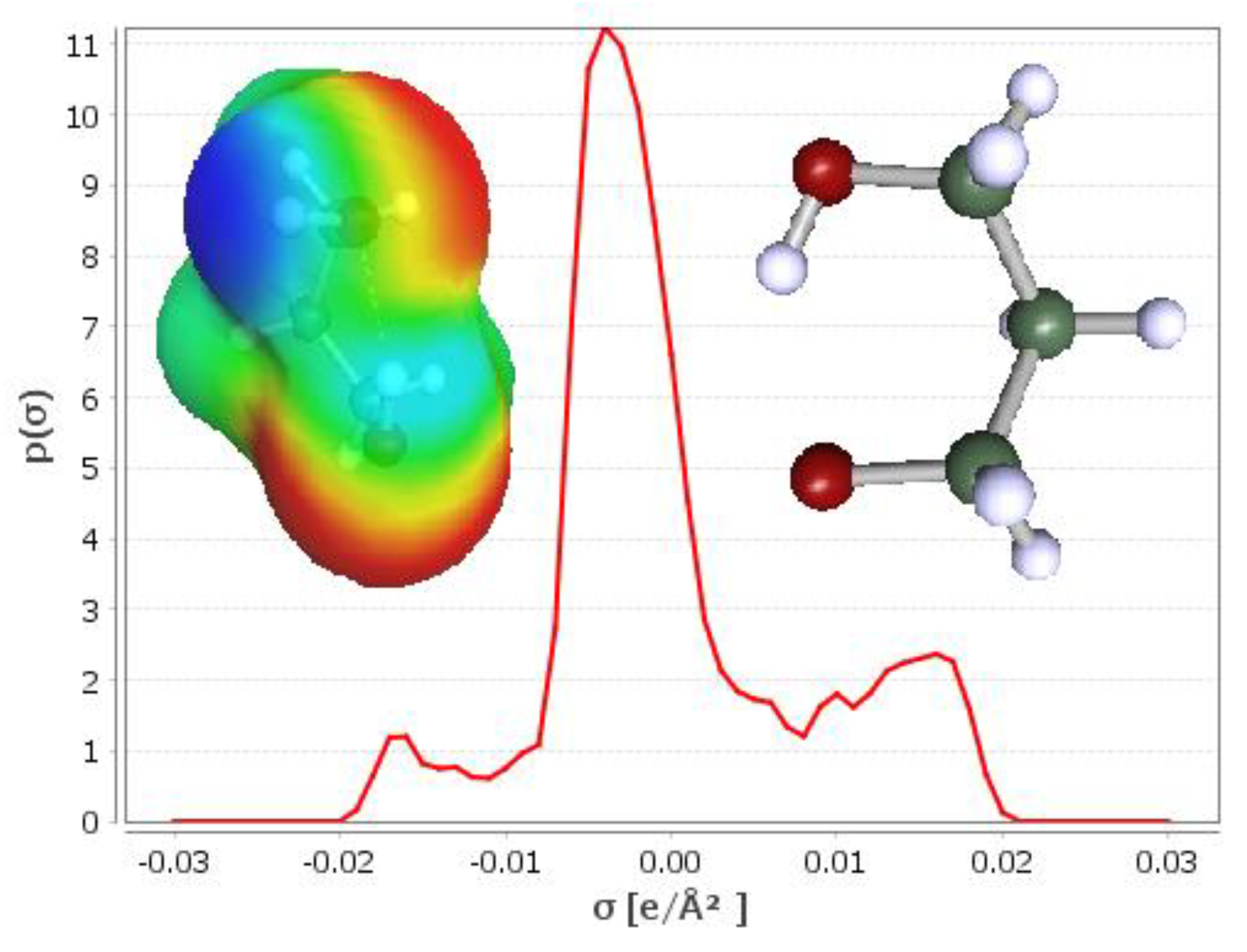
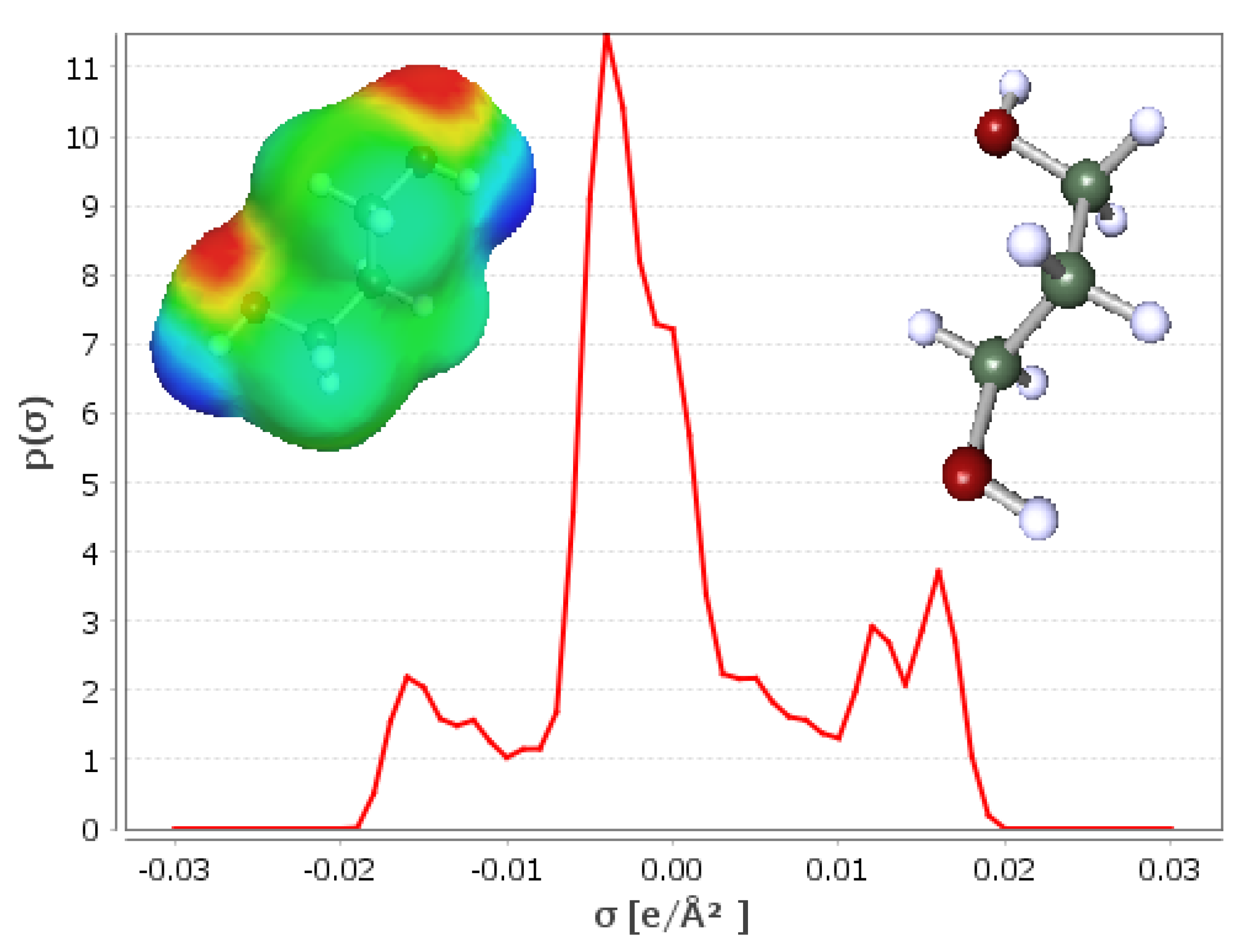
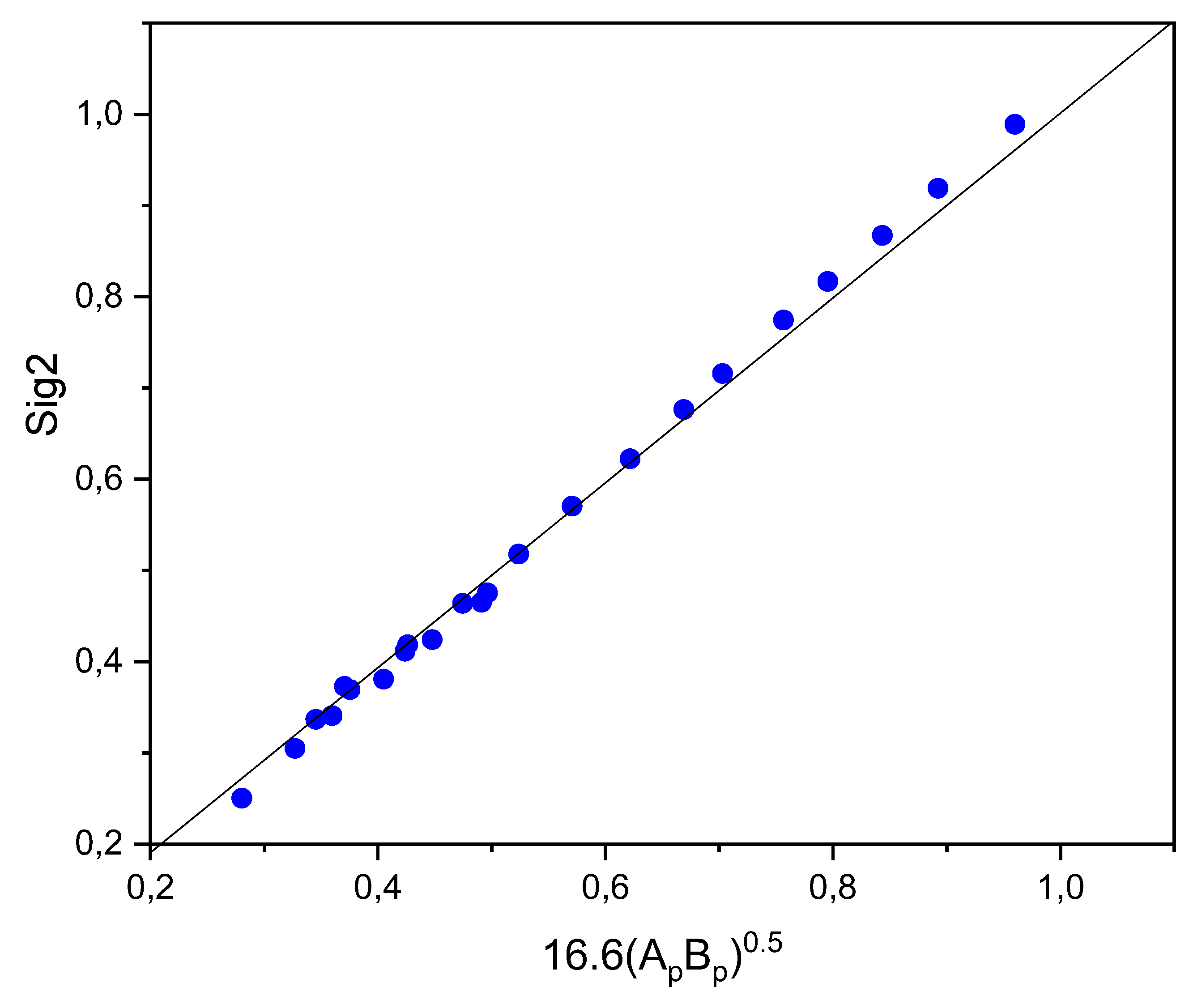
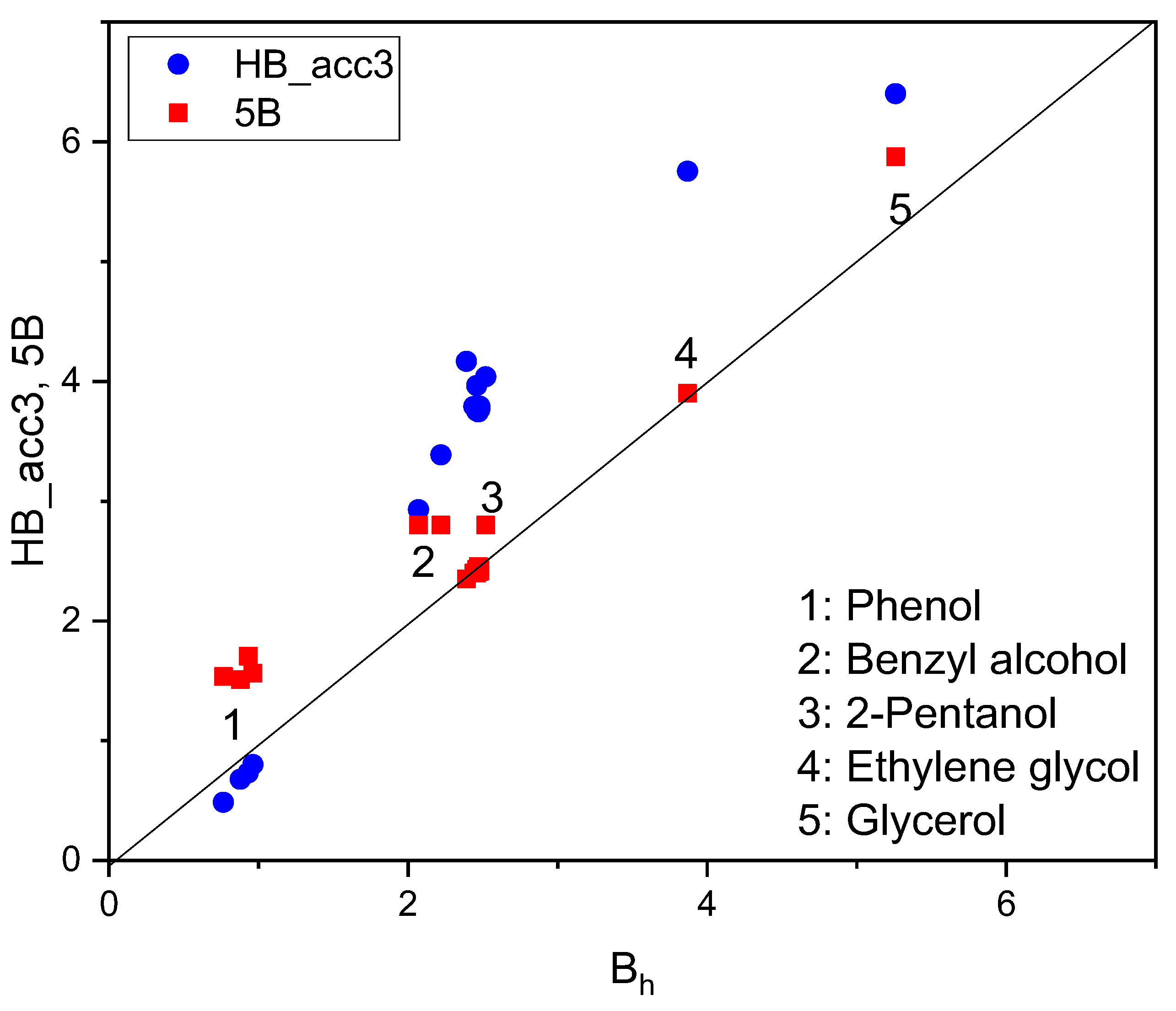
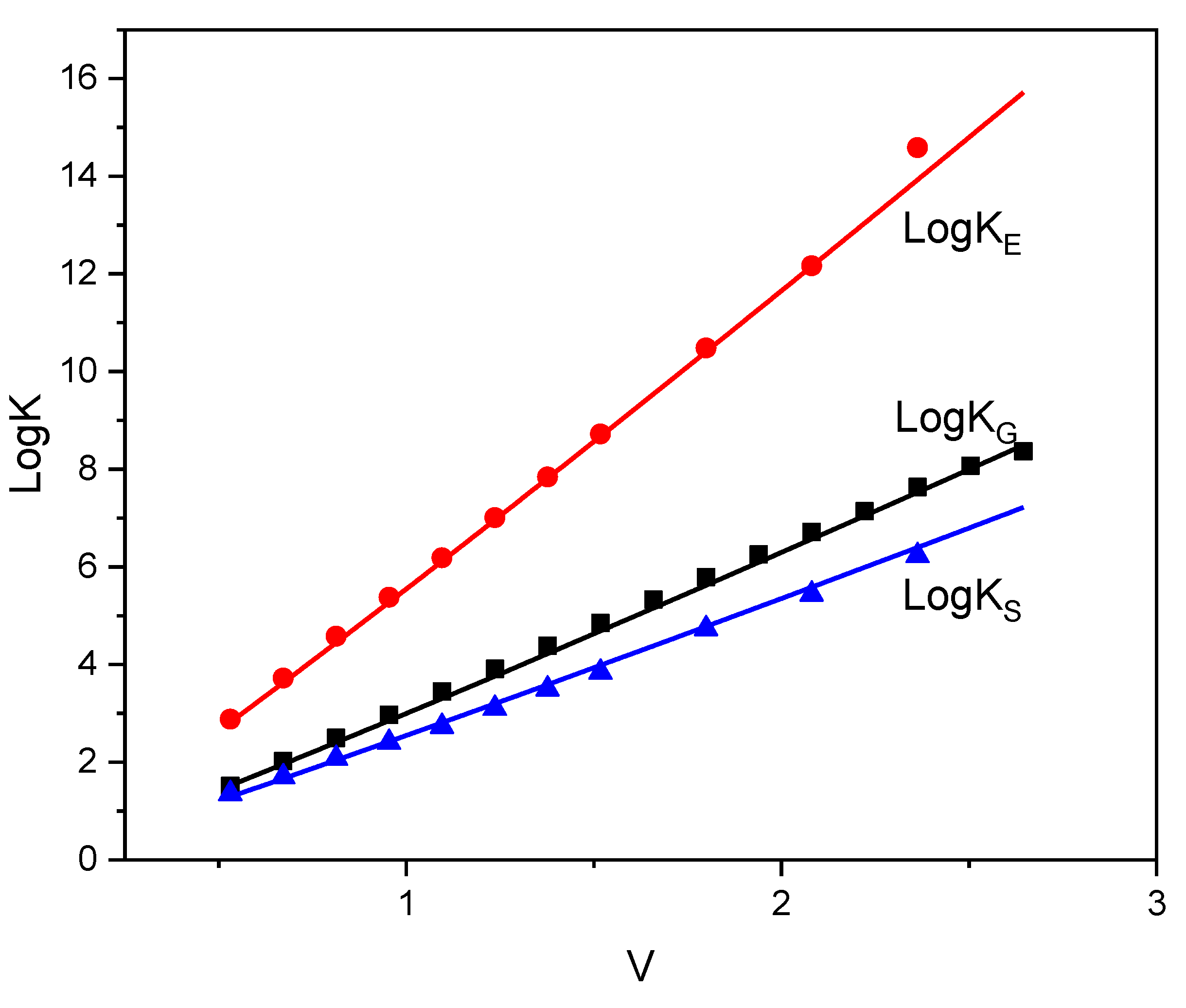

| SOLUTE | Ah | Ap | Bp | Bh |
|---|---|---|---|---|
| ETHANE | 0.00 | 0.28 | 0.23 | 0.00 |
| PROPANE | 0.00 | 0.32 | 0.29 | 0.00 |
| n-BUTANE | 0.00 | 0.37 | 0.37 | 0.00 |
| n-PENTANE | 0.00 | 0.42 | 0.40 | 0.00 |
| n-HEXANE | 0.00 | 0.47 | 0.46 | 0.00 |
| n-HEPTANE | 0.00 | 0.52 | 0.52 | 0.00 |
| n-OCTANE | 0.00 | 0.57 | 0.57 | 0.00 |
| n-NONANE | 0.00 | 0.62 | 0.63 | 0.00 |
| n-DECANE | 0.00 | 0.67 | 0.69 | 0.00 |
| n-UNDECANE | 0.00 | 0.71 | 0.73 | 0.00 |
| n-DODECANE | 0.00 | 0.77 | 0.78 | 0.00 |
| n-TRIDECANE | 0.00 | 0.80 | 0.84 | 0.00 |
| n-TETRADECANE | 0.00 | 0.85 | 0.89 | 0.00 |
| n-PENTADECANE | 0.00 | 0.90 | 0.94 | 0.00 |
| n-HEXADECANE | 0.00 | 0.96 | 1.02 | 0.00 |
| n-OCTADECANE | 0.00 | 1.05 | 1.11 | 0.00 |
| ISOBUTANE | 0.00 | 0.39 | 0.37 | 0.00 |
| ISOPENTANE | 0.00 | 0.44 | 0.41 | 0.00 |
| 2-METHYLPENTANE | 0.00 | 0.49 | 0.46 | 0.00 |
| 3-METHYLPENTANE | 0.00 | 0.49 | 0.45 | 0.00 |
| 3-METHYLHEXANE | 0.00 | 0.54 | 0.51 | 0.00 |
| 2,2-DIMETHYLHEXANE | 0.00 | 0.61 | 0.60 | 0.00 |
| 2,5-DIMETHYLHEXANE | 0.00 | 0.59 | 0.58 | 0.00 |
| CYCLOPENTANE | 0.00 | 0.35 | 0.33 | 0.00 |
| CYCLOHEXANE | 0.00 | 0.35 | 0.33 | 0.00 |
| CYCLOHEPTANE | 0.00 | 0.38 | 0.37 | 0.00 |
| CYCLOOCTANE | 0.00 | 0.42 | 0.42 | 0.00 |
| ETHYLENE | 0.00 | 0.67 | 0.73 | 0.02 |
| PROPYLENE | 0.00 | 0.67 | 0.80 | 0.06 |
| 1-BUTENE | 0.00 | 0.70 | 0.84 | 0.06 |
| 1-PENTENE | 0.00 | 0.74 | 0.90 | 0.07 |
| 1-HEXENE | 0.00 | 0.79 | 0.96 | 0.06 |
| 1-HEPTENE | 0.00 | 0.83 | 1.02 | 0.06 |
| 1-OCTENE | 0.00 | 0.86 | 1.07 | 0.05 |
