Submitted:
04 July 2024
Posted:
12 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results and Discussion
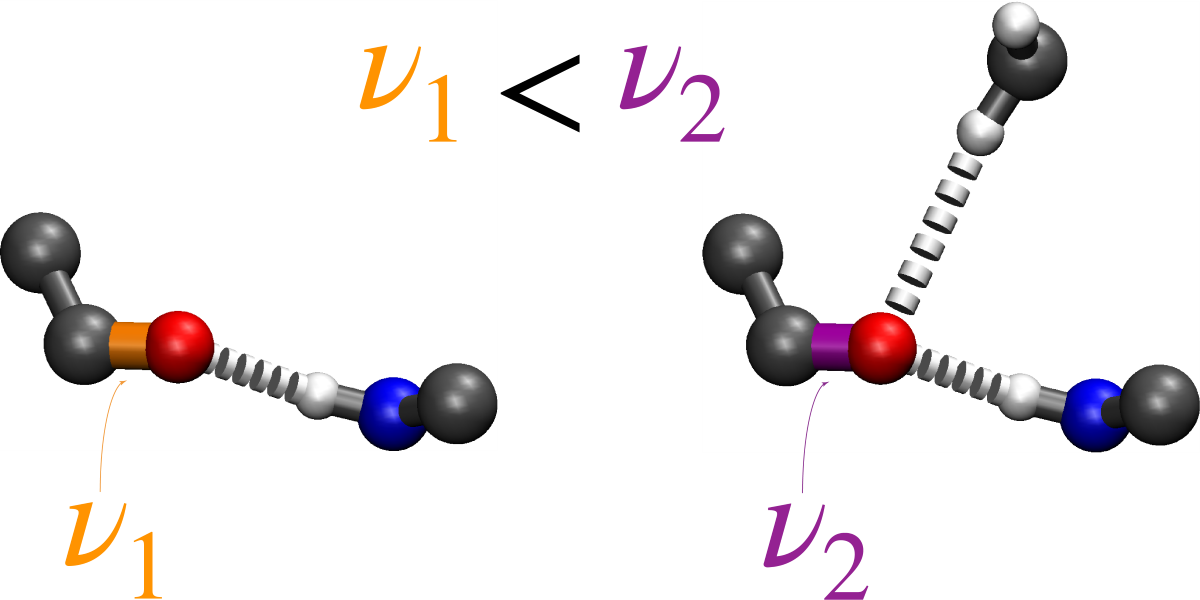
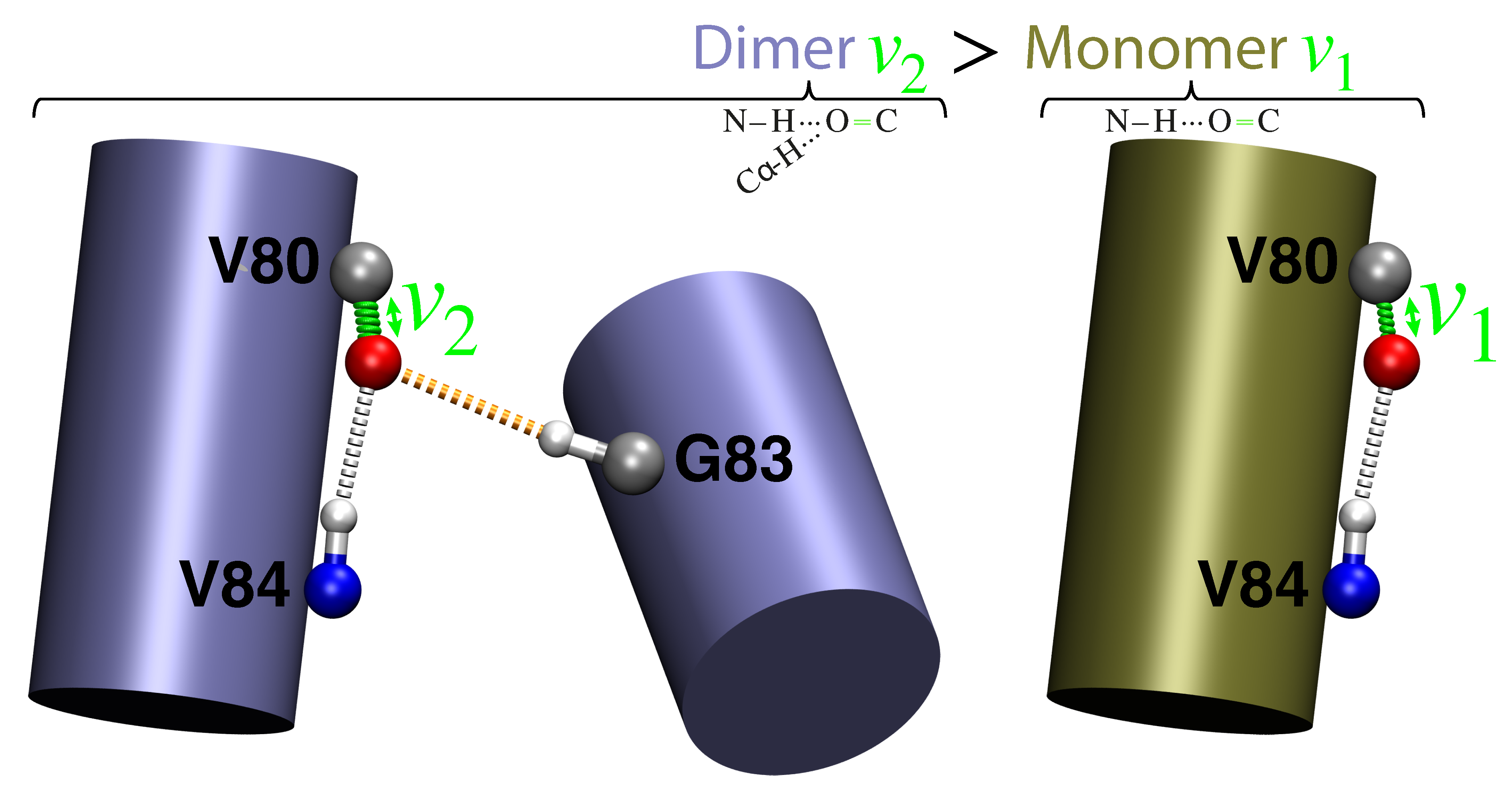
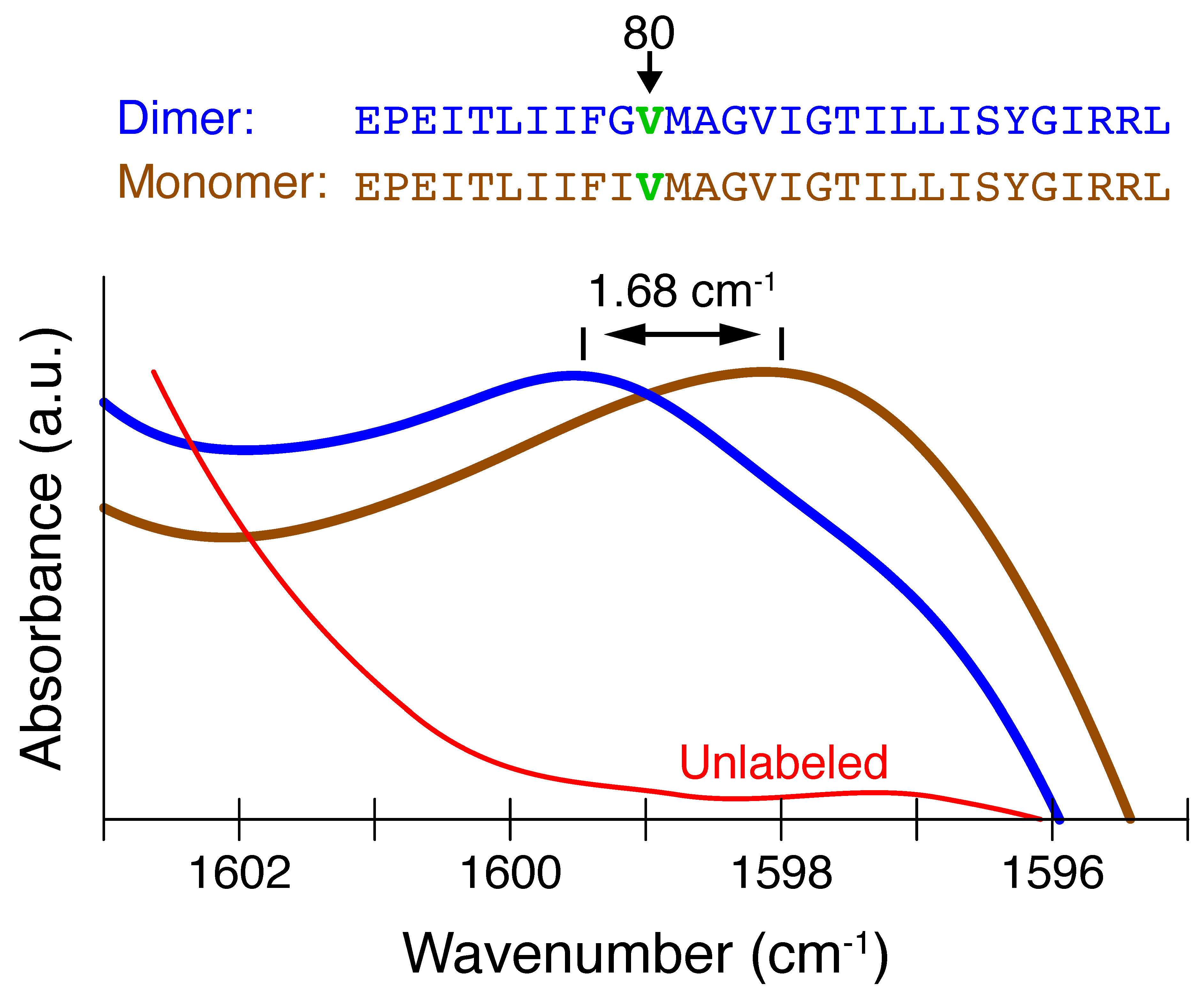
