In the domain of supervised classification, various methodologies have been proposed to tackle the challenges posed by batch data and to enhance the interpretability and adaptability of classifiers. Among these, feature interval learning (FIL) techniques have emerged as prominent approaches. FIL approaches offer a distinct paradigm for classification, wherein the feature space is partitioned into intervals for each feature. Each interval is associated with a specific class, enabling a more nuanced representation of feature patterns and enhancing robustness to noise and variability in the data [
2]. FIL methods encompass a range of techniques, including hypercuboid or hyperrectangle learning and prototype-based classification, which learn decision boundaries or representative prototypes to distinguish between classes. The concept of hypercuboid has been exploited for many years [
7]
Hyper-cuboid approaches represent one category of FIL methods, where the feature space is partitioned into hyper-cuboid regions based on the distribution of training data [
7]. These regions serve as decision boundaries, facilitating efficient classification of new instances. Hyper-cuboid approaches are particularly effective in handling high-dimensional data and are known for their computational efficiency and simplicity. Prototype-based FIL methods, on the other hand, generate prototypes for each class based on the distribution of feature intervals in the training data [
3,
8,
9]. These prototypes encapsulate the characteristic features of each class and are used to classify new instances by measuring their resemblance to the prototypes. Prototype-based FIL methods offer interpretability and transparency in decision-making, as the classification process is based on clear feature ranges. In [
2] describes a family of FIL algorithms. Some of FIL algorithms are voting feature intervals [
3], k-nearest neighbor on feature projections [
10] and Naive Bayes classifier (NBC) [
1]. The common idea of the FIL algorithms, learn the concept description in the form of a set of disjoint intervals separately for each feature. At the testing phase the FIL algorithms predict the class of new data point as the label of with the highest total weighted votes of the individual features. For example in
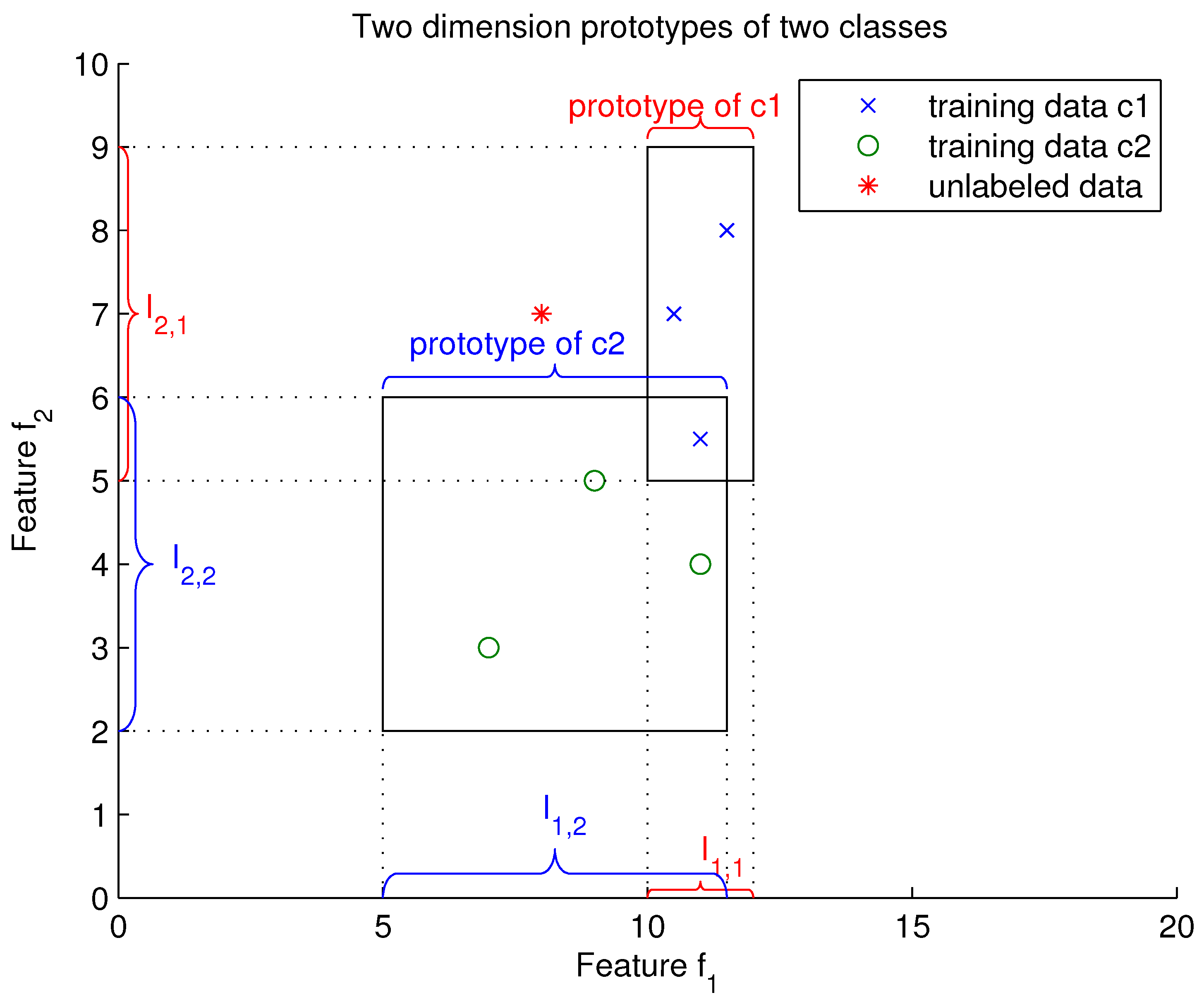
Figure 1, the horizontal axis is feature
and the vertical axis is the feature
. The two rectangles represent the two prototypes of classes
and
. The prototype of class
is represented by the interval
and
(intervals determined using the training sets of class
’the blue
x sign’. And the prototype of class
is represented by the intervals
and
(intervals calculated using the training sets of class
‘the green
o sign’. The objective is to assign the unlabeled data (the red star * in
Figure 1) to the closest rectangular.
