1. Introduction
The mitochondrial network is critical for many aspects of the subcellular survival mechanisms of organisms. Known to be the powerhouse of the cell, mitochondria are responsible for various aspects of energy homeostasis, oxidative stress, calcium handling, cell signalling, and, thus, cell survival [
1]. The dynamic nature of the mitochondria population is critical to the integrity of the subcellular network structures that these organelles maintain and to the control of the quality of mitochondrial proteins and other components [
2,
3,
4]. For example, the early dynamic events that occur during apoptosis include cristae remodeling, mitochondrial fragmentation, and membrane “blebbing” or zeiosis [
5]. Inhibition of these processes, either through downregulation directed by RNA-interference or by expression of a dominant-negative mutant form of
Drp1, can slow the rate of mitochondrial fragmentation and, in turn, the cascade of apoptotic events [
6,
7,
8]. The activity of the Drp1 protein promotes caspase-independent mitochondrial fission and cristae remodeling to amplify the process of apoptosis whether cell death is instigated by either the specific activity of the pro-apoptotic protein BID or by the general consequences of oxidative stress [
9]. Overall, the mechanics of mitochondrial fission plays a crucial role in the amplification of aspects of the essential cellular process of apoptosis.
Modifications to the mitochondrial network seem to differentially influence a number of signalling pathways. In response to a series of molecular cues [
10,
11,
12], the mitochondrial network participates in a delicate balance between continuous division and fusion processes [
13,
14]. Mitochondrial fusion helps compromised mitochondria, with likely bearing highly damaged DNA and proteins, to actively exchange components with other more healthy mitochondria to decrease the severity of heteroplasmy, and help with functional complementation [
15,
16]. Mitochondrial fission allows for the segregation of irreversibly damaged portions of the mitochondrial network and subsequent degradation [
17]. Mitochondrial fission necessarily requires
dynamin-related protein 1 (
Drp1) and
FIS1 [
18]. Nevertheless, an explicit understanding of the factors promoting fission and fusion remains limited.
The Bcl-2 family proteins interact with Drp1, and Drp1 can promote apoptosis in Bcl-2 protein-dependent and independent manners [
9,
19].The two Bcl-2 family homologues in
Drosophila melanogaster are
Buffy (anti-apoptotic) and
Debcl (pro-apoptotic). The Debcl protein can interacts with Drp1 in Drosophila to activate apoptosis via the JNK pathway [
20]. Drp1 is required for a standard rate of Cyt-c release and caspase activation during programmed cell death [
21]. Drp1 increases the mitochondrial fragmentation under nitrosative stress in primary cerebrocortical neuron culture [
22]. The Drp1 protein interacts with other proteins involved in a number of mitochondrial processes, such as protein product of
Bax [
10,
12,
22], the products of
Pink1 (PTEN-induced putative kinase 1) and
park in Cos-7 cells [
23]. Mutation of the
Pink1 and
park genes are among the most prominent causes of early onset of PD [
24]. The roles of
Pink1 and
park are vital to ubiquitin-dependent mitophagy [
25]. The
Pink and
parkin loss of function mutation increase the mitochondrial localization of Drp1 in flies [
26]. The parkin protein ubiquitinates the Drp1 for proteasomal degradation, incriminate the Drp1 for dysregulated mitochondrial dynamics in parkin loss of function induced mitochondrial morphology in HeLa and SH-SY5Y cells [
27]. Number of stresses can increase the mitochondrial translocation of Drp1 in neurons and initiate apoptosis or mitophagy [
28]. In a number of models of ALS, dephosphorylation of Drp1 through the activity of protein phosphatase 1, has been identified as causative of the disease-associated phenotypes [
29]. As well, Drp1-mediated mitochondrial fragmentation caused by the administration of rotenone has been identified in a rat model of PD-like changes to the olfactory bulb [
30]. As adjustment of Drp1 activity may be key to the inhibition of the pathology of ALS and PD, investigation of such alterations is essential to knowledge required for the development of therapies. Excessive activity of Drp1 increase mitochondrial fission and consequently promote cell death.
Here we propose that alteration of
Drp1 expression results in a degenerative phenotype. We utilize Drosophila to model neurodegenerative disease because it is an excellent model system to study the genes and proteins affected in ALS and PD [
31]. The anticipated role of mitochondria in PD pathogenesis has made the study of the interactions of Drp1 gene important to modelling this disease in Drosophila. In these experiments, we exploited the UAS-Gal4 system to direct the overexpression and inhibition of the genes of interest in selected neuronal tissues using the Ddc-Gal4 transgene [
32,
33]. The proposal is that the
Drp1 overexpression phenotype is due to excessive activities related to apoptosis and can be rescued by the appropriate regulation by anti-apoptotic Bcl-2 gene,
Buffy. In an established
park-RNAi model of PD [
34], we directed and inhibited the expression of the
Drp1 gene. The ND-like phenotypes of
Ddc-Gal4 park-RNAi were rescued by the expression of
Drp1-
RNAi transgenes. The phenotype produced in response to the expression of
Drp1-RNAi is due to the diminishment of mitochondrial integrity and maybe rescued through modification of the responsible signalling pathway. The careful regulation of mitochondrial dynamics is important to control mitochondrial induced defects. The strategy is to identify the basic mechanisms in the fly model to encourage further validation in mammalian model organisms.
4. Discussion
The protein product of the
Drp1 gene, along with the participation of other mitochondrial protection proteins, is involved in the processes of mitochondrial fission, apoptosis, and mitophagy. Excessive mitochondrial fragmentation can be associated with dysfunctional metabolic diseases whereas a “hyper-fused” mitochondrial network can serve to protect cells from metabolic insult and autophagy [
35]. In the skeletal muscle of mice,
Drp1 overexpression can cause a severe impairment of post-natal muscle growth as the production of protein may become attenuated and growth hormone pathways may be down regulated [
36]. Conditions of high fat and/or high glucose levels can cause excessive oxidative stress along with mitochondrial fragmentation as mediated by the Drp1 protein [
37,
38]. These phenotypes are similar to the increased activity of Drp1 as observed with Cos and PC12 cells [
39]. In humans, protein kinase A (or PKA) can phosphorylate and inactivate the pro-apoptotic Bcl-2 family member protein Bad [
40] and the Drp1 protein [
41] in a complex effort to promote cell survival. The effect of
Drp1 overexpression and consequently excessive mitochondrial fragmentation can be toxic to many physiological processes.
The Bcl-2 family proteins assist the pro-fission activity of the Drp1 protein during apoptosis in nematodes and mammals [
42]. However, in non-apoptotic cells of mammals, the Bcl-2 family proteins have both pro-fission and pro-fusion activities. The overexpression of
Drp1 in selected neurons along with the overexpression of
Buffy or the inhibition of
Debcl has resulted in an increase in median lifespan and of the ability to climb over the increased lifespan. In complementary experiments,
Buffy inhibition and
Debcl overexpression have resulted in reduced lifespans accompanied with impaired climbing abilities, consistent with the conclusion that
Buffy can function as the antithesis of
Debcl [
43]. The rescue of the
Drp1 expression phenotype is in accordance with the role of the Buffy protein as “the guardian of mitochondria”. As proteins, Buffy can interact with Debcl to inhibit
Debcl-induced cell death. As a mechanism, this process could be due to decreased activity of the Debcl protein to influence cooperation with Drp1 in the promotion of cell death [
20]. The pro-apoptotic Debcl protein acts to induces apoptosis through a caspase-independent mechanism that triggers the release of Cytochrome C [
44] in an activity that resembles the loss of Drp1 [
9]. The overexpression of
Debcl and
Drp1 together in selected neurons does not alter the phenotype generated by overexpression of
Drp1 without
Debcl. This may not be surprising as Drp1 and Debcl functions seem to cooperate to promote apoptosis [
20]. Indeed, this and earlier studies have demonstrated that Drp1 can play various roles in mitochondrial fragmentation and apoptosis, to act in concert with anti- and pro-survival proteins, dependent upon the stimuli.
