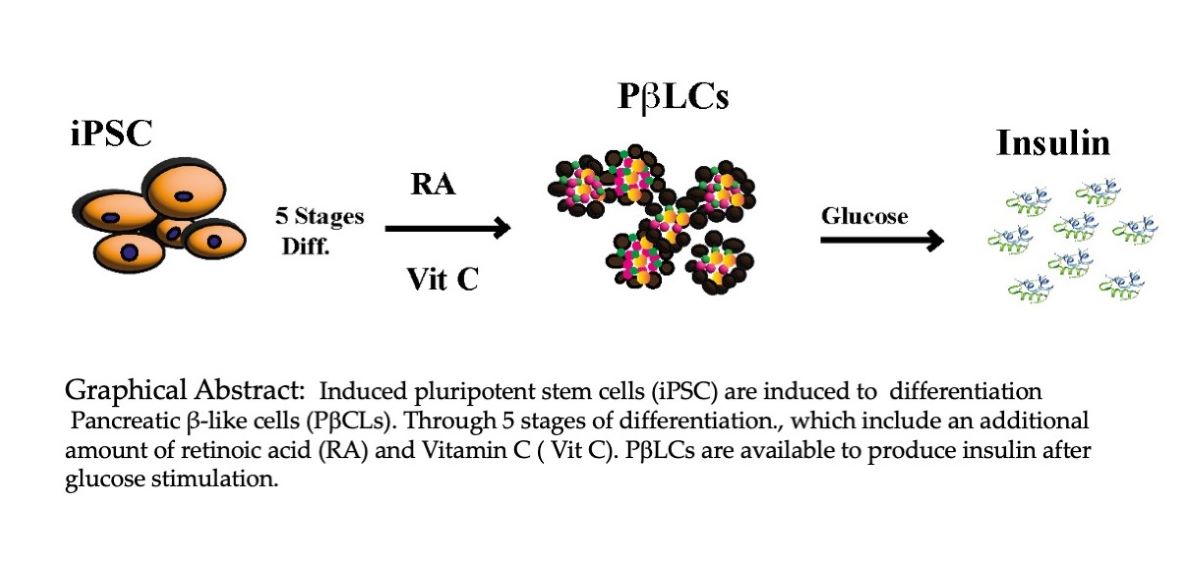
1. Introduction
Diabetes mellitus (DM) is a chronic, non-transmissible disease recognized as a leading cause of mortality globally, with high morbidity due to the chronic deterioration of insulin-producing cells[1]. It is estimated that diabetes affects one in ten individuals worldwide, with a projected global-specific health expenditure that rapidly approaches a trillion USD [2,3]. Furthermore, diabetes induces a series of vascular complications caused by different inflammatory, metabolic, and oxidation stress mechanisms [4,5,6]. Diabetes primarily manifests itself as type 1 diabetes, in which an autoimmune process destroys insulin-secreting cells[7,8], and type 2 diabetes mellitus, characterized by the progressive destruction of islets due to functional alteration of insulin [9]. Although medical therapy can delay complications in T2DM, curing diabetes is a desirable objective worldwide [10]. Given these challenges, regenerative medicine presents a promising solution. Recent years have seen remarkable in organ and tissue regeneration in vitro [11–13]. Endocrine pancreatic tissue regeneration has demonstrated encouraging results, restoring carbohydrate metabolism in primate models and patients with DM [14,15].
Pluripotent Stem Cells (PSCs) have been proposed as an encouraging medical treatment due to their high plasticity and multiple sources. PSCs can be obtained from embryonic tissues, umbilical cords, and some cellular "niches" in the adult tissue [16]. However, these sources of pluripotent cells may have ethical limitations or be restricted and difficult to extract. Reprogramming somatic cells into induced pluripotent stem cells (iPSCs) offers an alternative, increasing the limited supply of pluripotent cells extracted from adult tissue [17]. These cells can be derived from the patients or compatible donors and differentiated to replace affected tissue, thus avoiding immunological rejection and immunosuppressive therapies [18,19].
Several methods to differentiate β-pancreatic cells have been developed recently, utilizing various cofactors and small molecules to replicate pancreatic development in vitro. Molecules like activin A, fibroblast growth factor (bFGF), fibroblast growth factor-10 recombinant protein (FGF10), NOGGIN, and SANT-1 have stimulated the activation of different molecular pathways involved in pancreatic differentiation. However, many questions remain regarding how pancreatic development can be successfully achieved in vitro and how the optimal combination of molecules and cofactors guarantees a substantial number of functional β-cells [20–23].
The widespread nature of DM necessitates access to preventive and therapeutic resources for many patients. Current treatment options are limited, particularly for those with diminished beta cell numbers or complete functional decline, leading to the high cost of chronic treatment. This study aims to introduce a comprehensive protocol for the in vitro generation of insulin-producing pancreatic β cells. Further research into the molecular physiology of beta cells and insulin secretion is crucial for developing effective treatment strategies. We aim to establish a protocol that effectively produces mature insulin-secreting beta cells widely available at low costs.
2. Results
2.1. iPSCs Growth and Maintenance
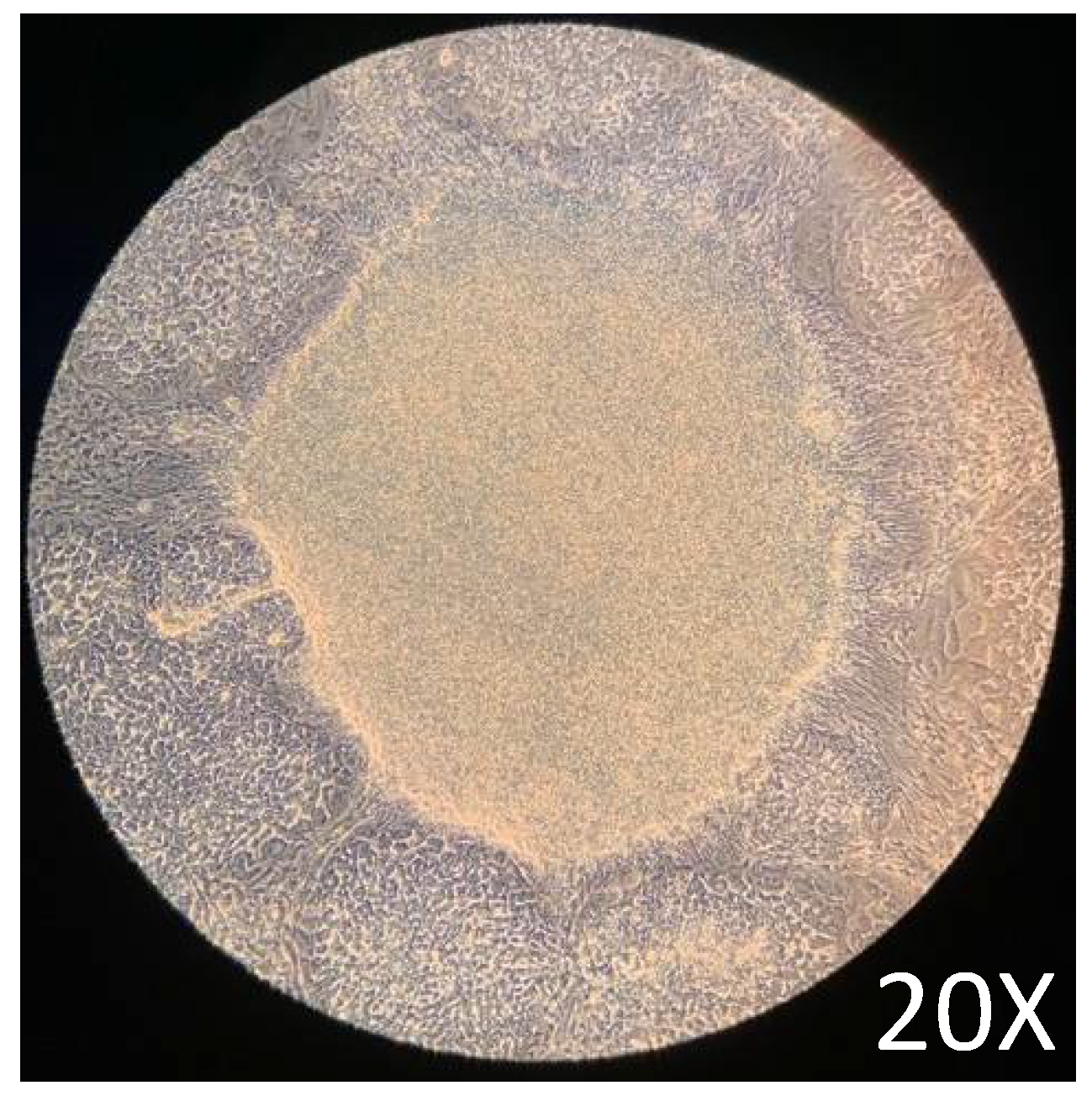
Maintenance of pluripotency before the differentiation assay is critical to guarantee the protocol's good performance. After 72 h of culture in a complete mTeSR1 medium, the cells showed the expected [25] growth characteristics, such as common growth with tightly packaged and distinct borders (
Figure 2). The growth characteristics obtained before the initiation of the differentiation assay were adequate according to previously reported protocols [25,26].
2.2. Differentiation of Definitive Endoderm from iPSCs
This study verified the effects of molecules such as (activin A, CHIR99021, bFGF, FGF10, EGF, SANT-1, retinoic acid, and ascorbic acid) during pancreatic in vitro development. These cytokines and small molecules are essential in activating and repressing signaling pathways such as Wnt, activin A, and Hedgehog, which are involved in pancreatic differentiation.
