1. Introduction
The study of cellulolytic microbial communities is a highly popular topic due to its applied importance and well-developed methodologies that allow for effective functional attribution and validation of results [
1]. It should be noted, however, that a considerable number of these studies concentrate on either the rumen microbiome or the construction of particular strain combinations [
2,
3]. Despite not being a rare topic of study, there is still a lack of representation of soil microorganisms with cellulolytic activity in both research studies and databases (CAZy [
4], PULDB [
5]) [
6]. Furthermore, the majority of these studies concentrate on the degradable substrate or the characterization of a particular cellulolytic ecotope [
7]. The microbial cellulolytic component of the substrate itself and the microorganisms from the surrounding microbial pool cannot be separated in the communities thus obtained. This, in turn, prevents us from discerning the role of biotic and abiotic components, and understanding the interaction of different trophic groups within the community, and thus determines the sustainability and efficiency of the microbial community [
8]. In this study, we employ a methodology that enables the investigation of the active soil microbiome through the analysis of the chronosequence involved in the colonization of a sterile substrate in nylon bags placed into soil. This approach represents a synthesis between traditional, culture-based techniques and standard procedures utilizing amplicon sequencing. In contrast to the simple mixing of soil with substrate, our methodology reduces the background of dominant soil microorganisms, which are frequently not active cellulolytic [
9].
In a prior experiment examining community succession on a sterile substrate [
10], soil microorganisms colonized oat straw, a material rich in simple sugars, proteins, and polysaccharides [
11]. Simple sugars stimulate bacterial growth in the initial stages of colonization, and proteins provide nitrogen, which is often a limiting resource in the decomposition of lignocellulosic substrates [
12]. In that study, community change was observed during the middle phases of substrate decomposition, when progressing from more accessible substrates (simple sugars) to polysaccharides.
Our study’s primary objective was to differentiate between two distinct and contrasting types of soil communities, which have not been previously accomplished. The utilization of crystalline cellulose as a substrate allowed the investigation of an isolated community with high catalytic potential for the development of highly efficient cellulolytic consortia based on it.
2. Materials and Methods
To set up the experiment, suspensions of sterile crystalline cellulose powder were placed in sterile nylon bags. Following this, the bags were placed at a depth of 5 mm in two types of soil: chernozem and sod-podzolic soil. The experiment was conducted at room temperature and a fixed humidity of 60% in 2-liter plastic containers for six months. At 7, 35, 70, and 98 days, the bags were removed from the soil. Three replicates were used for each data point, with each replicate corresponding to a different bag. At each sampling point, soil respiration was quantified compared to a control sample comprising an identical soil mixture lacking cellulose. The methodology has been previously described in detail [
13].
The soil DNA was isolated from the decomposed substrate, and the variable regions of the 16S rRNA gene and ITS2 fragment were sequenced on the Illumina MiSeq platform. The previously described routine methodology [
14] was employed for the processing of sequencing. Furthermore, additional filtering was employed, whereby ASVs that had not been defined to the phylum level were removed from the dataset. To investigate the network organization of communities, covariance networks were constructed for two soil types (sod-podzolic and chernozem). The networks demonstrate the co-occurrence of phylotypes for the chronoseries. Network analysis was conducted on the remaining ASVs, which were represented by more than five reads in no more than 10% of all samples. The SPIEC-EASI algorithm [
15], as part of the SpiecEasi package within the R language, was employed for the construction of the networks. Meinshausen-Buhlmann’s neighborhood selection method was used [
16]. The igraph package [
17] was employed to calculate clusters and modularity of the network structure, while the random walk algorithm was utilized to select clusters. The GGally package was used for visualization. The versions of the packages and the code used for post-processing can be found in the supplement (S1) and repository (
https://github.com/crabron/dyn_cell).
3. Results
This section may be divided by subheadings. It should provide a concise and precise description of the experimental results, their interpretation, as well as the experimental conclusions that can be drawn.
Two sets of amplicon libraries were obtained - for the variable region of the 16S rRNA gene (78400 reads) and ITS2 (117110 reads). The most represented phyla in the prokaryotic community were Pseudomonadota, Bacteroidota, Bacillota, Verrucomicrobiota, and Actinobacteriota. Chronoseries were similar at the phylum level for different soil types. The early stages were characterized by representatives of Gammaproteobacteria, Bacillota, and Actinobacteriota. For the late stages - alphaproteobacteria, Bacteroidota, Verrucomicrobiota, Planctomycetota, and Myxococcota.