The directed inhibition in
Drosophila melanogaster of
Drp1 in a subset of neurons results in an age-dependent loss in climbing ability, a phenotype strongly associated with the modelling of ND in flies. The overexpression of
Buffy in neurons that co-express
Drp1-RNAi led to a decrease in the median lifespan accompanied with a rescue of the impaired locomotor ability. The recovery in age-dependent climbing ability over time may be evidence of a complicated regulatory relationship. A study shows
Drp1 inhibition reduces the total accumulation of pro-apoptotic Bcl-2 protein, Bax, on mitochondria outer membrane in HeLa cell lines [
12]. This intermediate phenotype was not expected but may be important in the determination of the pathology of neurological diseases [
45,
46]. The inhibition of anti-apoptotic
Buffy or overexpression of pro-apoptotic
Debcl enhanced the loss of
Drp1-induced phenotype. The interaction of Bax with Drp1 in mammals seems to evolve from the Debcl and Drp1 protein interactions during the course of evolution. Drp1 protein interacts directly and indirectly with Bcl-2 family protein to facilitate MOMP in apoptotic cells [
42]. Overall, we believe that we have established that
Buffy confers survival advantage to flies overexpressing
Drp1 and provides a partially rescued intermediate phenotype in flies with a loss of
Drp1 function.
The
park gene is crucial to the function of
Pink1-dependent mitochondrial mitophagy. The loss of the parkin protein is a cause of great cellular stress as a major mechanism that controls mitophagy is compromised. Under normal circumstances, the
park-encoded ubiquitin ligase recruits the Drp1 protein to mediate mitochondrial fragmentation during mitophagy [
23,
47]. In mouse embryonic fibroblasts, loss of
park does not produce visible effects upon the net number of mitochondria [
48]. Furthermore, the loss of
park along with the loss of
Drp1 increases the number of mitochondria by threefold, which can be interpreted that
park controls mitochondria fragmentation in a
Drp1 knockout background. Therefore,
park may regulate, negatively, the
Drp1-independent mitochondrial division. Alternatively, the parkin protein may direct the ubiquitination of the mitochondrial localised Drp1 protein to lead its proteasomal mediated degradation [
26] and, hence, the activity of Drp1 protein on mitochondria is higher in the
Drp1-parkin co-inhibition state as compared to the inhibition of Drp1 alone. In these experiments, the critical class flies that have the directed co-expression of
Drp1-RNAi and
park-RNAi inhibitory transgenes live longer than those that express
park-RNAi and
Drp1-RNAi under the
Ddc-Gal4 transgene individually. This support the hypothesis that the basic mechanism of PD-like phenotypes may be conserved among mammals [
49,
50] and the Diptera. This could be due to the establishment of an altered mitochondrial network to enhance homeostasis and benefit cellular health. The inhibition of
Drp1 has been shown to suppress
parkin mutant phenotypes which suggests a role for this protein in 1) in a pathway that regulates mitochondrial health and integrity and 2) in interactions with mitochondrial proteins. Regardless of the underlying mechanism, this work provides strong evidence of
Drp1 as an important therapeutic target for maintaining mitochondrial health.
Figure 1.
Drp1 is evolutionarily conserved between Drosophila and humans. (a) Clustal Omega multiple sequence alignment of D. melanogaster Drp1 (NP_608694.2) protein with the H. sapiens (NP_001265392.1) shows evolutionarily conserved domains identified using the NCBI Conserved Domain Database (CDD) and further confirmed by the Eukaryotic Linear Motif (ELM) resource. The two well documented phosphorylation sites are identified, S606 and S627 in dynamin-1-like protein (DLP-1) isoform 4 of H. sapiens; and S616 and T637 in Drp1 of D. melanogaster. The asterisks indicate the residues that are identical; the colons indicate the conserved substitutions; and the dots indicates the semi-conserved substitutions. Colour differences indicate the chemical nature of amino acids: red indicates small hydrophobic (includes aromatic) residues; blue indicates acidic; magenta indicates basic; and green indicates basic with hydroxyl or amine groups. (bi) The original Dynamin-1 like protein (DLP-1) structure of H. sapiens (NP_001265392.1) from the NCBI structure database. (bii) The Phyre2 web portal for protein modelling, prediction and analysis mediated the development of a model of the Drp1 protein of D. melanogaster (NP_608694.2) from a 76% identical protein with a confidence of 100%. The N terminus is coloured in Magenta; C terminus is coloured in Red and a consensus ATG8 binding region at N terminus is coloured in orange.
Figure 1.
Drp1 is evolutionarily conserved between Drosophila and humans. (a) Clustal Omega multiple sequence alignment of D. melanogaster Drp1 (NP_608694.2) protein with the H. sapiens (NP_001265392.1) shows evolutionarily conserved domains identified using the NCBI Conserved Domain Database (CDD) and further confirmed by the Eukaryotic Linear Motif (ELM) resource. The two well documented phosphorylation sites are identified, S606 and S627 in dynamin-1-like protein (DLP-1) isoform 4 of H. sapiens; and S616 and T637 in Drp1 of D. melanogaster. The asterisks indicate the residues that are identical; the colons indicate the conserved substitutions; and the dots indicates the semi-conserved substitutions. Colour differences indicate the chemical nature of amino acids: red indicates small hydrophobic (includes aromatic) residues; blue indicates acidic; magenta indicates basic; and green indicates basic with hydroxyl or amine groups. (bi) The original Dynamin-1 like protein (DLP-1) structure of H. sapiens (NP_001265392.1) from the NCBI structure database. (bii) The Phyre2 web portal for protein modelling, prediction and analysis mediated the development of a model of the Drp1 protein of D. melanogaster (NP_608694.2) from a 76% identical protein with a confidence of 100%. The N terminus is coloured in Magenta; C terminus is coloured in Red and a consensus ATG8 binding region at N terminus is coloured in orange.