| BENZENE | 0.00 | 1.24 | 1.14 | 0.00 |
| TOLUENE | 0.00 | 1.22 | 1.23 | 0.00 |
| ETHYLBENZENE | 0.00 | 1.25 | 1.29 | 0.00 |
| n-PROPYLBENZENE | 0.00 | 1.29 | 1.36 | 0.00 |
| n-BUTYLBENZENE | 0.00 | 1.34 | 1.42 | 0.00 |
| o-XYLENE | 0.00 | 1.25 | 1.34 | 0.00 |
| m-XYLENE | 0.00 | 1.22 | 1.31 | 0.00 |
| p-XYLENE | 0.00 | 1.21 | 1.29 | 0.00 |
| 1-PENTYNE | 0.49 | 1.01 | 1.51 | 0.16 |
| 1-HEXYNE | 0.48 | 1.05 | 1.57 | 0.17 |
| 3-HEXYNE | 0.00 | 1.10 | 1.39 | 0.34 |
| DICHLOROMETHANE | 0.94 | 0.94 | 1.08 | 0.00 |
| CHLOROFORM | 1.14 | 0.59 | 0.71 | 0.00 |
| CARBON TETRACHLORIDE | 0.00 | 0.71 | 0.37 | 0.00 |
| 2,2-DICHLOROPROPANE | 0.00 | 1.48 | 1.43 | 0.00 |
| CARBON TETRAFLUORIDE | 0.00 | 0.33 | 0.14 | 0.00 |
| DIETHYL ETHER | 0.00 | 0.79 | 0.38 | 1.81 |
| DI-n-PROPYL ETHER | 0.00 | 0.85 | 0.47 | 1.79 |
| DI-n-BUTYL ETHER | 0.00 | 0.94 | 0.60 | 1.79 |
| FURAN | 0.24 | 1.40 | 1.24 | 0.13 |
| TETRAHYDROFURAN | 0.00 | 0.79 | 0.31 | 2.15 |
| 1,3-DIOXANE | 0.00 | 1.50 | 0.54 | 3.12 |
| 1,4-DIOXANE | 0.00 | 1.50 | 0.54 | 3.12 |
| TETRAHYDROPYRAN | 0.00 | 0.77 | 0.34 | 1.97 |
| METHYL FORMATE | 0.19 | 1.59 | 0.74 | 2.21 |
| ETHYL FORMATE | 0.15 | 1.56 | 0,73 | 2.36 |
| ETHYL ACETATE | 0.00 | 1.61 | 0.68 | 2.81 |
| n-PROPYL ACETATE | 0.00 | 1.60 | 0.72 | 2.80 |
| ISOPROPYL ACETATE | 0.00 | 1.65 | 0.68 | 2.68 |
| n-BUTYL ACETATE | 0.00 | 1.64 | 0.76 | 2.81 |
| ETHYL PROPIONATE | 0.00 | 1.53 | 0.73 | 2.66 |
| ETHYL n-BUTYRATE | 0.00 | 1.56 | 0.78 | 2.65 |
| n-PROPYL PROPIONATE | 0.00 | 1.54 | 0.74 | 2.69 |
| ACETONE | 0.02 | 1.47 | 0.38 | 2.95 |
| METHYL ETHYL KETONE | 0.00 | 1.41 | 0.43 | 2.78 |
| 2-PENTANONE | 0.00 | 1.43 | 0.47 | 2.81 |
| 3-PENTANONE | 0.00 | 1.35 | 0.51 | 2.58 |
| 3-HEXANONE | 0.00 | 1.37 | 0.53 | 2.62 |