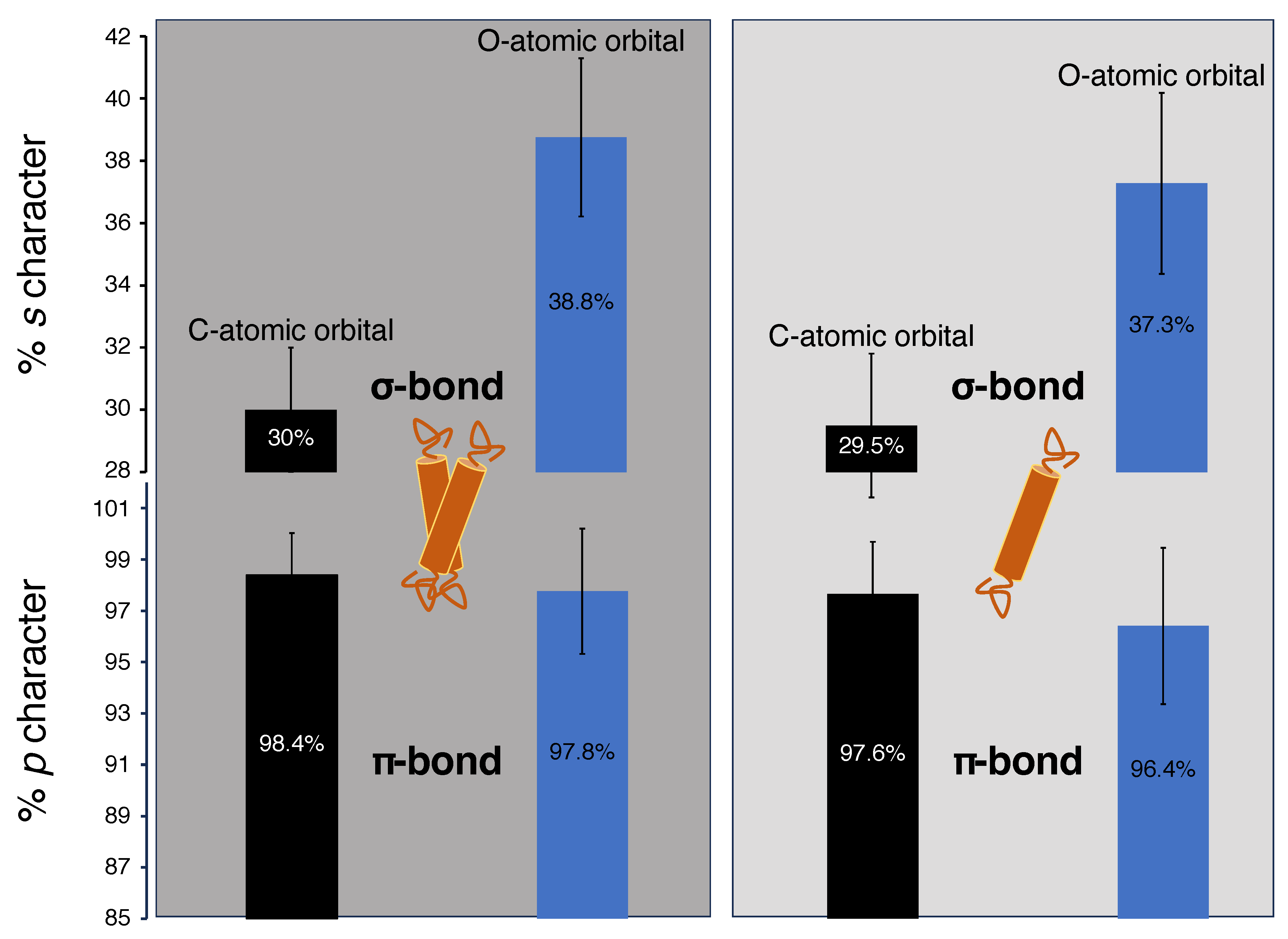
- Monomer (single H-bond):
- The canonical -helical H-bond between the carbonyl of valine 80 and the amide H of glycine 84 was mimicked by two N-methylacetamides.
- Dimer (two H-bonds):
- The inter-helical H-bonding system contained the canonical H-bond described above, with an additional non-canonical inter-helical hydrogen bond. The C−H of glycine 83 from the opposing helix is bonded to the same carbonyl of valine 80 that is also involved in the canonical hydrogen bond. This system was mimicked by two N-methylacetamides forming the canonical H-bond to which an acetylglycinemethylamide is hydrogen bonded.