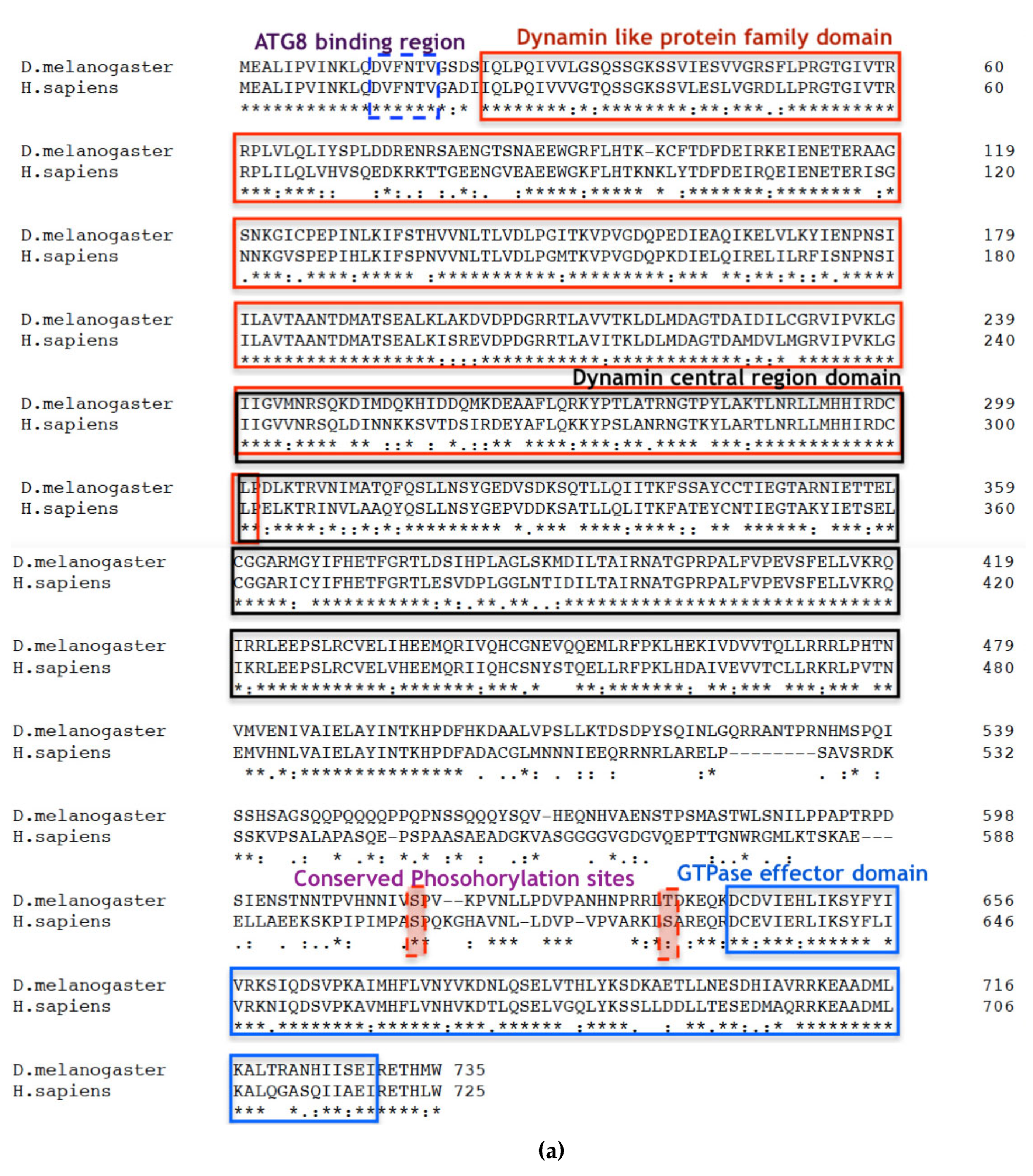
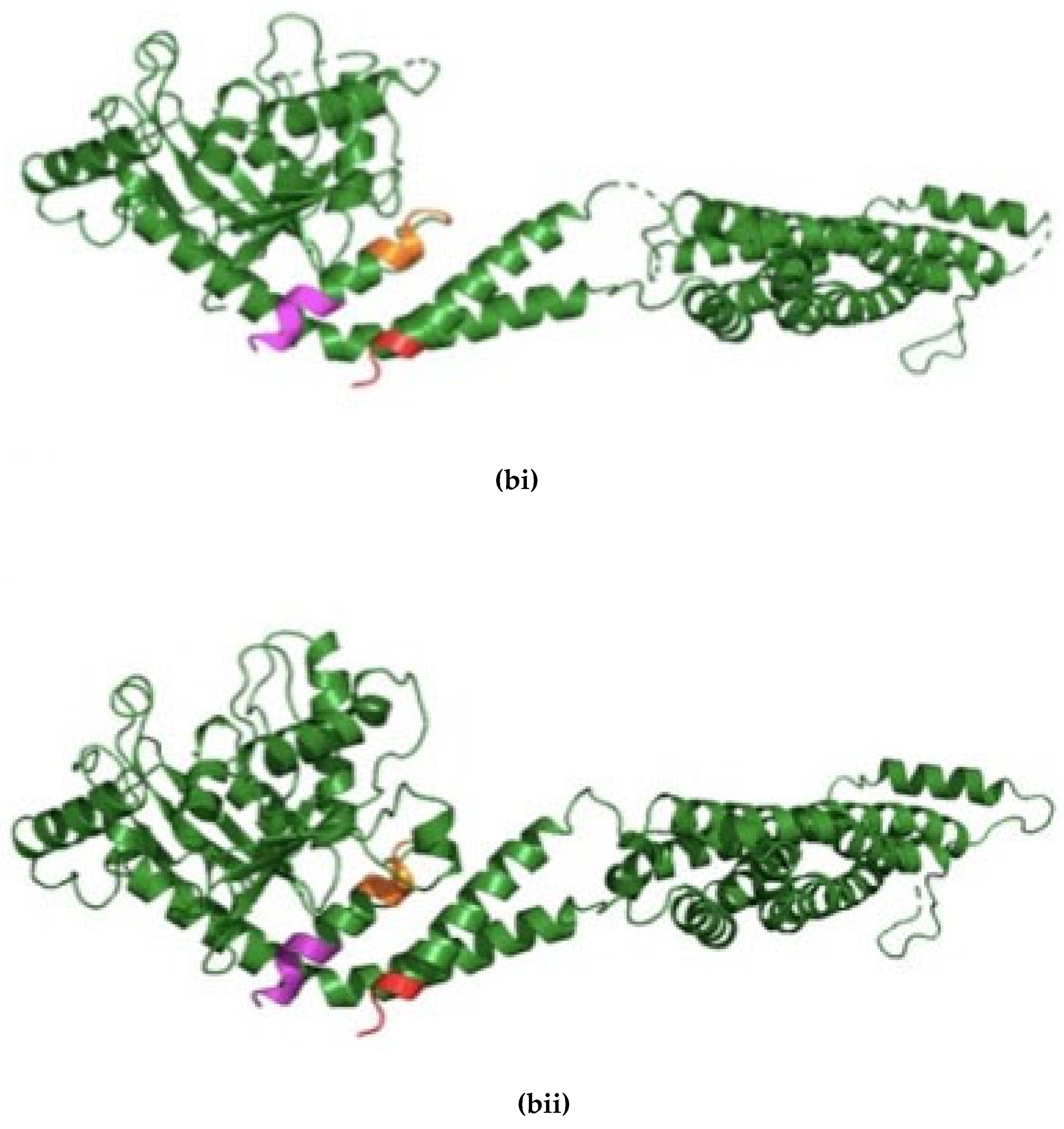
Figure 2.
Altered Drp1 expression under the control of Ddc-Gal44.3D influences the survival and climbing ability of flies. (a) The GraphPad prism8 generated graph of the longevity assay for the expression of Drp1, Drp1 RNAi’s under the control of Ddc-Gal4 transgene. The directed expression results in decreased median lifespan of 56 days compare to 68 days of control calculated by Log-rank Mantel Cox test, with Bonferroni correction. The inhibition of Drp1 under the control of Ddc-Gal4 transgene results in lifespan of 70 days with UAS-Drp1-RNAi1 and 72 days with UAS-Drp-RNAi2 compared to 68 days of control done by Log-rank Mantel Cox test, with Bonferroni correction. (b) The GraphPad prism8 generated graph of the climbing abilities of flies with overexpression of Drp1, Drp1 RNAi’s and control. The climbing ability of Drp1 overexpression and Drp1 RNAi’s flies have decreased compared to control as determined in nonlinear fitting of the climbing curve by 95% confidence interval (p-value <0.0001).
Figure 2.