| FORMALDEHYDE | 0.00 | 1.34 | 0.62 | 1.55 |
| ACETALDEHYDE | 0.00 | 1.39 | 0.47 | 2.36 |
| PROPANAL | 0.00 | 1.32 | 0.49 | 2.33 |
| BUTANAL | 0.00 | 1.34 | 0.54 | 2.32 |
| PENTANAL | 0.00 | 1.38 | 0.59 | 2.33 |
| HEXANAL | 0.00 | 1.43 | 0.66 | 2.32 |
| OCTANAL | 0.00 | 1.52 | 0.76 | 2.33 |
| METHANOL | 1.39 | 0.55 | 0.29 | 2.39 |
| ETHANOL | 1.23 | 0.66 | 0.35 | 2.46 |
| 1-PROPANOL | 1.23 | 0.70 | 0.41 | 2.44 |
| 1-BUTANOL | 1.22 | 0.74 | 0.44 | 2.47 |
| 1-PENTANOL | 1.22 | 0.79 | 0.51 | 2.46 |
| 1-HEXANOL | 1.22 | 0.83 | 0.55 | 2.48 |
| 1-HEPTANOL | 1.21 | 0.90 | 0.61 | 2.48 |
| 1-OCTANOL | 1.19 | 0.95 | 0.68 | 2.46 |
| 1-NONANOL | 1.21 | 0.99 | 0.73 | 2.47 |
| 1-DECANOL | 1.23 | 1.02 | 0.79 | 2.47 |
| ISOPROPANOL | 1.18 | 0.73 | 0.38 | 2.52 |
| 2-BUTANOL | 0.91 | 0.81 | 0.39 | 2.46 |
| 2-PENTANOL | 1.14 | 0.79 | 0.52 | 2.22 |
| GLYCEROL | 2.98 | 1.68 | 0.75 | 5.26 |
| ETHYLENE GLYCOL, c0 | 2.13 | 1.10 | 0.57 | 3.87 |
| ETHYLENE GLYCOL, c6 Econf = 8.98 |
2.97 | 0.86 | 0.86 | 4.11 |
| 1,3-PROPYLENE GLYCOL, c0 | 1.58 | 1.17 | 0.53 | 3.92 |
| 1,3-PROPYLENE GLYCOL, c3 Econf = 5.54 |
2.63 | 1.01 | 0.56 | 4.38 |
| 1,2-BUTANEDIOL | 1.73 | 1.20 | 0.64 | 3.66 |
| 1,3-BUTANEDIOL | 1.60 | 1.21 | 0.58 | 3.96 |
| 1,4-BUTANEDIOL | 1.63 | 1.08 | 0.55 | 3.98 |
| 2-METHOXYETHANOL c0 | 0.77 | 1.22 | 0.60 | 3.13 |
| 2-METHOXYETHANOL c7 Econf = 10.97 |
1.39 | 1.08 | 0.62 | 3.19 |
| 2-ETHOXYETHANOL c0 | 0.63 | 1.24 | 0.60 | 3.13 |
| 2-ETHOXYETHANOL c1 Econf = 2.59 |
1.35 | 1.20 | 0.47 | 4.23 |
| 2-ETHOXYETHANOL c9 Econf = 10.5 |
1.39 | 1.11 | 0.64 | 3.28 |
| PHENOL | 2.03 | 1.26 | 1.45 | 0.88 |
| o-CRESOL | 1.99 | 1.21 | 1.57 | 0.77 |
| m-CRESOL | 1.98 | 1.25 | 1.52 | 0.93 |
| p-CRESOL | 1.96 | 1.26 | 1.46 | 0.96 |