3. Conclusions
4. Materials and Methods
4.1. Sample Preparation
4.1.1. Isotopic Label Synthesis
4.1.2. Peptide Synthesis and Purification
4.1.3. Peptide Reconstitution
4.2. FTIR Spectroscopy
4.3. Computational Details
4.3.1. Molecular Dynamic Simulations
4.3.2. Quantum Mechanical Calculations
5. Patents
Supplementary Materials
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| DOAJ | Directory of open access journals |
| TLA | Three letter acronym |
| LD | Linear dichroism |
References
- van der Lubbe, S.C.; Fonseca Guerra, C. The Nature of Hydrogen Bonds: A Delineation of the Role of Different Energy Components on Hydrogen Bond Strengths and Lengths. Chemistry–An Asian Journal 2019, 14, 2760–2769. [Google Scholar] [CrossRef] [PubMed]
- Sheu, S.Y.; Yang, D.Y.; Selzle, H.L.; Schlag, E.W. Energetics of Hydrogen Bonds in Peptides. Proc Natl Acad Sci U S A 2003, 100, 12683–7. [Google Scholar] [CrossRef] [PubMed]
- Scheiner, S.; Kar, T.; Pattanayak, J. Comparison of Various Types of Hydrogen Bonds Involving Aromatic Amino Acids. J. Am. Chem. Soc. 2002, 124, 13257–13264. [Google Scholar] [CrossRef] [PubMed]
- Escobedo, A.; Topal, B.; Kunze, M.B.A.; Aranda, J.; Chiesa, G.; Mungianu, D.; Bernardo-Seisdedos, G.; Eftekharzadeh, B.; Gairi, M.; Pierattelli, R.; et al. Side Chain to Main Chain Hydrogen Bonds Stabilize a Polyglutamine Helix in a Transcription Factor. Nat. Commun. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Pauling, L.; Corey, R.B.; Branson, H.R. The Structure of Proteins; Two Hydrogen-Bonded Helical Configurations of the Polypeptide Chain. Proc Natl Acad Sci U S A 1951, 37, 205–11. [Google Scholar] [CrossRef] [PubMed]
- Brielle, E.S.; Arkin, I.T. Quantitative Analysis of Multiplex H-Bonds. J. Am. Chem. Soc. 2020, 142, 14150–14157. [Google Scholar] [CrossRef] [PubMed]
- Feldblum, E.S.; Arkin, I.T. Strength of a Bifurcated H Bond. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 4085–4090. [Google Scholar] [CrossRef] [PubMed]
- Manikandan, K.; Ramakumar, S. The Occurrence of C−H···O Hydrogen Bonds in -Helices and Helix Termini in Globular Proteins. Proteins 2004, 56, 768–81. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Chang, H.C.; Jiang, J.C.; Chen, J.C.C.; Kao, H.E.; Lin, S.H.; Lin, I.J.B. C−H···O Hydrogen Bonds in -Sheetlike Networks: Combined X-Ray Crystallography and High-Pressure Infrared Study. J Am Chem Soc 2003, 125, 12358–64. [Google Scholar] [CrossRef] [PubMed]
- Fabiola, G.F.; Krishnaswamy, S.; Nagarajan, V.; Pattabhi, V. C−H···O Hydrogen Bonds in Beta-Sheets. Acta Crystallogr D Biol Crystallogr 1997, 53, 316–20. [Google Scholar] [CrossRef] [PubMed]
- Bella, J.; Berman, H.M. Crystallographic Evidence for C−H···O=C Hydrogen Bonds in a Collagen Triple Helix. J. Mol. Biol. 1996, 264, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Derewenda, Z.S.; Lee, L.; Derewenda, U. The Occurrence of C−H···O Hydrogen Bonds in Proteins. J Mol Biol 1995, 252, 248–62. [Google Scholar] [CrossRef] [PubMed]
- Senes, A.; Ubarretxena-Belandia, I.; Engelman, D.M. The C−H···O=C Hydrogen Bond: A Determinant of Stability and Specificity in Transmembrane Helix Interactions. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 9056–9061. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, S.; Trievel, R.C. Carbon-Oxygen Hydrogen Bonding in Biological Structure and Function. J. Biol. Chem. 2012, 287, 41576–41582. [Google Scholar] [CrossRef] [PubMed]