Altered Drp1 expression under the control of Ddc-Gal44.3D influences the survival and climbing ability of flies. (a) The GraphPad prism8 generated graph of the longevity assay for the expression of Drp1, Drp1 RNAi’s under the control of Ddc-Gal4 transgene. The directed expression results in decreased median lifespan of 56 days compare to 68 days of control calculated by Log-rank Mantel Cox test, with Bonferroni correction. The inhibition of Drp1 under the control of Ddc-Gal4 transgene results in lifespan of 70 days with UAS-Drp1-RNAi1 and 72 days with UAS-Drp-RNAi2 compared to 68 days of control done by Log-rank Mantel Cox test, with Bonferroni correction. (b) The GraphPad prism8 generated graph of the climbing abilities of flies with overexpression of Drp1, Drp1 RNAi’s and control. The climbing ability of Drp1 overexpression and Drp1 RNAi’s flies have decreased compared to control as determined in nonlinear fitting of the climbing curve by 95% confidence interval (p-value <0.0001).
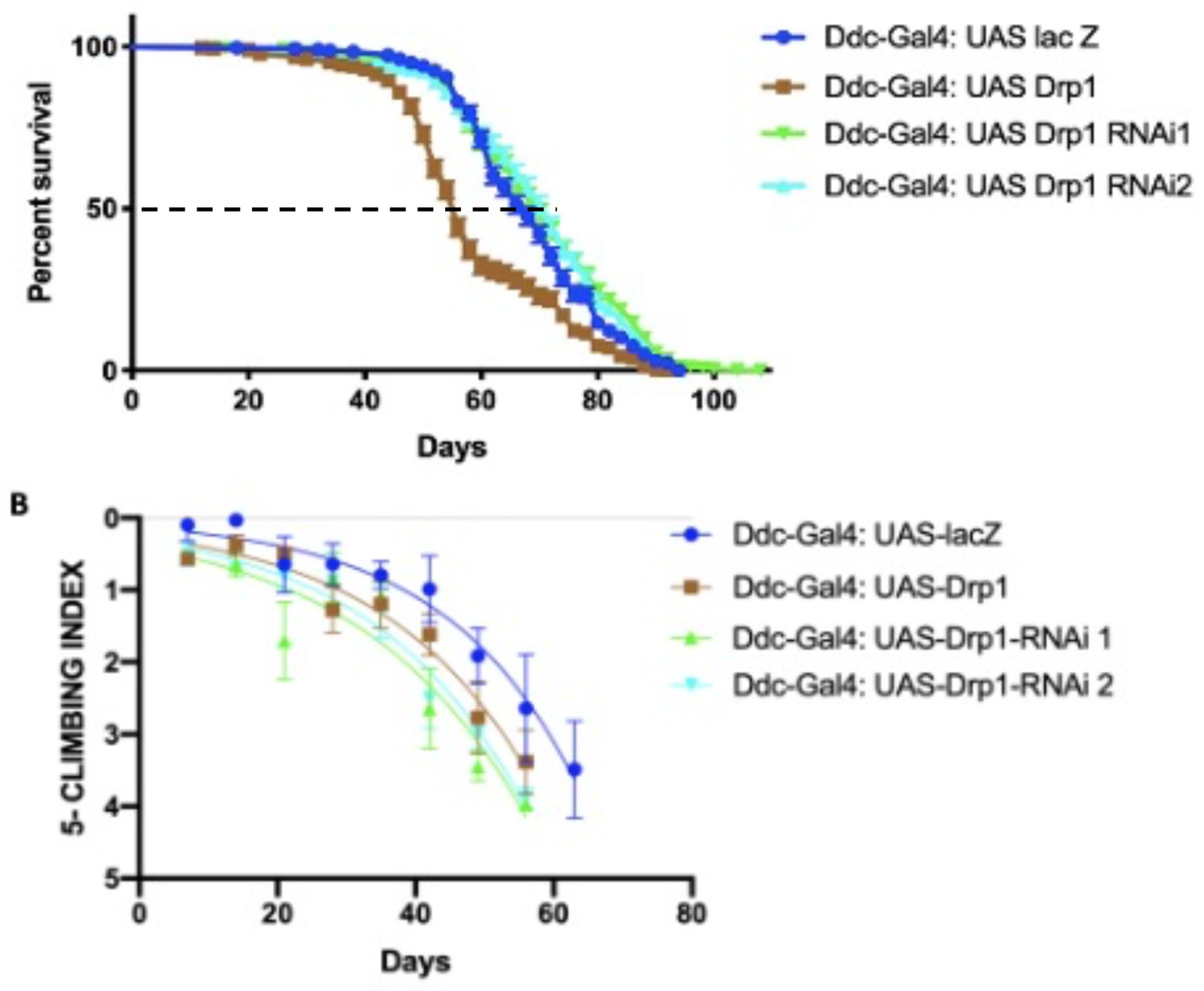
Figure 3.
The expression of Drp1-RNAi, directed by Ddc-Gal44.3D, can increase median lifespan and decrease climbing. (a) In control, Ddc-Gal44.3D UAS-lacZ critical class males resulted in a median life span of 62 days (n=308). Expression of Drp1 in Ddc-Gal44.3D resulted in a median life span of 56 days (n=310), much lower than the lacZ-expressing control; expression of Drp1 in Ddc-Gal44.3D UAS-Drp1-RNAi resulted in a median life span of 64 days (n=250) very similar to control (Ddc/lacZ) as determined by the Log-rank Mantel-Cox test (p value=0.0633) with Bonferroni correction. The graph of the longevity assay was generated by GraphPad prism8. (b) The Ddc-Gal44.3D flies express UAS-lacZ in control flies. The climbing abilities of Ddc-Gal44.3D UAS-Drp1 expressing flies have decreased compared to control as determined in the non-linear fitting of the climbing curve by a 95% confidence interval (p<0.0001). The flies' climbing ability expressing Drp1 in Ddc-Gal44.3D UAS-Drp1-RNAi transgene is similar to control as determined in the non-linear fitting of the climbing curve by a 95% confidence interval at P value=1.309. The graph of longevity assay was generated by GraphPad prism8 non-linear regression curve. (c) In control, Ddc-Gal44.3D UAS-lacZ critical class males resulted in a median life span of 62 days (n=308). Expression of Drp1-RNAi in Ddc-Gal44.3D resulted in a median life span of 70 days (n=321), much higher compared to the control; expression of Drp1-RNAi in Ddc-Gal44.3D UAS-Drp1 resulted in a median life span of 64 days (n=327), very similar to control (Ddc/lacZ) as determined by the Log-rank Mantel-Cox test (p value=0.0582) with Bonferroni correction. The graph of the longevity assay was generated by GraphPad prism8. (d) The Ddc-Gal44.3D flies express UAS-lacZ in control flies. The climbing abilities of Ddc-Gal44.3D UAS-Drp1-RNAi expressing flies have decreased compared to control as determined in the non-linear fitting of the climbing curve by a 95% confidence interval (p<0.0001). The flies' climbing ability expressing Drp1-RNAi in Ddc-Gal44.3D UAS-Drp1 transgene is similar to control (Ddc/lacZ) as determined in the non-linear fitting of the climbing curve by a 95% confidence interval at p value=0.0027. The graph of longevity assay was generated by GraphPad prism8 non-linear regression curve.
Figure 3.