| BENZYL ALCOHOL | 1.23 | 1.59 | 1.18 | 2.07 |
| ACETIC ACID | 2.04 | 1.11 | 0.76 | 2.70 |
| PROPIONIC ACID | 1.99 | 1.06 | 0.78 | 2.62 |
| n-BUTYRIC ACID | 1.98 | 1.07 | 0.83 | 2.62 |
| n-PENTANOIC ACID | 1.97 | 1.12 | 0.89 | 2.62 |
| ETHYLAMINE | 0.43 | 0.95 | 0.32 | 2.73 |
| n-BUTYLAMINE | 0.43 | 1.04 | 0.45 | 2.73 |
| DIETHYLAMINE | 0.20 | 0.84 | 0.40 | 2.02 |
| DI-n-PROPYLAMINE | 0.19 | 0.93 | 0.52 | 2.02 |
| TRIMETHYLAMINE | 0.00 | 0.65 | 0.35 | 1.49 |
| ANILINE | 1.62 | 1.41 | 1.82 | 0.78 |
| PYRIDINE | 0.00 | 1.60 | 0.56 | 2.11 |
| FORMAMIDE | 3.03 | 0.84 | 0.36 | 4.23 |
| DIMETHYL SULFOXIDE | 0.10 | 2.30 | 0.29 | 5.24 |
| ACETONITRILE | 0.31 | 1.67 | 0.91 | 2.00 |
| ACRYLONITRILE | 0.43 | 1.48 | 1.00 | 1.52 |
| PROPIONITRILE | 0.08 | 1.64 | 0.91 | 2.02 |
| ACETAMIDE | 2.61 | 1.03 | 0.34 | 4.74 |
| N,N-DIMETHYLFORMAMIDE | 0.00 | 1.66 | 0.25 | 4.17 |
| N-METHYL FORMAMIDE | 1.58 | 1.21 | 0.28 | 4.20 |
| WATER | 2.89 | 0.28 | 0.27 | 2.99 |
| Solute |
LogKS exp [19] |
LogKS Eq. 18 |
|---|---|---|
| ISOBUTANE | 1.57 | 1.63 |
| ISOPENTANE | 1.99 | 2.00 |
| 2-METHYLPENTANE | 2.25 | 2.38 |
| 3-METHYLPENTANE | 2.25 | 2.38 |
| 1-BUTENE | 1.62 | 1.65 |
| 1-PENTENE | 1.99 | 2.00 |
| 1-HEXENE | 2.38 | 2.35 |
| 3-HEXYNE | 2.40 | 2.40 |
| CYCLOPENTANE | 2.03 | 2.00 |
| CYCLOHEXANE | 2.25 | 2.33 |
| CYCLOHEPTANE | 2.64 | 2.67 |
| CYCLOOCTANE | 2.95 | 3.03 |
| BENZENE | 2.36 | 2.32 |
| TOLUENE | 2.58 | 2.61 |
| ETHYLBENZENE | 2.93 | 2.92 |
| n-PROPYLBENZENE | 3.19 | 3.25 |
| n-BUTYLBENZENE | 3.58 | 3.60 |
| o-XYLENE | 2.99 | 2.92 |
| m-XYLENE | 2.92 | 2.92 |
| p-XYLENE | 2.93 | 2.92 |
| CARBON TETRACHLORIDE | 2.23 | 2.23* |
| DIETHYL ETHER | 2.21 | 2.00 |
| DI-n-PROPYL ETHER | 2.67 | 2.69 |
| DI-n-BUTYL ETHER | 3.39 | 3.42 |