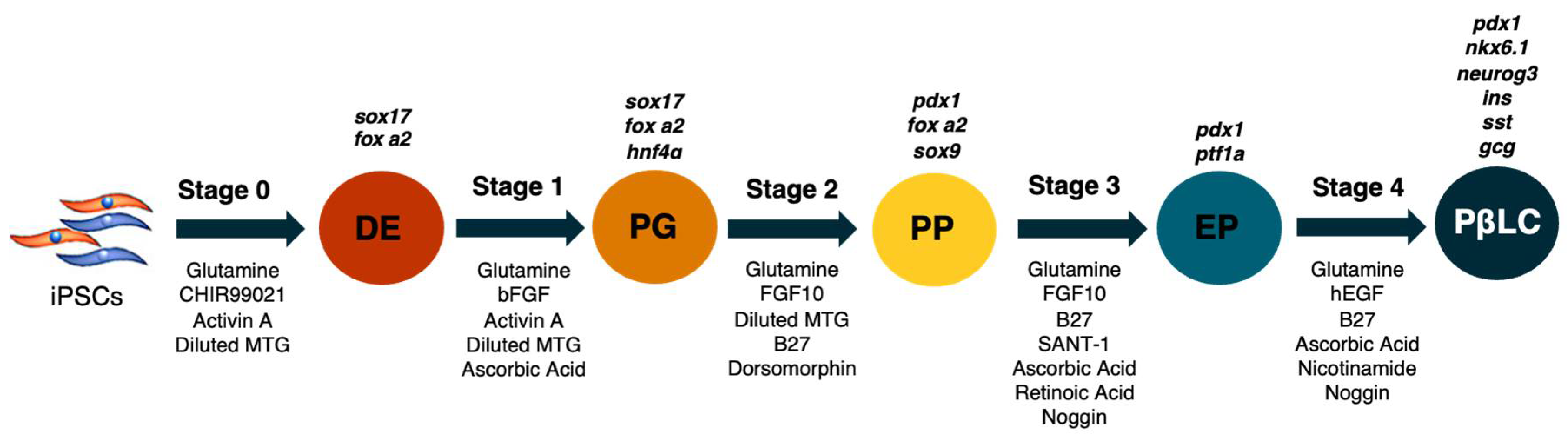
The strategy to generate PβLC in vitro from iPSCs is outlined in
Figure 1. The basal medium was mTeSR1 without specific medium supplementation; the growth medium was only supplemented with cytokines and small molecules, modified from Korytnikov and Nostro [21]. The protocol requires two weeks and is developed as a sequence in which each stage represents a different differentiation period. During each step, the mixture of cofactors and small molecules orchestrates the differentiation of iPSCs into PβLC. The differentiation is developed under serum-free/xeno-free (SF-XF) conditions to avoid any external cofactor that may interfere with the results, such as the fetal bovine serum (FBS) [27–29].
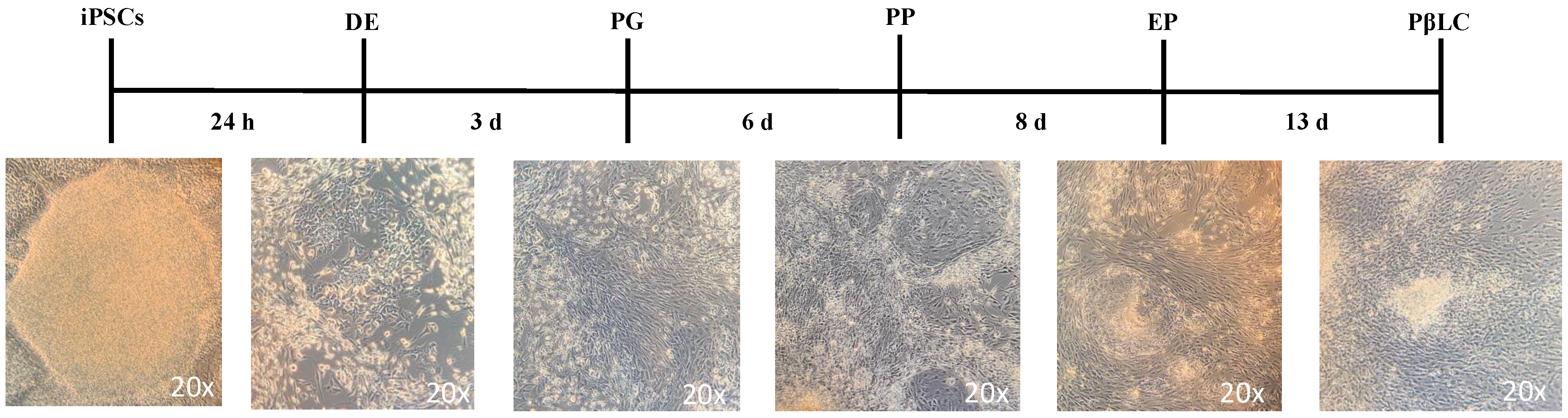
Figure 3.
Schematic representation of the protocol to differentiate iPSCs into PβLCs. iPSCs were seeded on vitronectin-coated cell wells and incubated with the cofactors and small molecules indicated in Fig 1 in a sequential protocol for 13 days. Images were captured with a phase contrast microscope (ZEISS AX10). Abbreviations: DE: Definitive endoderm; PG: Pancreatic gut tube; PP: Pancreatic progenitor; EP: Endocrine progenitor; PβLC: Pancreatic beta-like cell. h: hours, d: days.
Figure 3.
Schematic representation of the protocol to differentiate iPSCs into PβLCs. iPSCs were seeded on vitronectin-coated cell wells and incubated with the cofactors and small molecules indicated in Fig 1 in a sequential protocol for 13 days. Images were captured with a phase contrast microscope (ZEISS AX10). Abbreviations: DE: Definitive endoderm; PG: Pancreatic gut tube; PP: Pancreatic progenitor; EP: Endocrine progenitor; PβLC: Pancreatic beta-like cell. h: hours, d: days.
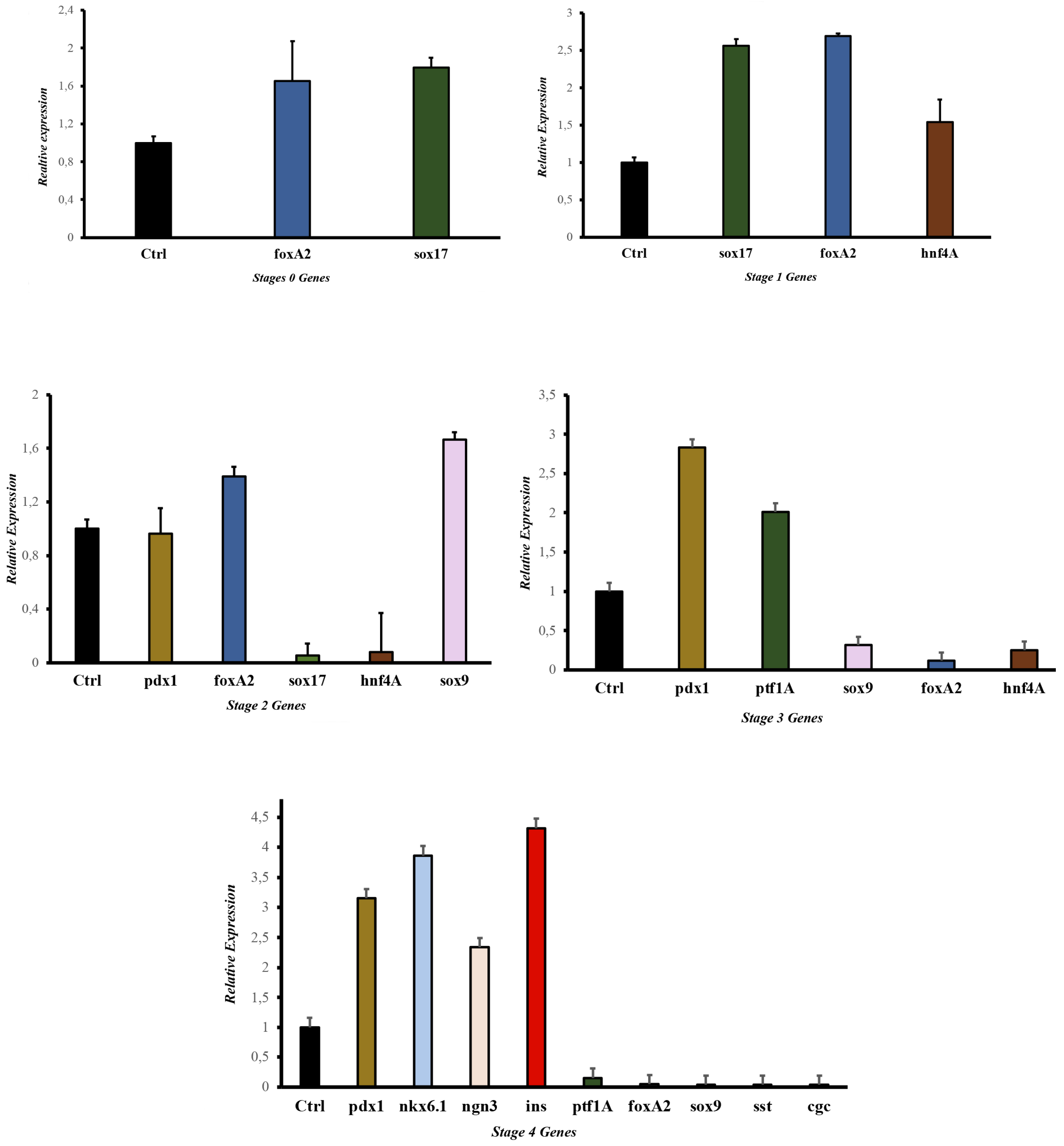
Some selected molecules for the development study have been previously reported responsible for endodermal differentiation, especially toward pancreas development [20,21,30–32]. The protocol allowed us to determine the proteins and genes expressed at each pancreatic differentiation stage. The gene expression and protein results (
Figure 4 and
Figure 5) showed that this methodology activated a process to obtain, in the beginning, cells of the definitive endoderm and, subsequently, PβLC that express characteristic pancreatic β cell markers. Notably, at the end of our protocol, only insulin expression and no other endocrine hormones, such as somatostatin (
sst) and glucagon (
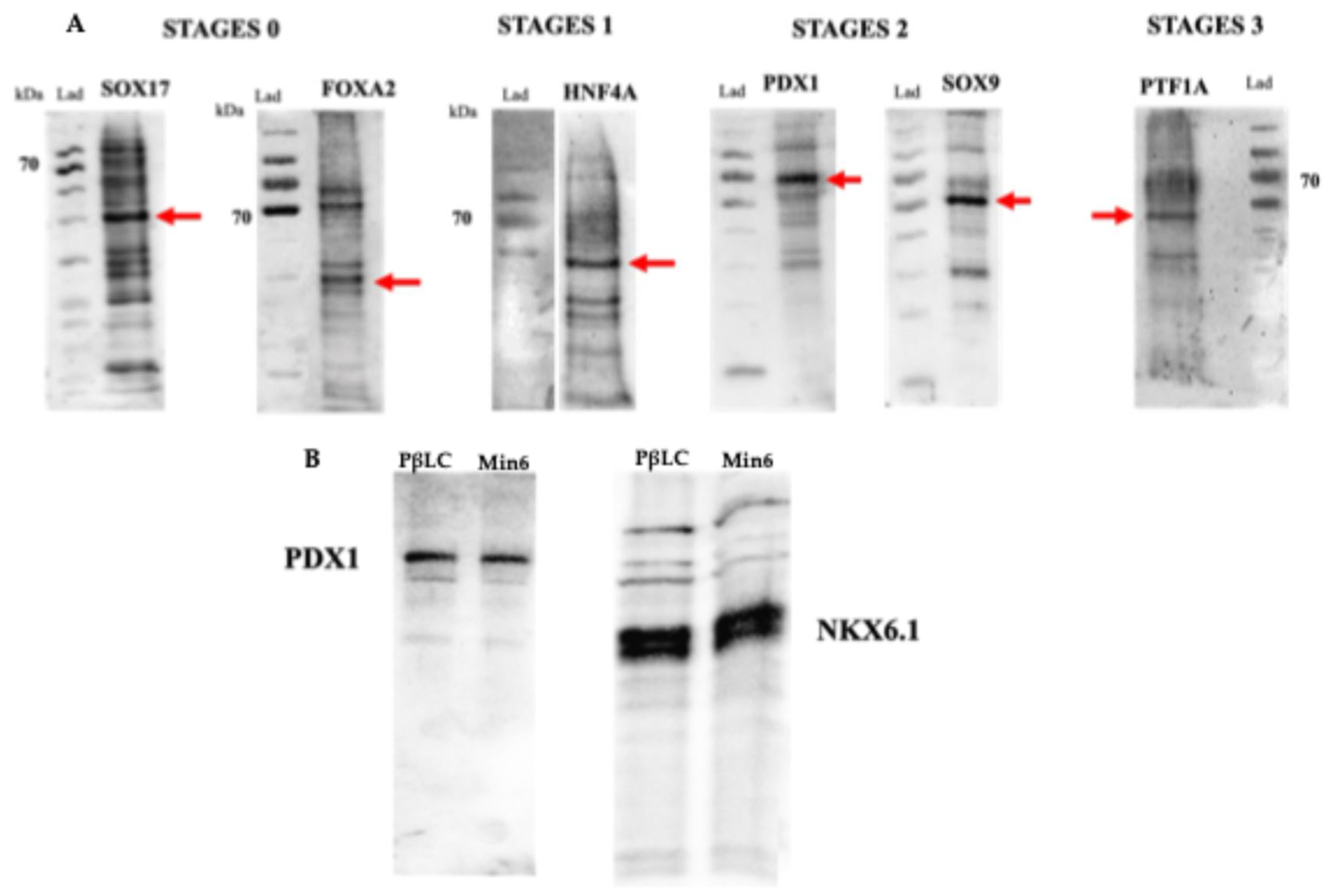
gcg) (detected by qPCR) (Fig 4 E), were obtained, showing that the cytokines and small molecules were adequate to ensure the production of PβLCs. Our results were confirmed by comparing protein expression between our differentiated cells (stage 4) and the insulin-secreting cells (Min6).
The generation of PβLCs was performed with the initial production of DE by incubating iPSCs with a mixture of activin A (100 ng/mL) and CHIR99021 (2 µM/mL) for 24 h. As previously reported, the iPSC cell-derived DE expressed the
sox17 and
foxA2 markers (
Figure 4 A and
Figure 5 A). Next, CHIR99021 was removed from the differentiation medium, and activin A (100 ng/mL) was used for another 24 h with bFGF and ascorbic acid, which allowed the DE-differentiated cells to maintain the lineage. The activin A concentration selected for stage 1 of differentiation was the same as that applied during stage 0 (100 ng/mL). We opted for this concentration due to the potential impact of lower levels of activin A during later stages of endodermal differentiation on the eventual yield of functional cells [31]. We also used a low concentration of CHIR99021 (2 µM/mL), which has been shown to induce endodermal differentiation. Otherwise, higher doses of CHIR99021 (5 mM/mL) can generate mesodermal differentiation [32–35].
2.3. Differentiation of Definitive Endoderm into Pancreatic Progenitors
After DE was achieved, pancreatic progenitors were observed using bFGF, ascorbic acid, and FGF10 molecules. The expression of the