- Derewenda, Z.S. CH groups as donors in hydrogen bonds: a historical overview and occurrence in proteins and nucleic acids. International Journal of Molecular Sciences 2023, 24, 13165. [Google Scholar] [CrossRef] [PubMed]
- Panigrahi, S.K.; Desiraju, G.R. Strong and Weak Hydrogen Bonds in the Protein-Ligand Interface. Proteins 2007, 67, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Blakeley, M.P.; Langan, P.; Niimura, N.; Podjarny, A. Neutron Crystallography: Opportunities, Challenges, and Limitations. Curr Opin Struct Biol 2008, 18, 593–600. [Google Scholar] [CrossRef] [PubMed]
- Derewenda, Z.S.; Derewenda, U.; Kobos, P.M. (His)C−H···O=C < Hydrogen Bond in the Active Sites of Serine Hydrolases. J Mol Biol 1994, 241, 83–93. [Google Scholar] [CrossRef]
- MacKenzie, K.R.; Prestegard, J.H.; Engelman, D.M. A Transmembrane Helix Dimer: Structure and Implications. Science 1997, 276, 131–3. [Google Scholar] [CrossRef] [PubMed]
- Teese, M.G.; Langosch, D. Role of GxxxG Motifs in Transmembrane Domain Interactions. Biochemistry 2015, 54, 5125–5135. [Google Scholar] [CrossRef] [PubMed]
- Arkin, I.T.; Brunger, A.T. Statistical Analysis of Predicted Transmembrane Alpha-Helices. Biochim Biophys Acta 1998, 1429, 113–28. [Google Scholar] [CrossRef] [PubMed]
- Yohannan, S.; Faham, S.; Yang, D.; Grosfeld, D.; Chamberlain, A.K.; Bowie, J.U. A C−H···O=C Hydrogen Bond in a Membrane Protein Is Not Stabilizing. J Am Chem Soc 2004, 126, 2284–5. [Google Scholar] [CrossRef] [PubMed]
- Arbely, E.; Arkin, I.T. Experimental Measurement of the Strength of a C−H···O=C Bond in a Lipid Bilayer. J. Am. Chem. Soc. 2004, 126, 5362–5363. [Google Scholar] [CrossRef] [PubMed]
- Tadesse, L.; Nazarbaghi, R.; Walters, L. Isotopically Enhanced Infrared-Spectroscopy - a Novel Method for Examining Secondary Structure at Specific Sites in Conformationally Heterogeneous Peptides. J. Am. Chem. Soc. 1991, 113, 7036–7037. [Google Scholar] [CrossRef]
- Torres, J.; Kukol, A.; Goodman, J.M.; Arkin, I.T. Site-Specific Examination of Secondary Structure and Orientation Determination in Membrane Proteins: The Peptidic (13)C=(18)O Group as a Novel Infrared Probe. Biopolymers 2001, 59, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Tomita, M.; Marchesi, V.T. Amino-acid sequence and oligosaccharide attachment sites of human erythrocyte glycophorin. Proc Natl Acad Sci U S A 1975, 72, 2964–8. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A.; Flanagan, J.M.; Hunt, J.F.; Adair, B.D.; Bormann, B.J.; Dempsey, C.E.; Engelman, D.M. Glycophorin a Dimerization Is Driven by Specific Interactions Between Transmembrane Alpha-Helices. J Biol Chem 1992, 267, 7683–9. [Google Scholar] [CrossRef] [PubMed]
- Krimm, S.; Bandekar, J. Vibrational Spectroscopy and Conformation of Peptides, Polypeptides, and Proteins. Adv Protein Chem 1986, 38, 181–364. [Google Scholar] [CrossRef] [PubMed]
- Bormann, B.J.; Knowles, W.J.; Marchesi, V.T. Synthetic Peptides Mimic the Assembly of Transmembrane Glycoproteins. J Biol Chem 1989, 264, 4033–7. [Google Scholar] [CrossRef] [PubMed]
- Bormann, B.J.; Engelman, D.M. Intramembrane Helix-Helix Association in Oligomerization and Transmembrane Signaling. Annu Rev Biophys Biomol Struct 1992, 21, 223–42. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, L.; Schlegel, H.B. On the Physical Origin of Blue-Shifted Hydrogen Bonds. J Am Chem Soc 2002, 124, 9639–47. [Google Scholar] [CrossRef] [PubMed]