Overall, while hypercuboid or hyperrectangle learning algorithms offer computational efficiency and simplicity, FIL methods provide greater flexibility, robustness, and interpretability in classifying new data points, especially in scenarios with complex data distributions and high-dimensional feature spaces. However, predicting class labels based on the highest total weighted votes of individual features is a straightforward approach in FIL algorithms, it may have limitations in handling feature importance, noise, feature relationships, imbalanced datasets, and the trade-off between interpretability and accuracy. Addressing these inconveniences requires careful consideration of feature selection, noise reduction techniques, feature engineering, and robust mechanisms for handling imbalanced data. Beyond traditional majority voting approaches, another advancements in FIL is the integration of outranking measures into the classification process. Outranking measures, rooted in preference learning principles, extend the classification paradigm by capturing subtle preferences and trade-offs among alternative solutions [
8,
11]. By conducting pairwise comparisons on each feature, outranking measures facilitate more nuanced decision-making and enhance adaptability to specific datasets [
5]. Several FIL classifiers based on outranking measures were developed since decades, among them we have PROAFTN [
9], PROCFTN [
12] and K Closest Resemblance (K-CR) or PROCTN [
13]. These classifiers offer several advantages in the context of classification tasks. By evaluating the data points based on their pairwise performance on each feature, outranking techniques can capture subtle differences in feature importance and contribution to class membership. Instead of blindly aggregating votes from all features, outranking considers the relative performance of alternatives on each feature, effectively filtering out noise and focusing on discriminative features that contribute most to the classification decision. This robustness to noise enhances the reliability and accuracy of classification outcomes. Outranking techniques can capture complex relationships between features by conducting pairwise comparisons on each feature individually. Unlike majority voting, which treats features independently, outranking takes into account the interactions and dependencies between features, allowing for more accurate and nuanced classification decisions. Outranking techniques provide clear and interpretable decision rules by quantifying the resemblance relationship between data points. Instead of relying on a simple count of votes, outranking assigns a degree of resemblance between alternatives based on their pairwise comparisons, making the classification process more transparent and understandable. This enhanced interpretability fosters trust and understanding in the classification model’s decision-making process. Outranking techniques can effectively handle imbalanced datasets by considering the relative performance of alternatives on each feature [
14]. Instead of biasing the classification decision towards the majority class, outranking evaluates alternatives based on their overall resemblance relationship, ensuring that all classes are appropriately considered in the classification process. This helps mitigate the impact of class imbalance and ensures more balanced and accurate predictions. Outranking techniques offer flexibility and adaptability to different types of data and classification tasks. By capturing subtle differences in feature importance and performance, outranking can accommodate various data distributions and class structures, making it suitable for a wide range of classification scenarios. The FIL classifiers based on outranking measures have been applied to the resolution on many real world practical problems such as medical diagnosis [
11,
15,
16,
17], image processing and classification [
14,
18], engineering design [
19], recommendation system [
20] and cybersecurity [
21]. Even though, these classifiers have several advantages, they are suffering from the computation complexity in their learning phase specifically when the number of features is very large. The PROAFTN and the K-CR build a set of prototypes in each class and then the inductive is based on the recursive approach, which is very complex. In this paper, we will introduce the FIL approach based on outranking measures for classification problems called Closest Resemblance
CR. The
CR algorithm is simple, effective, efficient and robust to irrelevant features. In the remaining paper, we will present the CR classifier, where each class is represented by one prototype and the classification rule is expressed as: "the data point is assigned to the class if the data point is resembled or roughly equivalent to the prototype of this class". Therefore, our proposed CR classifier is a prototype-based classification method that incorporates the FIL techniques to build the prototype of each class during the learning phase. The intervals in
CR are inspired from the reference intervals used in the era of evidence-based medicine [
22]. Unlike confidence intervals, which quantify uncertainty around a parameter estimate, prediction intervals capture the range within which a data point is likely to fall — they estimate the uncertainty associated with individual predictions.