hnf4A and
sox9 (
Figure 4 B, C, and
Figure 5 A) demonstrated the maintenance of pancreatic identity during differentiation. In the present protocol, CHIR99021 was removed after 24 h and replaced with ascorbic acid. This step allowed us to obtain a strong expression of
hnf4A and the continued expression of
sox17 and
foxA2 [20,27,28]. In addition, the concentration and exposure time of FGF10 used during our protocol conferred the cells an endocrine identity determined by the later expression of
pdx1 (subsequently detected) mediated by the reduction of the NOTCH signaling pathway (restricted to endocrine differentiation). Maintaining a PP differentiation stage is demonstrated by the later expression of characteristic genes such as
ptf1a (
Figure 4 D) [36–38].
Notably, our results demonstrate a process of pancreatic differentiation confirmed by the expression of
sox9 during the stage 3 protocol (
Figure 4 C).
sox9 is a characteristic marker of pancreatic differentiation. This molecule is exclusively expressed by the endocrine precursor cells during the expansion of the pancreatic epithelium, favoring the initial expression of
pdx1, as observed in the results (
Figure 4 C, D) [39].
Pancreatic differentiation from DE to PP is closely related to the activation and suppression of the NOTCH, Wnt, BMP, and FGF signaling pathways due to the expression of the
sox9 and
pdx1 genes related to pancreatic differentiation (
Figure 4 C, D, and E). However, during experimentation, hepatic differentiation genes such as
hnf4A and
foxA2 expression suggested a close association between the pancreatic and hepatic progenitors (
Figure 4 C, D) [40].
2.4. Differentiation of Pancreatic Progenitors into Pancreatic Beta-Like Cells
After obtaining PP, our protocol used a mix of cofactors and small molecules to minimize the expression of signaling pathways such as NOTCH, bone morphogenic proteins (BMP), and transforming growth factor beta (TGF-β) [41]. The cofactors and small molecules used during the S3 and the S4 allowed the expression of pancreatic β cell characteristic genes such as
ngn3, pdx1, nkx6.1, and ins (insulin) [42] (
Figure 4, and
Figure 5). However, it is noteworthy that signaling pathways such as NOTCH, BMP, and TGF-β are essential at the beginning of the pancreatic differentiation, where molecules such as activin A, glutamine, and CHIR99021 are used to favor and block their expression to allow hiPSCs to differentiate to a DE state. Nevertheless, with the development of the present protocol, those signaling pathways are suppressed, especially the BMP, because it is highly associated with hepatic differentiation. Therefore, we used Noggin (a BMP inhibitor) and SANT-1 to promote pancreas specification and stop the cells of a hepatic lineage differentiation [43].
The final step was to develop PβLC from PP. Notably, the
ngn3 gene must be expressed at the end of the protocol and not in other stages to avoid the differentiation of polyhormonal cells (alpha, gamma, etc.) (
Figure 4). Early expression of the
ngn3 gene will result in many polyhormonal cells [44]. As previously reported, our protocol used vitamin C at this differentiation stage (
Figure 1) to prevent polyhormonal cell formation [43].
During the last step of our protocol, our research group decided to add retinoic acid again due to its considerable importance in increasing the expression of
pdx1, which favors the differentiation and maturation of pancreatic β cells. However, to guarantee the success of this process, half the concentration of retinoic acid was used in the S4 medium compared to the S3 medium because using low concentrations of this cofactor during the final stages of differentiation is crucial to maintain constant
pdx1 expression (
Figure 4 C, D, and E) and favor the initiation of
nkx6.1 gene expression (
Figure 4 E), which seems to be restricted exclusively in pancreatic β cells [38,45,46].