- Boese, A.D. Density Functional Theory and Hydrogen Bonds: Are We There Yet? ChemPhysChem 2015, 16, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Mineev, K.S.; Bocharov, E.V.; Volynsky, P.E.; Goncharuk, M.V.; Tkach, E.N.; Ermolyuk, Y.S.; Schulga, A.A.; Chupin, V.V.; Maslennikov, I.V.; Efremov, R.G.; et al. Dimeric Structure of the Transmembrane Domain of Glycophorin a in Lipidic and Detergent Environments. Acta Naturae 2011, 3, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, R.; Dannenberg, J.J. H-bonding cooperativity and energetics of alpha-helix formation of five 17-amino acid peptides. J Am Chem Soc 2003, 125, 8124–9. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.P.; Singh, K.; Hazra, A.; Madhusudhan, M.S. Peptide bond planarity constrains hydrogen bond geometry and influences secondary structure conformations. Curr Res Struct Biol 2021, 3, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Morozov, A.V.; Kortemme, T. Potential functions for hydrogen bonds in protein structure prediction and design. Advances in protein chemistry 2005, 72, 1–38. [Google Scholar] [PubMed]
- Mandala, V.S.; McKay, M.J.; Shcherbakov, A.A.; Dregni, A.J.; Kolocouris, A.; Hong, M. Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat Struct Mol Biol 2020, 27, 1202–1208. [Google Scholar] [CrossRef] [PubMed]
- Alabugin, I.V.; Manoharan, M.; Peabody, S.; Weinhold, F. Electronic basis of improper hydrogen bonding: a subtle balance of hyperconjugation and rehybridization. Journal of the American Chemical Society 2003, 125, 5973–5987. [Google Scholar] [CrossRef] [PubMed]
- Alabugin, I.V.; Bresch, S.; Gomes, G.d.P. Orbital Hybridization: a Key Electronic Factor in Control of Structure and Reactivity. J. Phys. Org. Chem. 2015, 28, 147–162. [Google Scholar] [CrossRef]
- Bent, H.A. An Appraisal of Valence-bond Structures and Hybridization in Compounds of the First-row elements. Chemical Reviews 1961, 61, 275–311. [Google Scholar] [CrossRef]
- Grimme, S. Density functional theory with London dispersion corrections. Wiley Interdisciplinary Reviews: Computational Molecular Science 2011, 1, 211–228. [Google Scholar] [CrossRef]
- Dunning Jr, T.H. Gaussian basis sets for use in correlated molecular calculations. I. The atoms boron through neon and hydrogen. The Journal of chemical physics 1989, 90, 1007–1023. [Google Scholar] [CrossRef]
- Voth, A.R.; Khuu, P.; Oishi, K.; Ho, P.S. Halogen bonds as orthogonal molecular interactions to hydrogen bonds. Nat Chem 2009, 1, 74–9. [Google Scholar] [CrossRef] [PubMed]
- Seyfried, M.S.; Lauber, B.S.; Luedtke, N.W. Multiple-Turnover Isotopic Labeling of Fmoc- and Boc-Protected Amino Acids With Oxygen Isotopes. Organic Letters 2010, 12, 104–106. [Google Scholar] [CrossRef] [PubMed]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-Pdb Viewer: an environment for comparative protein modeling. electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef] [PubMed]
- Poger, D.; Mark, A.E. On the validation of molecular dynamics simulations of saturated and cis-monounsaturated phosphatidylcholine lipid bilayers: a comparison with experiment. Journal of Chemical Theory and Computation 2010, 6, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Kandt, C.; Ash, W.L.; Tieleman, D.P. Setting up and running molecular dynamics simulations of membrane proteins. Methods 2007, 41, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Bekker, H.; Berendsen, H.; Dijkstra, E.; Achterop, S.; Vondrumen, R.v.; Vanderspoel, D.; Sijbers, A.; Keegstra, H.; Renardus, M. Gromacs-a parallel computer for molecular-dynamics simulations. In Proceedings of the 4th international conference on computational physics (PC 92). World Scientific Publishing; 1993; pp. 252–256. [Google Scholar]