| ETHYL ACETATE | 2.71 | 2.59 |
| n-BUTYL ACETATE | 3.41 | 3.16 |
| ACETONE | 2.27 | 2.10 |
| METHYL ETHYL KETONE | 2.49 | 2.33 |
| 2-PENTANONE | 2.80 | 2.60 |
| 3-PENTANONE | 2.60 | 2.60 |
| ACETALDEHYDE | 2.01 | 1.96 |
| PROPANAL | 2.21 | 2.10 |
| BUTANAL | 2.34 | 2.33 |
| PENTANAL | 2.51 | 2.60 |
| HEXANAL | 2.88 | 2.90 |
| Solute |
LogKS exp [19] |
LogKS,nhb Eq. 18 |
LogKS,hb |
-ΔSHB/ JK-1 mol-1 |
-ΔHHB [57] kJmol-1 |
-ΔGHB/ kJmol-1 Eq. 20 |
-ΔGHB/ kJmol-1 LSER[62] |
|---|---|---|---|---|---|---|---|
| METHANOL | 2.95 | 0.88 | 2.07 | 39.58 | 23.08 | 11.28 | 13.14 |
| ETHANOL | 3.47 | 1.40 | 2.08 | 39.78 | 24.93 | 13.07 | 11.49 |
| 1-PROPANOL | 4.02 | 1.68 | 2.34 | 44.78 | 24.61 | 11.26 | 11.16 |
| 2-PROPANOL | 4.06 | 1.68 | 2.38 | 45.62 | 23.74 | 10.14 | 10.43 |
| 1-BUTANOL | 4.55 | 2.01 | 2.54 | 48.72 | 24.16 | 9.63 | 10.21 |
| 1-PENTANOL | 4.79 | 2.34 | 2.45 | 46.85 | 24.01 | 10.04 | 10.59 |
| 2-PENTANOL | 4.76 | 2.34 | 2.42 | 46.43 | 23.77 | 9.93 | 10.34 |
| 1-HEXANOL | 5.44 | 2.69 | 2.75 | 52.67 | 24.24 | 8.54 | 10.06 |
| 1-HEPTANOL | 6.01 | 3.05 | 2.96 | 56.64 | 23.64 | 6.75 | 9.77 |
| 1-OCTANOL | 6.14 | 3.42 | 2.72 | 52.10 | 23.43 | 7.90 | 9.40 |
| 1-NONANOL | 6.75 | 3.79 | 2.96 | 56.68 | 23.68 | 6.78 | |
| 1-DECANOL | 6.84 | 4.17 | 2.67 | 51.11 | 23.26 | 8.02 | 9.40 |
| ETHYLENE GLYCOL,c0 | 4.68 | 1.77 | 2.91 | 54.51 | 43.29 | 26.70 | 25.24 |
| 1,3-PROPYLENE GLYCOL,c0 | 4.74 | 2.02 | 2.72 | 51.10 | 50.21 | 34.70 | |
| GLYCEROL | 6.18 | 2.32 | 3.86 | 73.90 | 51.61 | 29.58 | |
| BENZYL ALCOHOL | 4.87 | 2.88 | 2.00 | 38.25 | 22.47 | 11.06 | 10.32 |
| PHENOL | 4.26 | 2.59 | 1.67 | 31.91 | 17.58 | 8.07 | |
| o-CRESOL | 4.55 | 2.88 | 1.67 | 31.98 | 13.22 | 3.69 | |
| m-CRESOL | 4.63 | 2.88 | 1.76 | 33.65 | 17.98 | 7.95 | |