2.5. Glucose-Stimulate Insulin Secretion
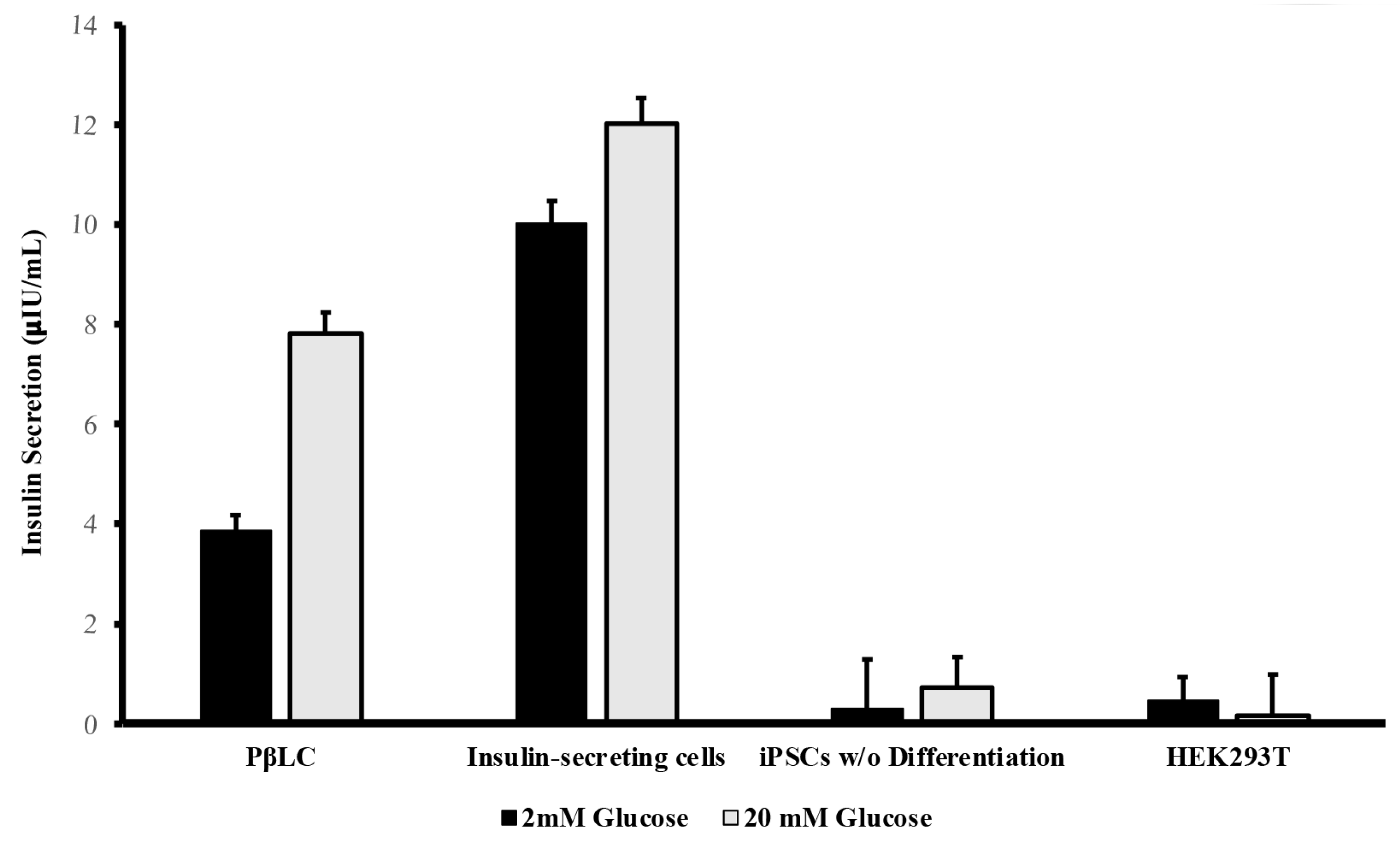
PβLCs underwent a glucose challenge to assess their glucose-stimulated insulin secretion (GSIS) capability.
Figure 6 shows the results obtained from the insulin Elisa test for the PβLC, the insulin-secreting cells (Min6), and two negative controls (iPSCs w/o differentiation and HEK293T cell line).
The insulin secretion by the differentiated cells underscores the effectiveness of the protocol developed by our group. While the insulin levels from PβLC were not as high as those observed from the insulin-secreting cells, their capacity to respond to glucose by measurably secreting insulin in vitro suggests potential functionality. However, further research is essential to fully determine the differentiated cells' maturation status and functionality in future rodent models. Generally, as previously reported by insulin-producing pdx1+/nkx6.1+ cells, they will show functionality and maturation during in vivo graft experiments.
3. Discussion
In the current study, iPSC cells were differentiated into insulin-producing Pancreatic β-like cells using small molecules and cofactors that mimicked embryonic development through activation/inhibition of cell-specific signaling [47]. The insulin secretion by the PβLCs was subject to glucose concentration in the medium, making it a good indicator of real insulin-producing beta-like cells. Our protocol is based on previous protocols, with some modifications to reduce the effect of uncontrolled external factors and to stimulate the differentiation of insulin-producing cells [48–50].
The morphological changes detected during the protocol demonstrate that the combined cofactors and small molecules selected during the study effectively differentiated into iPSCs. However, some apoptotic cells are present during the differentiation protocol, likely due to changes in mitochondrial function triggering typical apoptotic behavior (Kim & Hyun, 2016).
During differentiation, apoptosis in iPSCs is mainly attributed to p53 mitochondrial translocations, cytochrome c release, and reactive oxygen species (ROS) production.
Figure 3 illustrates a reduction in apoptotic cells by the end of the protocol. This reduction may result from the maturation process, which decreases the cell proliferation rate and regulates p53, thereby inhibiting apoptosis [51,52].
Initially, iPSCs must differentiate towards definitive endoderm (DE), with sox17 and foxa2 being relevant markers in this early state. Activation of the Wnt pathway and TGF is crucial for this differentiation process. Various concentrations of Activin A and CHIR99021 have been employed. In the current protocol, we observed that using Activin A for 72 hours at a concentration of 2 μM/mL and 100 ng/mL of CHIR99021 during 24 hours was suitable for expressing DE markers (
Figure 4 A and
Figure 5 A) [43,53,54].
After obtaining the endodermal lineage, we directed the cells to the pancreatic gut tube (PG) (stage 1 of our differentiation protocol), which is considered the period in which the cell fate is determined to be the liver or pancreatic lineage, so the mixture of cofactors and small molecules should be used appropriately [47] [43,55]. The induction of the pancreatic lineage was performed by bFGF 5 ng/mL, a cofactor responsible for the development of the pancreatic epithelium [37]. It is important to consider that lower doses of FGF have been used for hepatic-hepatic-like differentiation [56]. In this stage, the expression of
hnf4a and the preservation of
foxA2 and sox17 expression are good indicators of pancreatic gut differentiation (
Figure 4 A, B) [20,28].
FGF10 and dosomorphin are added to develop the pancreatic lineage, which initiates the unfolding of pancreatic epithelium. At this stage, pdx1 and sox9 are reliable indicators of pancreatic progenitors (
Figure 4 C and
Figure 5 A) associated with the definitive establishment of the pancreatic lineage. The presence of the transcription factor
pdx1 manifests the identity of the future cells [57], which will later transform into insulin-producing β cells. FGF 10 is a fibroblast growth factor that induces epithelial branching, and the addition of B27 as a supplement increases the development of the cells. We decided to maintain the use of B27 to promote and preserve the viability of our cells during the assay and reduce the stress and possible apoptosis caused by the continuous change of the medium [58,59]. FGF10 significantly impacts differentiation since it helps develop the pancreatic epithelium, restricting premature endocrine differentiation and maintaining the progenitor state of the cells through the activation of the notch signaling pathway [60]. Sox9 is essential in specifying and maintaining pancreatic progenitor cells and differentiation towards the beta cell lineage, which later differentiate into endocrine progenitors expressing
pdx1, ngn3, and ptf1a [61,62]. Moreover, the cells' expression of the
sox9 gene during the differentiation protocol is a clear signal of the expansion of the pancreatic epithelium [39].
Subsequently, we achieved endocrine precursor cells characterized by the expression of markers such as
ptf1A (
Figure 4 D). It is essential to highlight the use of cofactors such as FGF10, SANT-1, ascorbic acid, retinoic acid, and Noggin, which contribute to establishing pancreatic β identity during development. Notably, the continued use of FGF10 is needed during these final stages (PP and EP) because a deficiency of this cofactor can trigger hepatic differentiation [62–64]. Noggin (BMP inhibitor) and SANT-1 (inhibitor of sonic hedgehog (signaling) also prevented hepatic differentiation [48,65]. Likewise, ascorbic and retinoic acids became fundamental during this step, contributing to cell identity's final establishment. Ascorbic acid prevents the formation of polyhormonal cells, while retinoic acid acts as an activator of the
pdx1 gene, increasing its expression and thus favoring future insulin expression [43]. Ascorbic acid was earlier used (
Figure 1, S1) to reduce the expression of ngn3 and thus prevent the appearance of acinar cells [66].