The expression of Drp1-RNAi, directed by Ddc-Gal44.3D, can increase median lifespan and decrease climbing. (a) In control, Ddc-Gal44.3D UAS-lacZ critical class males resulted in a median life span of 62 days (n=308). Expression of Drp1 in Ddc-Gal44.3D resulted in a median life span of 56 days (n=310), much lower than the lacZ-expressing control; expression of Drp1 in Ddc-Gal44.3D UAS-Drp1-RNAi resulted in a median life span of 64 days (n=250) very similar to control (Ddc/lacZ) as determined by the Log-rank Mantel-Cox test (p value=0.0633) with Bonferroni correction. The graph of the longevity assay was generated by GraphPad prism8. (b) The Ddc-Gal44.3D flies express UAS-lacZ in control flies. The climbing abilities of Ddc-Gal44.3D UAS-Drp1 expressing flies have decreased compared to control as determined in the non-linear fitting of the climbing curve by a 95% confidence interval (p<0.0001). The flies' climbing ability expressing Drp1 in Ddc-Gal44.3D UAS-Drp1-RNAi transgene is similar to control as determined in the non-linear fitting of the climbing curve by a 95% confidence interval at P value=1.309. The graph of longevity assay was generated by GraphPad prism8 non-linear regression curve. (c) In control, Ddc-Gal44.3D UAS-lacZ critical class males resulted in a median life span of 62 days (n=308). Expression of Drp1-RNAi in Ddc-Gal44.3D resulted in a median life span of 70 days (n=321), much higher compared to the control; expression of Drp1-RNAi in Ddc-Gal44.3D UAS-Drp1 resulted in a median life span of 64 days (n=327), very similar to control (Ddc/lacZ) as determined by the Log-rank Mantel-Cox test (p value=0.0582) with Bonferroni correction. The graph of the longevity assay was generated by GraphPad prism8. (d) The Ddc-Gal44.3D flies express UAS-lacZ in control flies. The climbing abilities of Ddc-Gal44.3D UAS-Drp1-RNAi expressing flies have decreased compared to control as determined in the non-linear fitting of the climbing curve by a 95% confidence interval (p<0.0001). The flies' climbing ability expressing Drp1-RNAi in Ddc-Gal44.3D UAS-Drp1 transgene is similar to control (Ddc/lacZ) as determined in the non-linear fitting of the climbing curve by a 95% confidence interval at p value=0.0027. The graph of longevity assay was generated by GraphPad prism8 non-linear regression curve.
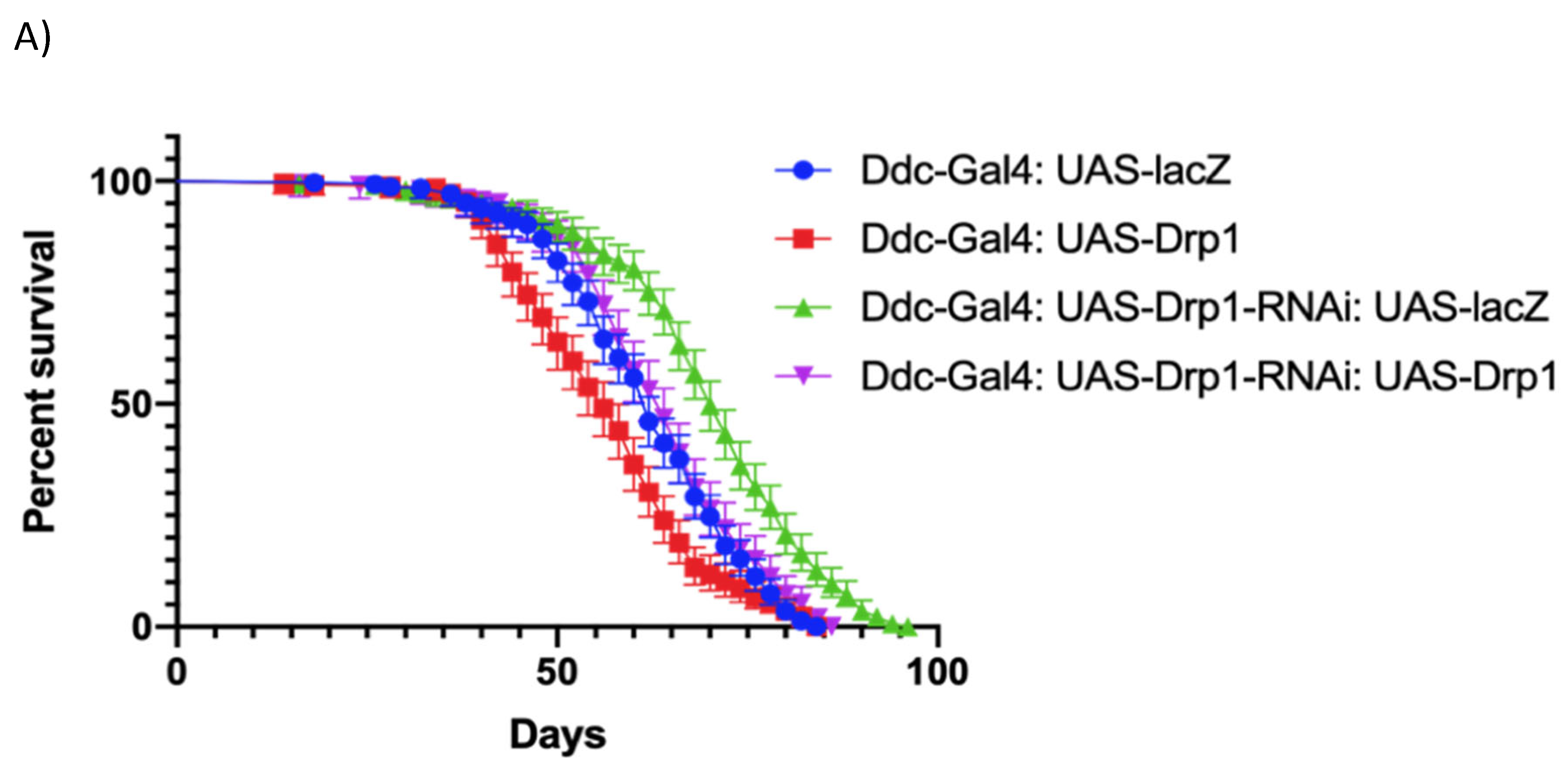
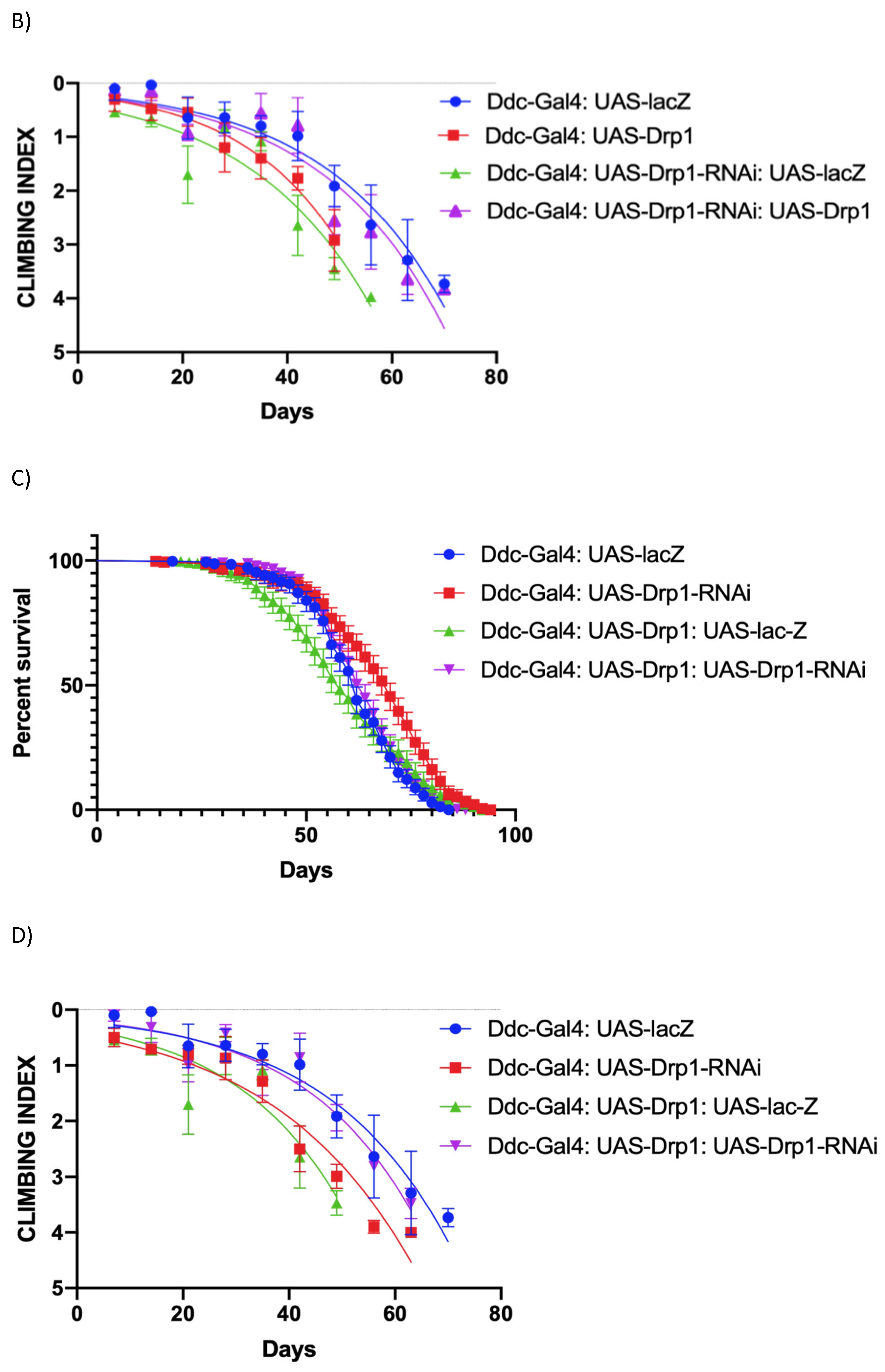
Figure 4.