| p-CRESOL | 4.96 | 2.88 | 2.09 | 39.92 | 18.59 | 6.69 | |
| 2-METHOXY-ETHANOL c0 | 3.28 | 2.03 | 1.25 | 23.99 | 19.53 | 12.38 | 9.06 |
| 2-ETHOXY-ETHANOL c0 | 3.93 | 2.32 | 1.61 | 30.89 | 19.21 | 10.00 | 9.45 |
| ACETIC ACID | 2.69 | 1.84 | 0.85 | 16.27 | 25.31 | 20.46 | 14.95 |
| PROPIONIC ACID | 3.43 | 2.05 | 1.38 | 26.43 | 25.39 | 17.51 | |
| n-BUTYRIC ACID | 4.09 | 2.32 | 1.77 | 33.84 | 25.37 | 15.28 | |
| n-PENTANOIC ACID | 4.85 | 2.62 | 2.23 | 42.71 | 24.89 | 12.16 | |
| n-BUTYLAMINE | 2.69 | 2.00 | 0.69 | 13.19 | 2.84 | -1.10 | |
| DIETHYLAMINE | 2.38 | 2.00 | 0.38 | 7.33 | 2.69 | 0.50 | |
| WATER | 2.72 | 0.38 | 2.35 | 44.77 | 41.15 | 27.80 | 27.37 |
| SOLUTE | LSER [62,63] | This Work / COSMO-RS [57] | |||
|
-A1a2/ kJ/mol |
-B1b2/ kJ/mol |
ch12 |
-ch12Ah1Bh2 / kJ/mol |
-ch12BBh1Ah2 / kJ/mol |
|
| Solvent: WATER | |||||
| WATER | 24.41 | 15.79 | 0,42 | 20,56 | 20,56 |
| TETRAHYDROFURAN | 0.00 | 20.03 | 0.69 | 0.00 | 24.52 |
| TETRAHYDROPYRAN | 0.00 | 21.43 | 0.72 | 0.00 | 23.26 |
| ETHYL ACETATE | 0.00 | 18.82 | 0.38 | 0.00 | 17.59 |
| n-PROPYL ACETATE | 0.00 | 18.82 | 0.38 | 0.00 | 17.61 |
| n-BUTYL ACETATE | 0.00 | 18.79 | 0.39 | 0.00 | 18.03 |
| ACETONE | 1.28 | 20.62 | 0.40 | 0.14 | 19.52 |
| METHYL ETHYL KETONE | 0.00 | 21.31 | 0.41 | 0.00 | 18.64 |
| 2-PENTANONE | 0.00 | 21.30 | 0.42 | 0.00 | 19.64 |
| ACETALDEHYDE | 0.00 | 18.82 | 0.35 | 0.00 | 13.60 |
| PROPANAL | 0.00 | 18.82 | 0.35 | 0.00 | 13.48 |
| BUTANAL | 0.00 | 18.82 | 0.35 | 0.00 | 13.51 |
| DMSO | 0.00 | 38.05 | 0.32 | 0.57 | 27.81 |
| METHANOL | 13.76 | 19.65 | 0.49 | 11.53 | 19.17 |
| ETHANOL | 11.84 | 20.07 | 0.55 | 11.63 | 22.48 |
| 1-PROPANOL | 11.84 | 20.07 | 0.56 | 11.61 | 22.35 |
| 1-BUTANOL | 11.80 | 20.16 | 0.55 | 11.51 | 22.52 |