Finally, we obtained PβLCs from EP due to cofactors such as hEGF, nicotinamide, ascorbic acid, and retinoic acid. During this last step of our protocol, the production of insulin and the absence of other endocrine hormones, such as SST and GCG (
Figure 4 E), may be considered a positive result due to the absence of polyhormonal cells. Once differentiated cells obtain pancreatic identity, retinoic acid is crucial due to the production of diverse retinoic acid derivates produced by the enzyme retinaldehyde dehydrogenase type II [67] that promotes pancreatic development at an early stage [68] and later supports beta cell differentiation [69,70].
The determination of
nkx6.1 expression (
Figure 4 E) indicates the progression of a pancreatic progenitor state to a PβLC.
nkx6.1 is considered a transcription factor exclusively expressed by adult β-pancreatic cells (
Figure 4 E and
Figure 5 B) because it is directly related to the glucose metabolism of the Glut 2 gene [57].
The last step of our protocol was characterized by the effects of hEGF, which promoted the expansion of pancreatic cells, improving the number of
pdx1-positive cells [71]. Likewise, using nicotinamide and noggin induces the differentiation and maturation of cells that coexpress
pdx1/nkx6.1 [72,73], which may subsequently mature into functional β-cells [74], (
Figure 4 E and
Figure 5 B).
In this study, we employed a protocol utilizing two key compounds, ascorbic acid and retinoic acid, to enhance the differentiation of pancreatic beta cells [20,21,30,75,76]. The rationale behind ascorbic acid and retinoic acid is their epigenetic influence, particularly their ability to increase gene expression through activating DNA demethylation proteins, such as the TET (ten-eleven translocation) enzymes [77,78]. Ascorbic acid is believed to significantly impact the differentiation and maintenance of pancreatic β-cell identity by influencing insulin release [79]. This effect is attributed to acid ascorbic's potential to modify the conductivity of ion channels, which in turn affects glucose metabolism and cellular uptake.
During our study, we reintroduced retinoic acid at the final stage of differentiation due to its crucial role in regulating the expression of the transcription factor Pdx1, which is essential for pancreatic development. However, it was necessary to adjust the concentration of retinoic acid carefully[66,80]. While it benefits pancreatic differentiation, an excessive amount can inhibit endocrine differentiation and disrupt insulin expression [81,82]. Therefore, maintaining an optimal concentration of retinoic acid is vital to promote pancreatic differentiation and β-cell maturation while preventing the formation of undesired cell phenotypes within the pancreatic islets[41,83].
4. Materials and Methods
4.1. Materials
The 20b iPSCs were acquired from the Harvard Stem Cell Institute (HSCI) (Boston, United States). Vitronectin (VTN-N; A14700) Recombinant human protein, truncated, Dulbecco´s phosphate-buffered saline without calcium and magnesium (DPBS; 14190250), L-Glutamine (25030081), Activin A recombinant human protein (PHC9564), heat-stable recombinant human basic bFGF, heat stable FGF10 (PHG0375), B27 supplement minus vitamin A (12587010), human Noggin recombinant protein (PHC1506), human epidermal growth factor recombinant protein (EGF), Dulbecco´s modified Eagle’s medium (DMEM) plus GlutaMax10569010) and, fetal bovine serum (FBS) (A5256701) were obtained from Thermo Scientific, United States; mTeSR1 (85850) medium was acquired from StemCell Technologies (Canada). CHIR99021 (SML1046), L-Ascorbic acid (A4544), dorsomorphin (P5499), nicotinamide (N0636), retinoic acid (R2625), monothioglycerol (MTG6145) and penicillin/streptomycin (P4333) were obtained from Sigma-Aldrich, United States; SANT-1 (559303-5MG) was acquired from Merck-Millipore, Germany.
Mammalian protein extraction reagent (M-PER 78501), Halt protease, phosphatase inhibitor cocktail, and Tween™ 20 Surfact-Amps™ detergent solution were obtained from Thermo Scientific, United States. A bicinchoninic acid kit (BCA) was acquired from GBioscience (United States). Laemmli sample buffer (2x, 161-0737) was acquired from (Bio-Rad, United States). Primary antibodies against forkhead box A2 (FOXA2) (710730), SRY-box transcription factor 17 (SOX17) (PA5-72815), hepatocyte nuclear factor alpha (HNF4A) (PA5-82159), SRY-box transcription factor 9 (SOX9) (PA5-81966), pancreas associated transcription factor 1a (PTF1A) (PA5-112677), and Neurogenin 3 (NeuroG3) (703206), goat anti-rabbit IgG (H+L), secondary antibodies, and horseradish peroxidase conjugate (G21234) were obtained from Invitrogen, United States. A recombinant pancreatic and duodenal homeobox 1 (PDX1) (ab219207) and a recombinant NK6.1 homeobox 1 (NKX6.1) antibodies (ab221549) were obtained from Abcam, United Kingdom. Immobilon-P PVDF membrane (IPVH00010) and Luminata Crescendo Western HRP Substrate were obtained from Merck Millipore, Darmstadt, Germany. A QuickExtract™ RNA extraction kit (QER090150) was obtained from Biosearch Technologies (United States), and OneScript® Plus Reverse Transcriptase (G237) was obtained from ABN (Canada).
4.2. iPSC Culture and Maintenance
The 20b iPSCs were cultured in 6-well plates coated with vitronectin and mTeSR1 medium. The plates were incubated at 37°C and 5% CO2 during all expansion and differentiation stages. Min6 cells (mouse insulinoma six cells) were kindly donated and used as an insulin-secreting result for β-cell differentiation [24]. Min-6 cells were cultured in DMEM with 20% fetal bovine serum, 1% Ciprofloxacin (Sigma-Aldrich, United States), and 1% penicillin/streptomycin. All the plates were incubated at 37°C and 5% CO2 during all the cultures, including expansion. Protein extraction was performed on Min6 cells.
The iPSC culture was fed following this procedure daily: the culture media was aspirated, and 2 mL of fresh mTeSR1 medium per well was added. The culture medium was changed, and the cells were washed with 2 mL of DPBS until they reached a growth confluency of 85-90%. Once the iPSC culture was 85-90% confluent, the cells were transferred to new pre-coated vitronectin 12-well plates.
4.3. Differentiation of iPSCs
The differentiation protocol was defined in 5 stages (
Figure 1). In each stage, the mTeSR1 medium was supplemented with a diverse mixture of cytokines and small molecules to achieve a step-by-step differentiation process. Each step was carried out in triplicate under the same incubation conditions at 37°C and 5% CO
2.
Figure 1.
Representative scheme of the differentiation process. Abbreviations: DE: Definitive endoderm; PG: Pancreatic gut tube; PP: Pancreatic progenitor; EP: Endocrine progenitor; PβLC: Pancreatic beta-like cell. Gene expression is represented in black at the top of each differentiation stage. The arrows describe the cytokines and small molecules required to perform the protocol. Modified from Korytnikov & Nostro (2016).
Figure 1.
Representative scheme of the differentiation process. Abbreviations: DE: Definitive endoderm; PG: Pancreatic gut tube; PP: Pancreatic progenitor; EP: Endocrine progenitor; PβLC: Pancreatic beta-like cell. Gene expression is represented in black at the top of each differentiation stage. The arrows describe the cytokines and small molecules required to perform the protocol. Modified from Korytnikov & Nostro (2016).
Standard feed differentiation medium (SFD) and diluted MTG medium were prepared according to Korytnikov and Nostro [21]. Once a stage was performed, protein extraction, total RNA extraction, western blot assays, and RT-PCR were carried out. Different stages were performed as follows:
- (1)
Cells in 12-well plates were cultured until 85-90% confluency. mTeSR1 was changed daily, while DPBS w/o Ca++ and Mg++ was used to wash the wells [20,21].
- (2)
Day 0: Stage 0 medium (S0) was supplemented with glutamine (1%), CHIR99021 (2 µM, activin A (100 ng/mL), and diluted MTG (3 µL/mL). The cells were incubated with an S0 medium for 24 hours.
- (3)
Days 1 - 2: The S1 medium was removed, and the cells were washed twice with DPBS w/o Ca++ and Mg++. S1 medium was supplemented with glutamine (1%), Activin A (100 ng/mL), diluted MTG (3 µL/mL), heat-stable recombinant human basic fibroblast growth factor (bFGF) (5 ng/mL), and ascorbic acid (50 µg/mL). The medium was changed each day during stage 1.
- (4)
Days 3 - 5: The S2 medium was removed, and the cells were rewashed with DPBS w/o Ca++ and Mg++ twice. S2 medium was supplemented with glutamine (1%), diluted MTG (3 µL/mL), dorsomorphin (0,75 µM), human fibroblastic growth factor-10 (FGF10) (50 ng/mL), and B27 supplement minus vitamin A (1%). Subsequently, the cells were incubated with the S2 medium, and the medium was changed each day during stage 2.
- (5)
Days 6 - 7: The S3 medium was removed, and the cells were washed twice. S3 medium was supplemented with glutamine (1%), ascorbic acid (50 µg/mL), FGF10 (50 ng/mL), B27 (1%), SANT-1 (0,25 µM), retinoic acid (2 µM) and recombinant human noggin (Noggin) (50 ng/mL). Cells during stage 3 were incubated at 37°C, 5% and CO2, and the medium was changed daily.
- (6)
Finally, on days 8-12, the cells were cultivated with S4 medium supplemented with glutamine (1%), ascorbic acid (50 µg/mL), B27 (1%), Noggin (50 ng/mL), recombinant human epidermal growth factor (hEGF) (100 ng/mL), retinoic acid (1 µM), and nicotinamide (10 mM).