Altered expression of Buffy and Debcl can enhance and suppress climbing ability in Drp1 over-expression flies. (a) In control, Ddc-Gal44.3D; UAS-Drp1 UAS-lacZ critical class males resulted in a median life span of 58 days (n=282). The overexpression of Buffy results in a median lifespan of 68 days (n=375) compares to 58 days of control (P value=0.0002); the inhibition of Buffy directed by the Ddc-Gal44.3D UAS-Drp1 transgene results in the median lifespan of 52 (n=274), much less compared to control, determined by Log-rank Mantel-Cox test at P-value <0.0001, with Bonferroni correction. The overexpression of DebclEY05743 results in a median lifespan of 60 days (n=331) similar to 58 days of control determined by Log-rank Mantel-Cox test at P-value 0.3293; the inhibition of Debcl directed by the Ddc-Gal4 UAS-Drp1 transgene result in the median lifespan of 66 (n=303); much higher than control, determined by Log-rank Mantel-Cox test at P value 0.0057, with Bonferroni correction. (b) The GraphPad prism8 generated graph of the climbing abilities of Ddc-Gal4 -Drp1 flies with the expression of Buffy, Buffy-RNAi, DebclEY05743, Debcl-RNAiv47515 and control. The climbing abilities of flies overexpressing Buffy have rescued compared to control as determined in the climbing curve's non-linear fitting by a 95% confidence interval (p<0.0001). The climbing ability of the flies was further weakened by the expression of UAS-Buffy-RNAi as determined in the non-linear fitting of the climbing curve by a 95% confidence interval at a p-value 0.0125 and 0.03293 respectively (n=50). The climbing abilities of flies expressing Debcl-RNAiv47515has rescued compared to control as determined by the non-linear fitting of the climbing curve by a 95% confidence interval (p value=0.0057). The graph of longevity assay was generated by GraphPad prism8 non-linear regression curve.
Figure 4.
Altered expression of Buffy and Debcl can enhance and suppress climbing ability in Drp1 over-expression flies. (a) In control, Ddc-Gal44.3D; UAS-Drp1 UAS-lacZ critical class males resulted in a median life span of 58 days (n=282). The overexpression of Buffy results in a median lifespan of 68 days (n=375) compares to 58 days of control (P value=0.0002); the inhibition of Buffy directed by the Ddc-Gal44.3D UAS-Drp1 transgene results in the median lifespan of 52 (n=274), much less compared to control, determined by Log-rank Mantel-Cox test at P-value <0.0001, with Bonferroni correction. The overexpression of DebclEY05743 results in a median lifespan of 60 days (n=331) similar to 58 days of control determined by Log-rank Mantel-Cox test at P-value 0.3293; the inhibition of Debcl directed by the Ddc-Gal4 UAS-Drp1 transgene result in the median lifespan of 66 (n=303); much higher than control, determined by Log-rank Mantel-Cox test at P value 0.0057, with Bonferroni correction. (b) The GraphPad prism8 generated graph of the climbing abilities of Ddc-Gal4 -Drp1 flies with the expression of Buffy, Buffy-RNAi, DebclEY05743, Debcl-RNAiv47515 and control. The climbing abilities of flies overexpressing Buffy have rescued compared to control as determined in the climbing curve's non-linear fitting by a 95% confidence interval (p<0.0001). The climbing ability of the flies was further weakened by the expression of UAS-Buffy-RNAi as determined in the non-linear fitting of the climbing curve by a 95% confidence interval at a p-value 0.0125 and 0.03293 respectively (n=50). The climbing abilities of flies expressing Debcl-RNAiv47515has rescued compared to control as determined by the non-linear fitting of the climbing curve by a 95% confidence interval (p value=0.0057). The graph of longevity assay was generated by GraphPad prism8 non-linear regression curve.