| 1-PENTANOL | 11.80 | 20.16 | 0.56 | 11.56 | 22.53 |
| 1-HEXANOL | 11.76 | 20.29 | 0.55 | 11.48 | 22.55 |
| 1-HEPTANOL | 11.78 | 20.19 | 0.55 | 11.40 | 22.59 |
| 1-OCTANOL | 11.71 | 20.31 | 0.56 | 11.33 | 22.64 |
| 1-NONANOL | 11.76 | 20.53 | 0.56 | 11.51 | 22.71 |
| 1-DECANOL | 11.66 | 20.37 | 0.56 | 11.70 | 22.71 |
| ISOPROPANOL | 9.81 | 23.42 | 0.60 | 11.99 | 24.74 |
| 2-PENTANOL | 10.44 | 23.42 | 0.64 | 12.38 | 23.31 |
| PHENOL | 19.50 | 12.63 | 0.50 | 17.17 | 7.18 |
| ETHYLENE GLYCOL, c0 | 18.57 | 32.62 | 0.63 | 23.08 | 40.54 |
| ETHYLENE GLYCOL, c6 | 18.57 | 32.62 | 0.54 | 27.23 | 36.39 |
| 2-ETHOXYETHANOL c0 | 9.72 | 34.10 | 0.89 | 9.56 | 45.97 |
| 2-ETHOXYETHANOL c1 | 9.72 | 34.10 | 0.60 | 13.74 | 41.79 |
| 2-ETHOXYETHANOL c9 | 9.72 | 34.10 | 0.71 | 16.90 | 38.63 |
| 1,2-BUTANEDIOL | 18.57 | 35.13 | 0,65 | 19,28 | 39,46 |
| 1,3-BUTANEDIOL | 20.65 | 35.13 | 0,71 | 19,52 | 46,63 |
| ACETIC ACID | 19.71 | 18.46 | 0,42 | 14,65 | 18,74 |
| PROPIONIC ACID | 19.42 | 18.54 | 0,44 | 15,12 | 19,23 |
| n-BUTYLAMINE | 5.12 | 25.51 | 0,41 | 2,98 | 18,35 |
| DIETHYLAMINE | 2.56 | 28.73 | 0,65 | 2,22 | 21,53 |
| PYRIDINE | 0.00 | 21.42 | 0,47 | 0,00 | 16,23 |
| Solvent: ETHANOL | |||||
| ETHANOL | 18.08 | 5.53 | 0.73 | 12.58 | 12.58 |
| 1-PROPANOL | 18.08 | 5.53 | 0.72 | 12.28 | 12.36 |
| 2-PROPANOL | 14.97 | 6.45 | 0.77 | 12.56 | 13.55 |
| 1-OCTANOL | 17.87 | 5.59 | 0.74 | 12.17 | 12.71 |
| ETHYLENE GLYCOL, c0 | 28.34 | 8.98 | 0.70 | 20.75 | 19.05 |
| ETHYLENE GLYCOL, c6 | 28.34 | 8.98 | 0.57 | 23.43 | 16.37 |
| 2-ETHOXYETHANOL c0 | 14.83 | 9.39 | 0.86 | 7.53 | 18.93 |
| 2-ETHOXYETHANOL c1 | 14.83 | 9.39 | 0.55 | 10.22 | 16.24 |
| 2-ETHOXYETHANOL c9 | 14.83 | 9.39 | 0.63 | 12.05 | 14.41 |