4.4. Western Blot
Once each differentiation stage was completed, the differentiation medium was collected, and the cells were washed twice with DPBS. Subsequently, the cells were homogenized in a mammalian protein extraction buffer (M-PER, ThermoScientific) with a protease and phosphatase inhibitor cocktail (Halt, ThermoScientific). The bicinchoninic acid assay (BCA) was used to determine the extracted protein content. Differentiation-related protein analysis was carried out using SDS electrophoresis and western blotting. Briefly, equal protein amounts (20-30 µg/mL) were mixed with an equal volume of Laemmli sample buffer, denatured at 70 °C for 10 min, separated by 12% SDS-PAGE with a constant voltage of 100V for 2h, and then transferred to a PVDF membrane. The membrane was incubated in blocking buffer (TBST 1X + Tween-20 (0,1% + 5% skim milk powder) for 1h at room temperature. The membrane was then incubated overnight at 4°C with primary antibodies against SOX17 (5 µg/mL), FOXA2 (4 µg/mL), HNF4A (0,4 µg/mL), recombinant anti-PDX1 antibody (5:500) and recombinant anti-NKX6.1 antibody (5:500), SOX9 (0,4 µg/mL), PTF1A (1:5000), and NGN3 (1:1000). Subsequently, the membranes were washed three times with 1X TBST and incubated with HRP goat anti-rabbit IgG (H+L) secondary antibody (1:5000) for 2 h at room temperature. Finally, the membrane was exposed for 5 minutes at room temperature to Luminata™ Crescendo Western HRP Substrate and visualized by chemiluminescence using a myECL imager (ThermoScientific, United States).
4.5. Total RNA Extraction RT-PCR and Quantitative PCR (qPCR)
Total RNA extraction was performed using a QuickExtract™ RNA extraction kit according to the manufacturer´s protocol. Total RNA was quantified using a NanoDrop system (Invitrogen, United States). The cDNA was obtained using OneScript® Plus Reverse transcriptase and oligo (dT) primers. qPCR was performed in triplicate on a CFX Opus 96 Real-Time PCR System (Bio-Rad, United States) with Luna® Universal qPCR Master Mix (M3003S) (New England Biolabs, United States). Expression data were normalized relative to the B-Actin transcript level. The fold change for each gene was calculated using the 2^
-ΔΔCt method. The qPCR conditions were as follows: initial denaturation at 95°C for 1 min followed by 40 cycles of 15 s at 95°C, 30 s at 60°C, and 15 s at 72°C. Primer sequences are shown in
Table 1.
The qPCR conditions were as follows: initial denaturation at 95°C for 1 min followed by 40 cycles of 5 s at 95°C, 10 s at 60°C, and 15s at 72°C. Primer sequences are shown in
Table 1.
4.6. Glucose Stimulation Insulin Secretion (GSIS)
Human insulin levels were determined using the Abcam Human Insulin ELISA kit (AB100578), UK. In summary, Pancreatic beta-like cells (PβLC) underwent a double Krebs-Ringer buffer (Krb) wash and a 2-hour pre-incubation in 2.5 mM glucose Krb. This was followed by a 1-hour incubation in low-glucose Krb (2 mM) and supernatant collection. After another double wash in Krb, the cells were exposed to high-glucose Krb (20 mM) for 1 hour, with subsequent supernatant collection. All measurements were conducted by triplicate.
4.7. Statistical Analysis
All the experiments were carried out at least three times. Data were displayed as mean ± SD. A two-tailed, unpaired Student’s t-test was performed to assess statistical significance, and P < 0.05 was considered significant.
5. Conclusions
In the present protocol, we successfully produced insulin-producing beta cells derived from iPSCs through four differentiation stages. We believe that the epigenetic regulation induced by ascorbic acid and retinoic acid plays a critical role in the differentiation of beta cells from iPSCs. A significant challenge in beta cell differentiation protocols is simplifying the production of mature beta cells and making them affordable for the general population worldwide. We consider that epigenetic regulation may play a principal role in these differentiation protocols, and it is possible that other small molecules can further enhance the differentiation of beta cells. Various protocols are currently under development, but the main goal remains to obtain pancreatic beta cells that are functionally active and capable of sustained insulin release in response to physiological cues.
Author Contributions
Conceptualization, FL, and FA.; methodology, FA, and FL.; formal analysis, FL.; investigation, FA, YU.; resources, FA and FL.; data curation, FL.; writing—original draft preparation, FA.; writing—review and editing, FL.; visualization, FL; supervision, FL.; project administration, FA and FL.; funding acquisition, FA and FL. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Direccion General de Investigacion (DIN) from Universidad de La Sabana, grant number MED 255-2019 and MED 292-2020.
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
The authors especially thank Lina Mendez for her technical support and cooperation and Yazmin Lopez for acquiring the financial funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Arroyave, F.; Montano, D.; Lizcano, F. Diabetes Mellitus Is a Chronic Disease that Can Benefit from Therapy with Induced Pluripotent Stem Cells. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Saadi, H.; Nagelkerke, N.; Carruthers, S.G.; Benedict, S.; Abdulkhalek, S.; Reed, R.; Lukic, M.; Nicholls, M.G. Association of TCF7L2 polymorphism with diabetes mellitus, metabolic syndrome, and markers of beta cell function and insulin resistance in a population-based sample of Emirati subjects. Diabetes Res Clin Pract 2008, 80, 392–398. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.; Karuranga, S.; Malanda, B.; Saeedi, P.; Basit, A.; Besancon, S.; Bommer, C.; Esteghamati, A.; Ogurtsova, K.; Zhang, P.; et al. Global and regional estimates and projections of diabetes-related health expenditure: Results from the International Diabetes Federation Diabetes Atlas, 9th edition. Diabetes Res Clin Pract 2020, 162, 108072. [Google Scholar] [CrossRef] [PubMed]
- Zinman, B.; Skyler, J.S.; Riddle, M.C.; Ferrannini, E. Diabetes Research and Care Through the Ages. Diabetes Care 2017, 40, 1302–1313. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, N.; Sahay, R.; Kalra, S.; Bajaj, S.; Dasgupta, A.; Shrestha, D.; Dhakal, G.; Tiwaskar, M.; Sahay, M.; Somasundaram, N.; et al. Consensus on Medical Nutrition Therapy for Diabesity (CoMeND) in Adults: A South Asian Perspective. Diabetes Metab Syndr Obes 2021, 14, 1703–1728. [Google Scholar] [CrossRef]
- Finucane, M.M.; Stevens, G.A.; Cowan, M.J.; Danaei, G.; Lin, J.K.; Paciorek, C.J.; Singh, G.M.; Gutierrez, H.R.; Lu, Y.; Bahalim, A.N.; et al. National, regional, and global trends in body-mass index since 1980: systematic analysis of health examination surveys and epidemiological studies with 960 country-years and 9.1 million participants. Lancet 2011, 377, 557–567. [Google Scholar] [CrossRef]
- American Diabetes, A. Diagnosis and classification of diabetes mellitus. Diabetes Care 2010, 33 Suppl 1, S62–69. [Google Scholar] [CrossRef]
- Boland, B.B.; Rhodes, C.J.; Grimsby, J.S. The dynamic plasticity of insulin production in beta-cells. Mol Metab 2017, 6, 958–973. [Google Scholar] [CrossRef] [PubMed]
- Garber, A.J.; Abrahamson, M.J.; Barzilay, J.I.; Blonde, L.; Bloomgarden, Z.T.; Bush, M.A.; Dagogo-Jack, S.; DeFronzo, R.A.; Einhorn, D.; Fonseca, V.A.; et al. Consensus Statement by the American Association of Clinical Endocrinologists and American College of Endocrinology on the Comprehensive Type 2 Diabetes Management Algorithm - 2019 Executive Summary. Endocr Pract 2019, 25, 69–100. [Google Scholar] [CrossRef]
- Phillip, M.; Bergenstal, R.M.; Close, K.L.; Danne, T.; Garg, S.K.; Heinemann, L.; Hirsch, I.B.; Kovatchev, B.P.; Laffel, L.M.; Mohan, V.; et al. The Digital/Virtual Diabetes Clinic: The Future Is Now-Recommendations from an International Panel on Diabetes Digital Technologies Introduction. Diabetes Technol Ther 2021, 23, 146–154. [Google Scholar] [CrossRef]
- Isobe, K.; Cheng, Z.; Nishio, N.; Suganya, T.; Tanaka, Y.; Ito, S. iPSCs, aging and age-related diseases. N Biotechnol 2014, 31, 411–421. [Google Scholar] [CrossRef]
- Stirban, A.O.; Tschoepe, D. Cardiovascular complications in diabetes: targets and interventions. Diabetes Care 2008, 31 Suppl 2, S215–221. [Google Scholar] [CrossRef]