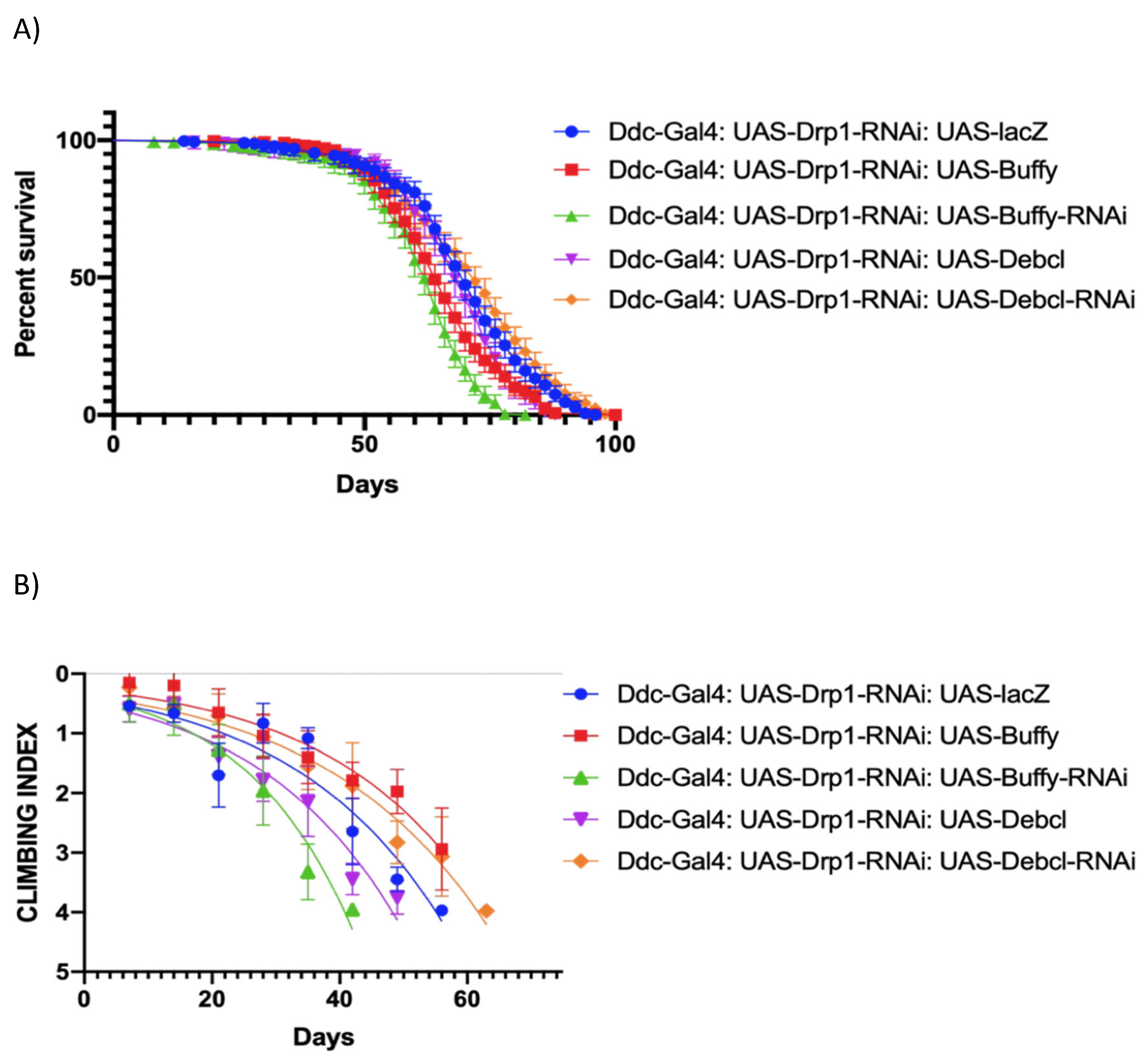
Figure 5.
Altered expression of Buffy and Debcl can enhance and suppress climbing ability in Drp1 loss of function flies. (a) In control, Ddc-Gal44.3D; UAS-Drp1 transgene results in the median lifespan of 62 (n=273), determined by Log-rank Mantel-Cox test at P-value <0.0001, with Bonferroni correction. The overexpression of DebclEY05743 results in a median lifespan of 68 days (n=331) much higher compared to control as determined by Log-rank Mantel-Cox test at P-value 0.0003; the inhibition of Debcl directed by the Ddc-Gal4; UAS-Drp1 transgene result in the median lifespan of 72 (n=303); similar to control, determined by Log-rank Mantel-Cox test at p-value 0.021, with Bonferroni correction. (b) The GraphPad prism8 generated graph of the climbing abilities of Ddc-Gal4 UAS-Drp1 flies with the expression of Buffy, Buffy-RNAi, DebclEY05743, Debcl-RNAiv47515 and control. The climbing abilities of flies overexpressing Buffy have rescued compared to control as determined in the climbing curve's non-linear fitting by a 95% confidence interval (p<0.0001). The climbing ability of the flies has further diminished through the expression of UAS-Buffy-RNAi and UAS-DebclEY05743 as determined in the non-linear fitting of the climbing curve by a 95% confidence interval at a p-value 0.0004 and 0.0002 respectively (n=50). The climbing abilities of flies expressing Debcl-RNAiv47515has rescued compared to control as determined by the non-linear fitting of the climbing curve by a 95% confidence interval (p value<0.0001). The graph of longevity assay was generated by GraphPad prism8 non-linear regression curve.
Figure 5.
Altered expression of Buffy and Debcl can enhance and suppress climbing ability in Drp1 loss of function flies. (a) In control, Ddc-Gal44.3D; UAS-Drp1 transgene results in the median lifespan of 62 (n=273), determined by Log-rank Mantel-Cox test at P-value <0.0001, with Bonferroni correction. The overexpression of DebclEY05743 results in a median lifespan of 68 days (n=331) much higher compared to control as determined by Log-rank Mantel-Cox test at P-value 0.0003; the inhibition of Debcl directed by the Ddc-Gal4; UAS-Drp1 transgene result in the median lifespan of 72 (n=303); similar to control, determined by Log-rank Mantel-Cox test at p-value 0.021, with Bonferroni correction. (b) The GraphPad prism8 generated graph of the climbing abilities of Ddc-Gal4 UAS-Drp1 flies with the expression of Buffy, Buffy-RNAi, DebclEY05743, Debcl-RNAiv47515 and control. The climbing abilities of flies overexpressing Buffy have rescued compared to control as determined in the climbing curve's non-linear fitting by a 95% confidence interval (p<0.0001). The climbing ability of the flies has further diminished through the expression of UAS-Buffy-RNAi and UAS-DebclEY05743 as determined in the non-linear fitting of the climbing curve by a 95% confidence interval at a p-value 0.0004 and 0.0002 respectively (n=50). The climbing abilities of flies expressing Debcl-RNAiv47515has rescued compared to control as determined by the non-linear fitting of the climbing curve by a 95% confidence interval (p value<0.0001). The graph of longevity assay was generated by GraphPad prism8 non-linear regression curve.