- Berendsen, H.J.; van der Spoel, D.; van Drunen, R. GROMACS: A message-passing parallel molecular dynamics implementation. Computer physics communications 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Lindahl, E.; Hess, B.; Van Der Spoel, D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. Molecular modeling annual 2001, 7, 306–317. [Google Scholar] [CrossRef]
- Pronk, S.; Páll, S.; Schulz, R.; Larsson, P.; Bjelkmar, P.; Apostolov, R.; Shirts, M.R.; Smith, J.C.; Kasson, P.M.; Van Der Spoel, D.; et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 2013, 29, 845–854. [Google Scholar] [CrossRef] [PubMed]
- Oostenbrink, C.; Villa, A.; Mark, A.E.; Van Gunsteren, W.F. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force-field parameter sets 53A5 and 53A6. Journal of computational chemistry 2004, 25, 1656–1676. [Google Scholar] [CrossRef] [PubMed]
- Hess, B.; Bekker, H.; Berendsen, H.J.; Fraaije, J.G. LINCS: A linear constraint solver for molecular simulations. Journal of computational chemistry 1997, 18, 1463–1472. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Physical review A 1985, 31, 1695. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied physics 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Nosé, S.; Klein, M. Constant pressure molecular dynamics for molecular systems. Molecular Physics 1983, 50, 1055–1076. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N · log (N) method for Ewald sums in large systems. The Journal of chemical physics 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Berendsen, H.J.; Grigera, J.R.; Straatsma, T.P. The missing term in effective pair potentials. Journal of Physical Chemistry 1987, 91, 6269–6271. [Google Scholar] [CrossRef]
- Shao, Y.; Gan, Z.; Epifanovsky, E.; Gilbert, A.T.; Wormit, M.; Kussmann, J.; Lange, A.W.; Behn, A.; Deng, J.; Feng, X.; et al. Advances in molecular quantum chemistry contained in the Q-Chem 4 program package. Molecular Physics 2015, 113, 184–215. [Google Scholar] [CrossRef]
- Becke, A.D. Density-functional thermochemistry. I. The effect of the exchange-only gradient correction. The Journal of chemical physics 1992, 96, 2155–2160. [Google Scholar] [CrossRef]
- Lee, C.Y.; Parr, W. RG Phys. Rev. B 1988, 37, 785–789. b) Becke, AD. Physical Review A 1988, 38, 3098–3100. [Google Scholar]
- Vosko, S.; Wilk, L.; Nusair, M. Development of the Colle–Salvetti correlation–energy formula into a functional of the electron density. Can J Phys 1980, 58, 1200–1211. [Google Scholar] [CrossRef]
- Stephens, P.J.; Devlin, F.J.; Chabalowski, C.F.; Frisch, M.J. Ab initio calculation of vibrational absorption and circular dichroism spectra using density functional force fields. The Journal of physical chemistry 1994, 98, 11623–11627. [Google Scholar] [CrossRef]
- Woon, D.; Dunning Jr, T. Electron Affinities of the First-Row Atoms Revisited. Systematic Basis Sets and Wave Functions. Journal of Chemical Physics 1993, 98, 1358. [Google Scholar] [CrossRef]
- Witte, J.; Neaton, J.B.; Head-Gordon, M. Effective Empirical Corrections for Basis Set Superposition Error in the def2-SVPD Basis: gCP and DFT-C. J Chem Phys 2017, 146, 234105. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