- Suchy, F.; Yamaguchi, T.; Nakauchi, H. iPSC-Derived Organs In Vivo: Challenges and Promise. Cell Stem Cell 2018, 22, 21–24. [Google Scholar] [CrossRef]
- Rickels, M.R.; Robertson, R.P. Pancreatic Islet Transplantation in Humans: Recent Progress and Future Directions. Endocr Rev 2019, 40, 631–668. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Liang, Z.; Wang, S.; Sun, D.; Wang, X.; Liew, S.Y.; Lu, S.; Wu, S.; Jiang, Y.; Wang, Y.; et al. Human pluripotent stem-cell-derived islets ameliorate diabetes in non-human primates. Nat Med 2022, 28, 272–282. [Google Scholar] [CrossRef]
- Zhou, Q.; Melton, D.A. Pancreas regeneration. Nature 2018, 557, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Wert, K.J.; Shvartsman, D.; Melton, D.A.; Jaenisch, R. Establishment of human pluripotent stem cell-derived pancreatic beta-like cells in the mouse pancreas. Proc Natl Acad Sci U S A 2018, 115, 3924–3929. [Google Scholar] [CrossRef]
- Apostolou, E.; Stadtfeld, M. Cellular trajectories and molecular mechanisms of iPSC reprogramming. Curr Opin Genet Dev 2018, 52, 77–85. [Google Scholar] [CrossRef]
- Malik, N.; Rao, M.S. A review of the methods for human iPSC derivation. Methods Mol Biol 2013, 997, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Pagliuca, F.W.; Millman, J.R.; Gurtler, M.; Segel, M.; Van Dervort, A.; Ryu, J.H.; Peterson, Q.P.; Greiner, D.; Melton, D.A. Generation of functional human pancreatic beta cells in vitro. Cell 2014, 159, 428–439. [Google Scholar] [CrossRef]
- Korytnikov, R.; Nostro, M.C. Generation of polyhormonal and multipotent pancreatic progenitor lineages from human pluripotent stem cells. Methods 2016, 101, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Leite, N.C.; Sintov, E.; Meissner, T.B.; Brehm, M.A.; Greiner, D.L.; Harlan, D.M.; Melton, D.A. Modeling Type 1 Diabetes In Vitro Using Human Pluripotent Stem Cells. Cell Rep 2020, 32, 107894. [Google Scholar] [CrossRef] [PubMed]
- Sim, E.Z.; Shiraki, N.; Kume, S. Recent progress in pancreatic islet cell therapy. Inflamm Regen 2021, 41, 1. [Google Scholar] [CrossRef]
- Miyazaki, J.; Araki, K.; Yamato, E.; Ikegami, H.; Asano, T.; Shibasaki, Y.; Oka, Y.; Yamamura, K. Establishment of a pancreatic beta cell line that retains glucose-inducible insulin secretion: special reference to expression of glucose transporter isoforms. Endocrinology 1990, 127, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, C.; Hale, C.; Mukhopadhyay, S. A Simple Multistep Protocol for Differentiating Human Induced Pluripotent Stem Cells into Functional Macrophages. Methods Mol Biol 2018, 1784, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekaran, V.; Carta, G.; da Costa Pereira, D.; Gupta, R.; Murphy, C.; Feifel, E.; Kern, G.; Lechner, J.; Cavallo, A.L.; Gupta, S.; et al. Generation and characterization of iPSC-derived renal proximal tubule-like cells with extended stability. Sci Rep 2021, 11, 11575. [Google Scholar] [CrossRef] [PubMed]
- Kunisada, Y.; Tsubooka-Yamazoe, N.; Shoji, M.; Hosoya, M. Small molecules induce efficient differentiation into insulin-producing cells from human induced pluripotent stem cells. Stem Cell Res 2012, 8, 274–284. [Google Scholar] [CrossRef] [PubMed]
- Kroon, E.; Martinson, L.A.; Kadoya, K.; Bang, A.G.; Kelly, O.G.; Eliazer, S.; Young, H.; Richardson, M.; Smart, N.G.; Cunningham, J.; et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol 2008, 26, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Andreasson, L.; Evenbratt, H.; Mobini, R.; Simonsson, S. Differentiation of induced pluripotent stem cells into definitive endoderm on Activin A-functionalized gradient surfaces. J Biotechnol 2021, 325, 173–178. [Google Scholar] [CrossRef]
- Millman, J.R.; Xie, C.; Van Dervort, A.; Gurtler, M.; Pagliuca, F.W.; Melton, D.A. Generation of stem cell-derived beta-cells from patients with type 1 diabetes. Nat Commun 2016, 7, 11463. [Google Scholar] [CrossRef]
- D'Amour, K.A.; Bang, A.G.; Eliazer, S.; Kelly, O.G.; Agulnick, A.D.; Smart, N.G.; Moorman, M.A.; Kroon, E.; Carpenter, M.K.; Baetge, E.E. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol 2006, 24, 1392–1401. [Google Scholar] [CrossRef] [PubMed]
- Cuesta-Gomez, N.; Verhoeff, K.; Jasra, I.T.; Pawlick, R.; Dadheech, N.; Shapiro, A.M.J. Characterization of stem-cell-derived islets during differentiation and after implantation. Cell Rep 2022, 40, 111238. [Google Scholar] [CrossRef]
- Jaremko, K.L.; Marikawa, Y. Regulation of developmental competence and commitment towards the definitive endoderm lineage in human embryonic stem cells. Stem Cell Res 2013, 10, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Chen, C.; Randolph, L.N.; Ye, S.; Zhang, X.; Bao, X.; Lian, X.L. Generation of pancreatic progenitors from human pluripotent stem cells by small molecules. Stem Cell Reports 2021, 16, 2395–2409. [Google Scholar] [CrossRef]
- Payne, C.; King, J.; Hay, D. The role of activin/nodal and Wnt signaling in endoderm formation. Vitam Horm 2011, 85, 207–216. [Google Scholar] [CrossRef]
- Ndlovu, R.; Deng, L.C.; Wu, J.; Li, X.K.; Zhang, J.S. Fibroblast Growth Factor 10 in Pancreas Development and Pancreatic Cancer. Front Genet 2018, 9, 482. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.; Francavilla, C. Regulation of FGF10 Signaling in Development and Disease. Front Genet 2018, 9, 500. [Google Scholar] [CrossRef]
- Jin, W.; Jiang, W. Stepwise differentiation of functional pancreatic beta cells from human pluripotent stem cells. Cell Regen 2022, 11, 24. [Google Scholar] [CrossRef]
- Diedisheim, M.; Oshima, M.; Albagli, O.; Huldt, C.W.; Ahlstedt, I.; Clausen, M.; Menon, S.; Aivazidis, A.; Andreasson, A.C.; Haynes, W.G.; et al. Modeling human pancreatic beta cell dedifferentiation. Mol Metab 2018, 10, 74–86. [Google Scholar] [CrossRef] [PubMed]
- Jennings, R.E.; Berry, A.A.; Gerrard, D.T.; Wearne, S.J.; Strutt, J.; Withey, S.; Chhatriwala, M.; Piper Hanley, K.; Vallier, L.; Bobola, N.; et al. Laser Capture and Deep Sequencing Reveals the Transcriptomic Programmes Regulating the Onset of Pancreas and Liver Differentiation in Human Embryos. Stem Cell Reports 2017, 9, 1387–1394. [Google Scholar] [CrossRef]
- Lorberbaum, D.S.; Kishore, S.; Rosselot, C.; Sarbaugh, D.; Brooks, E.P.; Aragon, E.; Xuan, S.; Simon, O.; Ghosh, D.; Mendelsohn, C.; et al. Retinoic acid signaling within pancreatic endocrine progenitors regulates mouse and human beta cell specification. Development 2020, 147. [Google Scholar] [CrossRef]
- Davis, J.C.; Alves, T.C.; Helman, A.; Chen, J.C.; Kenty, J.H.; Cardone, R.L.; Liu, D.R.; Kibbey, R.G.; Melton, D.A. Glucose Response by Stem Cell-Derived beta Cells In Vitro Is Inhibited by a Bottleneck in Glycolysis. Cell Rep 2020, 31, 107623. [Google Scholar] [CrossRef] [PubMed]
- Thakur, G.; Lee, H.J.; Jeon, R.H.; Lee, S.L.; Rho, G.J. Small Molecule-Induced Pancreatic beta-Like Cell Development: Mechanistic Approaches and Available Strategies. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Russ, H.A.; Parent, A.V.; Ringler, J.J.; Hennings, T.G.; Nair, G.G.; Shveygert, M.; Guo, T.; Puri, S.; Haataja, L.; Cirulli, V.; et al. Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. EMBO J 2015, 34, 1759–1772. [Google Scholar] [CrossRef]
- Bastidas-Ponce, A.; Roscioni, S.S.; Burtscher, I.; Bader, E.; Sterr, M.; Bakhti, M.; Lickert, H. Foxa2 and Pdx1 cooperatively regulate postnatal maturation of pancreatic beta-cells. Mol Metab 2017, 6, 524–534. [Google Scholar] [CrossRef]
- Bastidas-Ponce, A.; Scheibner, K.; Lickert, H.; Bakhti, M. Cellular and molecular mechanisms coordinating pancreas development. Development 2017, 144, 2873–2888. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S.; Sagrac, D.; Sahin, F. Differentiation Potential of Mesenchymal Stem Cells into Pancreatic beta-Cells. Adv Exp Med Biol 2020, 1247, 135–156. [Google Scholar] [CrossRef] [PubMed]
- Hashemitabar, M.; Heidari, E. Redefining the signaling pathways from pluripotency to pancreas development: In vitro beta-cell differentiation. J Cell Physiol 2019, 234, 7811–7827. [Google Scholar] [CrossRef]