| ACETIC ACID | 30.08 | 5.08 | 0.47 | 13.39 | 8.96 |
| n-BYTYL AMINE | 7.82 | 7.02 | 0.82 | 4.87 | 15.69 |
| DMSO | 0.00 | 10.48 | 0.54 | 0.77 | 19.88 |
| WATER | 37.25 | 4.35 | 0.60 | 24.22 | 12.66 |
| Solvent: 1-OCTANOL | |||||
| 1-OCTANOL | 19.74 | 4.37 | 0.70 | 11.69 | 11.69 |
| ETHANOL | 19.98 | 4.32 | 0.69 | 11.91 | 11.52 |
| 1-PROPANOL | 19.98 | 4.32 | 0.73 | 12.55 | 12.09 |
| 2-PROPANOL | 16.54 | 5.03 | 0.74 | 12.19 | 12.59 |
| ETHYLENE GLYCOL, c0 | 31.31 | 7.01 | 0.65 | 19.52 | 17.15 |
| ETHYLENE GLYCOL, c6 | 31.31 | 7.01 | 0.53 | 21.97 | 14.70 |
| 2-ETHOXYETHANOL c0 | 16.39 | 7.33 | 0.77 | 6.80 | 16.37 |
| 2-ETHOXYETHANOL c1 | 16.39 | 7.33 | 0.49 | 9.19 | 13.98 |
| 2-ETHOXYETHANOL c9 | 16.39 | 7.33 | 0.56 | 10.81 | 12.36 |
| ACETIC ACID | 33.24 | 3.97 | 0.47 | 13.37 | 8.56 |
| n-BYTYL AMINE | 8.64 | 5.48 | 0.83 | 4.98 | 15.35 |
| DMSO | 0.00 | 8.18 | 0.54 | 0.77 | 19.06 |
| WATER | 41.17 | 3.39 | 0.59 | 23.80 | 11.91 |
| Solvent: DMSO | |||||
| DMSO (Dimethyl Sulfoxide) | 0.00 | 5.33 | 0.004 | 0.01 | 0.01 |
| ETHANOL | 17.55 | 2.81 | 0.36 | 13.17 | 0.52 |
| 1-PROPANOL | 17.55 | 2.81 | 0.36 | 13.08 | 0.51 |
| 1-OCTANOL | 17.34 | 2.85 | 0.36 | 12.94 | 0.53 |
| 2-PROPANOL | 14.53 | 3.28 | 0.36 | 12.54 | 0.53 |
| ETHYLENE GLYCOL, c0 | 27.50 | 4.57 | 0.41 | 25.79 | 0.92 |
| ETHYLENE GLYCOL, c6 | 27.50 | 4.57 | 0.29 | 26.00 | 0.71 |
| 2-ETHOXYETHANOL c0 | 14.39 | 4.78 | 0.62 | 11.62 | 1.14 |
| 2-ETHOXYETHANOL c1 | 14.39 | 4.78 | 0.30 | 12.02 | 0.74 |
| 2-ETHOXYETHANOL c9 | 14.39 | 4.78 | 0.29 | 12.19 | 0.57 |
| ACETIC ACID | 29.20 | 2.59 | 0.31 | 18.81 | 0.49 |
| n-BYTYL AMINE | 7.59 | 3.58 | 0.04 | 0.50 | 0.06 |
| WATER | 36.16 | 2.21 | 0.33 | 28.28 | 0.58 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