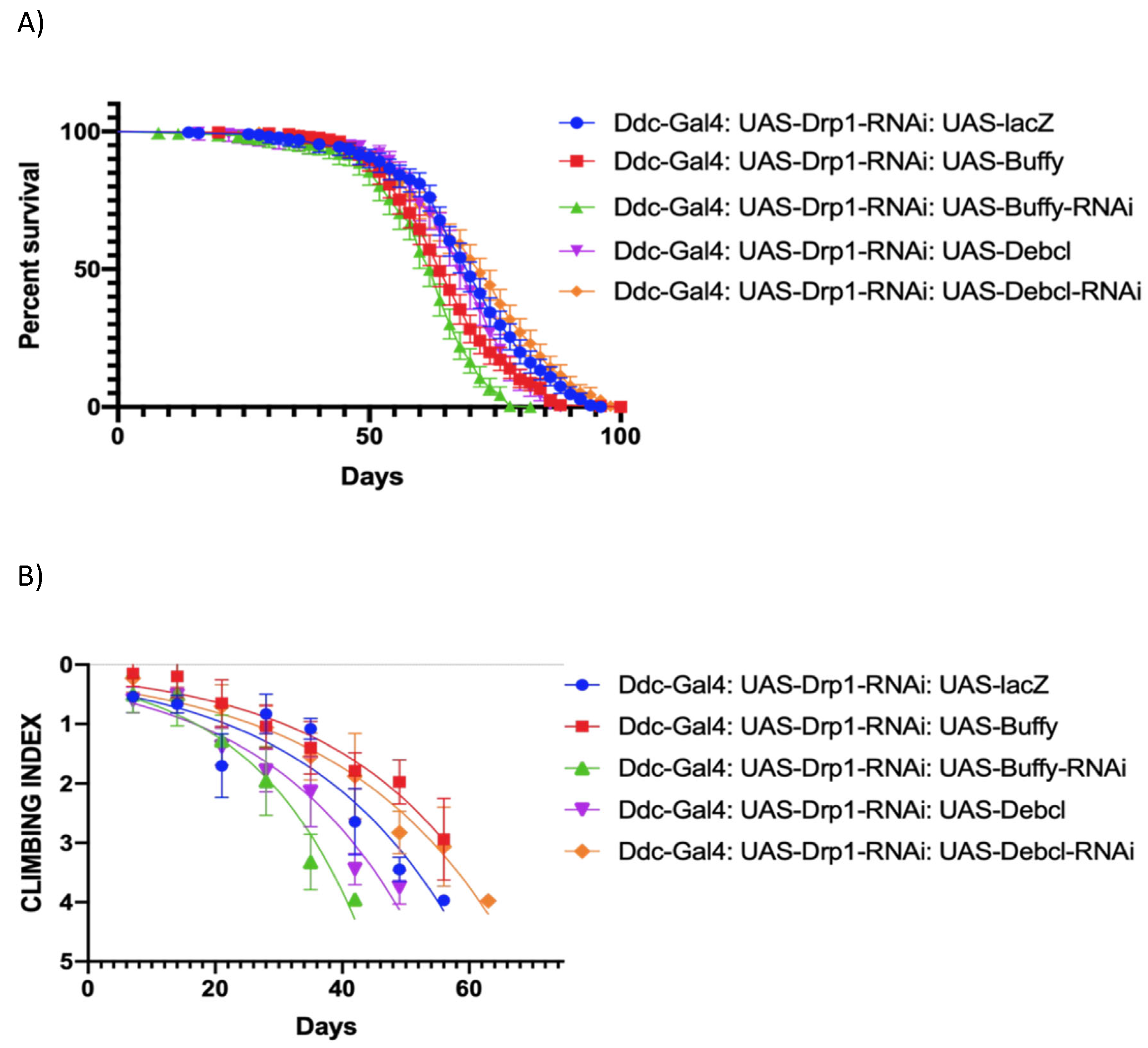
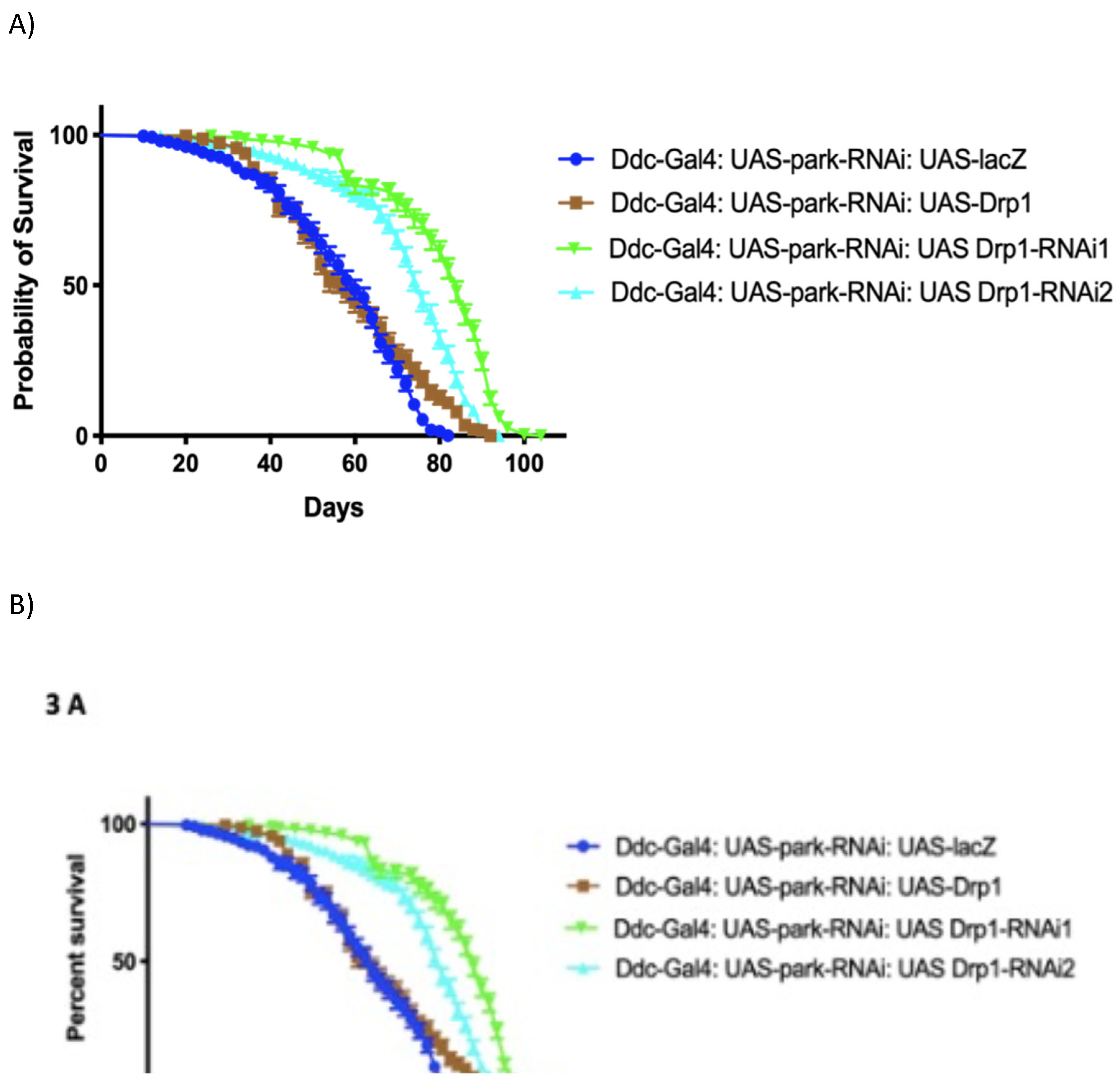
Figure 6.
Inhibition of Drp1 expression enhances lifespan and climbing ability of parkin loss of function in Ddc-Gal44.3D park-RNAi flies. (a) The graph of longevity assay generated by GraphPad prism8 with altered Drp1 expression in Ddc-Gal4 park-RNAi expressing flies. The overexpression results in median lifespan of 58 days similar to 60 days of control (lacZ/park-RNAi) determined by Log-rank Mantel-Cox test, with Bonferroni correction. The inhibition of Drp1 in neurons using Ddc-Gal4 transgene along with park-RNAi results in increased lifespan of 84 days with UAS-Drp1-RNAi1 and lifespan of 76 days with UAS-Drp1-RNAi2 compare to 60 days of control done by Log-rank Mantel Cox test, with Bonferroni correction. (b) The GraphPad prism8 generated graph of the climbing abilities of flies with overexpression of Drp1, Drp1 RNAi and control. The climbing abilities of and Drp1 RNAi flies has rescued compared to control as determined in non-linear fitting of the climbing curve by 95% confidence interval.
Figure 6.
Inhibition of Drp1 expression enhances lifespan and climbing ability of parkin loss of function in Ddc-Gal44.3D park-RNAi flies. (a) The graph of longevity assay generated by GraphPad prism8 with altered Drp1 expression in Ddc-Gal4 park-RNAi expressing flies. The overexpression results in median lifespan of 58 days similar to 60 days of control (lacZ/park-RNAi) determined by Log-rank Mantel-Cox test, with Bonferroni correction. The inhibition of Drp1 in neurons using Ddc-Gal4 transgene along with park-RNAi results in increased lifespan of 84 days with UAS-Drp1-RNAi1 and lifespan of 76 days with UAS-Drp1-RNAi2 compare to 60 days of control done by Log-rank Mantel Cox test, with Bonferroni correction. (b) The GraphPad prism8 generated graph of the climbing abilities of flies with overexpression of Drp1, Drp1 RNAi and control. The climbing abilities of and Drp1 RNAi flies has rescued compared to control as determined in non-linear fitting of the climbing curve by 95% confidence interval.