- Vegas, A.J.; Veiseh, O.; Gurtler, M.; Millman, J.R.; Pagliuca, F.W.; Bader, A.R.; Doloff, J.C.; Li, J.; Chen, M.; Olejnik, K.; et al. Long-term glycemic control using polymer-encapsulated human stem cell-derived beta cells in immune-competent mice. Nat Med 2016, 22, 306–311. [Google Scholar] [CrossRef]
- Marotta, D.; Rao, C.; Fossati, V. Human Induced Pluripotent Stem Cell (iPSC) Handling Protocols: Maintenance, Expansion, and Cryopreservation. Methods Mol Biol 2022, 2454, 1–15. [Google Scholar] [CrossRef]
- Sola, S.; Morgado, A.L.; Rodrigues, C.M. Death receptors and mitochondria: two prime triggers of neural apoptosis and differentiation. Biochim Biophys Acta 2013, 1830, 2160–2166. [Google Scholar] [CrossRef] [PubMed]
- Demine, S.; Schiavo, A.A.; Marin-Canas, S.; Marchetti, P.; Cnop, M.; Eizirik, D.L. Pro-inflammatory cytokines induce cell death, inflammatory responses, and endoplasmic reticulum stress in human iPSC-derived beta cells. Stem Cell Res Ther 2020, 11, 7. [Google Scholar] [CrossRef] [PubMed]
- Naujok, O.; Diekmann, U.; Lenzen, S. The generation of definitive endoderm from human embryonic stem cells is initially independent from activin A but requires canonical Wnt-signaling. Stem Cell Rev Rep 2014, 10, 480–493. [Google Scholar] [CrossRef]
- Yabe, S.G.; Fukuda, S.; Takeda, F.; Nashiro, K.; Shimoda, M.; Okochi, H. Efficient generation of functional pancreatic beta-cells from human induced pluripotent stem cells. J Diabetes 2017, 9, 168–179. [Google Scholar] [CrossRef] [PubMed]
- Sui, L.; Bouwens, L.; Mfopou, J.K. Signaling pathways during maintenance and definitive endoderm differentiation of embryonic stem cells. Int J Dev Biol 2013, 57, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Maehr, R.; Chen, S.; Snitow, M.; Ludwig, T.; Yagasaki, L.; Goland, R.; Leibel, R.L.; Melton, D.A. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc Natl Acad Sci U S A 2009, 106, 15768–15773. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Bader, T.N.; Jin, S. Signaling Molecules Regulating Pancreatic Endocrine Development from Pluripotent Stem Cell Differentiation. Int J Mol Sci 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Salisbury, R.J.; Blaylock, J.; Berry, A.A.; Jennings, R.E.; De Krijger, R.; Piper Hanley, K.; Hanley, N.A. The window period of NEUROGENIN3 during human gestation. Islets 2014, 6, e954436. [Google Scholar] [CrossRef] [PubMed]
- Hussein, S.M.; Batada, N.N.; Vuoristo, S.; Ching, R.W.; Autio, R.; Narva, E.; Ng, S.; Sourour, M.; Hamalainen, R.; Olsson, C.; et al. Copy number variation and selection during reprogramming to pluripotency. Nature 2011, 471, 58–62. [Google Scholar] [CrossRef]
- Bray, S.J. Notch signalling in context. Nat Rev Mol Cell Biol 2016, 17, 722–735. [Google Scholar] [CrossRef]
- Apelqvist, A.; Li, H.; Sommer, L.; Beatus, P.; Anderson, D.J.; Honjo, T.; Hrabe de Angelis, M.; Lendahl, U.; Edlund, H. Notch signalling controls pancreatic cell differentiation. Nature 1999, 400, 877–881. [Google Scholar] [CrossRef]
- Hart, A.; Papadopoulou, S.; Edlund, H. Fgf10 maintains notch activation, stimulates proliferation, and blocks differentiation of pancreatic epithelial cells. Dev Dyn 2003, 228, 185–193. [Google Scholar] [CrossRef]
- Mossahebi-Mohammadi, M.; Quan, M.; Zhang, J.S.; Li, X. FGF Signaling Pathway: A Key Regulator of Stem Cell Pluripotency. Front Cell Dev Biol 2020, 8, 79. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Browning, V.L.; Odorico, J.S. Activin, BMP and FGF pathways cooperate to promote endoderm and pancreatic lineage cell differentiation from human embryonic stem cells. Mech Dev 2011, 128, 412–427. [Google Scholar] [CrossRef]
- Apelqvist, A.; Ahlgren, U.; Edlund, H. Sonic hedgehog directs specialised mesoderm differentiation in the intestine and pancreas. Curr Biol 1997, 7, 801–804. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Ma, X.; Zhu, S. Recent advances and potential applications of human pluripotent stem cell-derived pancreatic beta cells. Acta Biochim Biophys Sin (Shanghai) 2020, 52, 708–715. [Google Scholar] [CrossRef]
- Serup, P. Signaling pathways regulating murine pancreatic development. Semin Cell Dev Biol 2012, 23, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.; Gallego-Llamas, J.; Ribes, V.; Kedinger, M.; Niederreither, K.; Chambon, P.; Dolle, P.; Gradwohl, G. Dorsal pancreas agenesis in retinoic acid-deficient Raldh2 mutant mice. Dev Biol 2005, 284, 399–411. [Google Scholar] [CrossRef]
- Tulachan, S.S.; Doi, R.; Kawaguchi, Y.; Tsuji, S.; Nakajima, S.; Masui, T.; Koizumi, M.; Toyoda, E.; Mori, T.; Ito, D.; et al. All-trans retinoic acid induces differentiation of ducts and endocrine cells by mesenchymal/epithelial interactions in embryonic pancreas. Diabetes 2003, 52, 76–84. [Google Scholar] [CrossRef]
- Shen, C.N.; Marguerie, A.; Chien, C.Y.; Dickson, C.; Slack, J.M.; Tosh, D. All-trans retinoic acid suppresses exocrine differentiation and branching morphogenesis in the embryonic pancreas. Differentiation 2007, 75, 62–74. [Google Scholar] [CrossRef]
- Zhang, D.; Jiang, W.; Liu, M.; Sui, X.; Yin, X.; Chen, S.; Shi, Y.; Deng, H. Highly efficient differentiation of human ES cells and iPS cells into mature pancreatic insulin-producing cells. Cell Res 2009, 19, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Ren, Z.; Xu, F.; Zhou, X.; Song, C.; Wang, V.Y.; Liu, W.; Lu, L.; Thomson, J.A.; Chen, G. Nicotinamide Promotes Cell Survival and Differentiation as Kinase Inhibitor in Human Pluripotent Stem Cells. Stem Cell Reports 2018, 11, 1347–1356. [Google Scholar] [CrossRef] [PubMed]
- Otonkoski, T.; Beattie, G.M.; Mally, M.I.; Ricordi, C.; Hayek, A. Nicotinamide is a potent inducer of endocrine differentiation in cultured human fetal pancreatic cells. J Clin Invest 1993, 92, 1459–1466. [Google Scholar] [CrossRef] [PubMed]
- Aigha, II; Abdelalim, E.M. NKX6.1 transcription factor: a crucial regulator of pancreatic beta cell development, identity, and proliferation. Stem Cell Res Ther 2020, 11, 459. [Google Scholar] [CrossRef]
- Nostro, M.C.; Sarangi, F.; Yang, C.; Holland, A.; Elefanty, A.G.; Stanley, E.G.; Greiner, D.L.; Keller, G. Efficient generation of NKX6-1+ pancreatic progenitors from multiple human pluripotent stem cell lines. Stem Cell Reports 2015, 4, 591–604. [Google Scholar] [CrossRef] [PubMed]
- Veres, A.; Faust, A.L.; Bushnell, H.L.; Engquist, E.N.; Kenty, J.H.; Harb, G.; Poh, Y.C.; Sintov, E.; Gurtler, M.; Pagliuca, F.W.; et al. Charting cellular identity during human in vitro beta-cell differentiation. Nature 2019, 569, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.H.; Chang, C.C. Vitamin C's essential role in DNA and histone demethylation and a preclinical rationale for its therapeutic high-dose potential in renal cell carcinoma. Ann Transl Med 2019, 7, S117. [Google Scholar] [CrossRef]
- Lee Chong, T.; Ahearn, E.L.; Cimmino, L. Reprogramming the Epigenome With Vitamin C. Front Cell Dev Biol 2019, 7, 128. [Google Scholar] [CrossRef] [PubMed]
- Bergsten, P.; Moura, A.S.; Atwater, I.; Levine, M. Ascorbic acid and insulin secretion in pancreatic islets. J Biol Chem 1994, 269, 1041–1045. [Google Scholar] [CrossRef]
- Agrawal, A.; Narayan, G.; Gogoi, R.; Thummer, R.P. Recent Advances in the Generation of beta-Cells from Induced Pluripotent Stem Cells as a Potential Cure for Diabetes Mellitus. Adv Exp Med Biol 2021, 1347, 1–27. [Google Scholar] [CrossRef]
- Cardenas-Diaz, F.L.; Osorio-Quintero, C.; Diaz-Miranda, M.A.; Kishore, S.; Leavens, K.; Jobaliya, C.; Stanescu, D.; Ortiz-Gonzalez, X.; Yoon, C.; Chen, C.S.; et al. Modeling Monogenic Diabetes using Human ESCs Reveals Developmental and Metabolic Deficiencies Caused by Mutations in HNF1A. Cell Stem Cell 2019, 25, 273–289. [Google Scholar] [CrossRef] [PubMed]
- Rovira, M.; Huang, W.; Yusuff, S.; Shim, J.S.; Ferrante, A.A.; Liu, J.O.; Parsons, M.J. Chemical screen identifies FDA-approved drugs and target pathways that induce precocious pancreatic endocrine differentiation. Proc Natl Acad Sci U S A 2011, 108, 19264–19269. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, M.; Loffler, K.A.; Edfalk, S.; Selander, L.; Dahl, U.; Ricordi, C.; Jeon, J.; Correa-Medina, M.; Diez, J.; Edlund, H. Retinoic acid promotes the generation of pancreatic endocrine progenitor cells and their further differentiation into beta-cells. PLoS One 2008, 3, e2841. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).