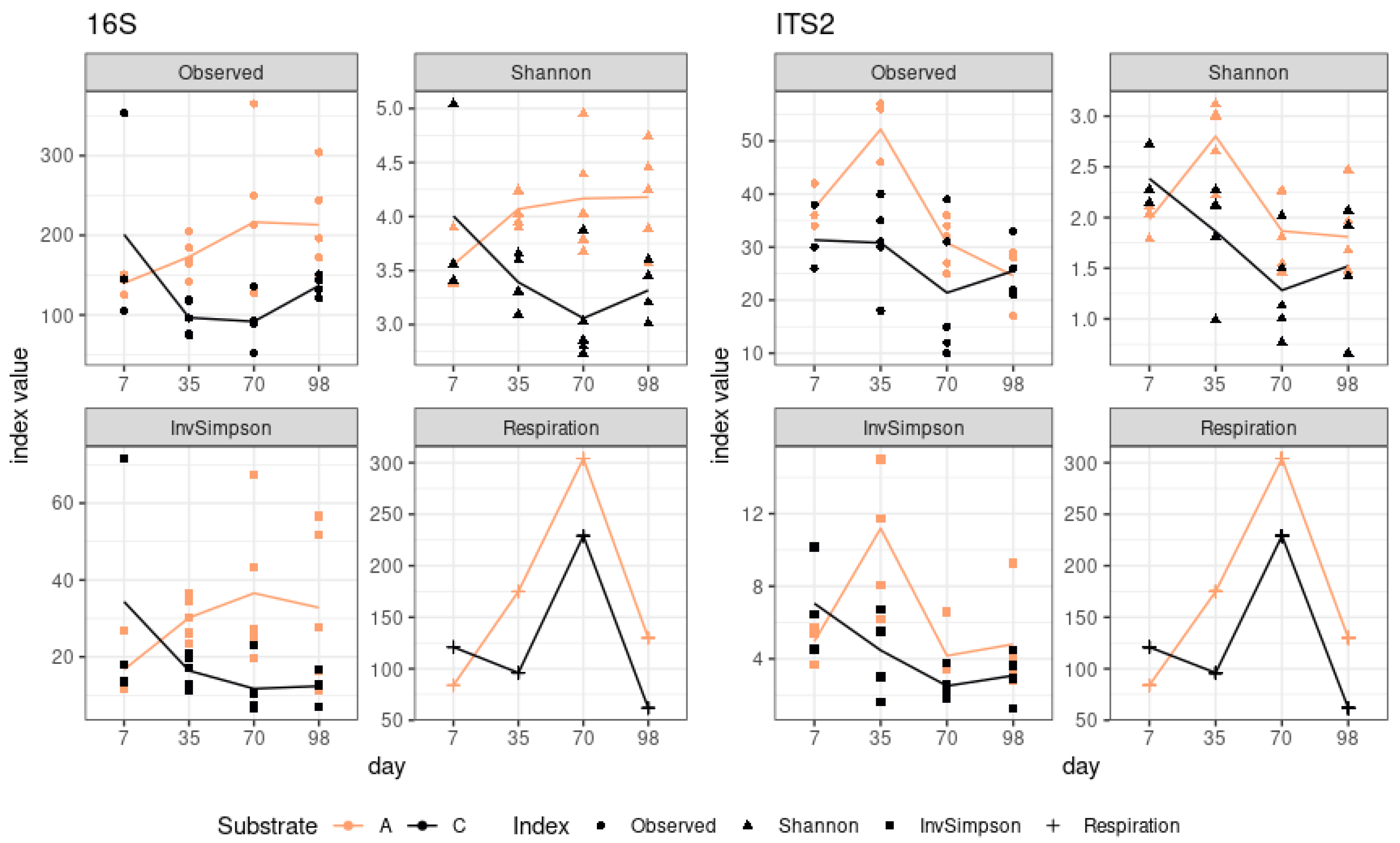
The growth of alpha-diversity indices was a characteristic feature of sod-podzolic soil, whereas a decline was observed in chernozem (
Figure 1). While alpha-diversity indices for different soil types demonstrated opposing dynamics, the respiration profile exhibited a consistent pattern, displaying a growth phase up to the third point and a subsequent slight decline. The lack of significant shifts in respiration and diversity dynamics can be attributed to the absence of a change in the community’s substrate preference, from simple sugars to complex substrates. Conversely, there is no evidence to suggest that the increase in respiration at day 70 is linked to the taxonomic shift of the community.
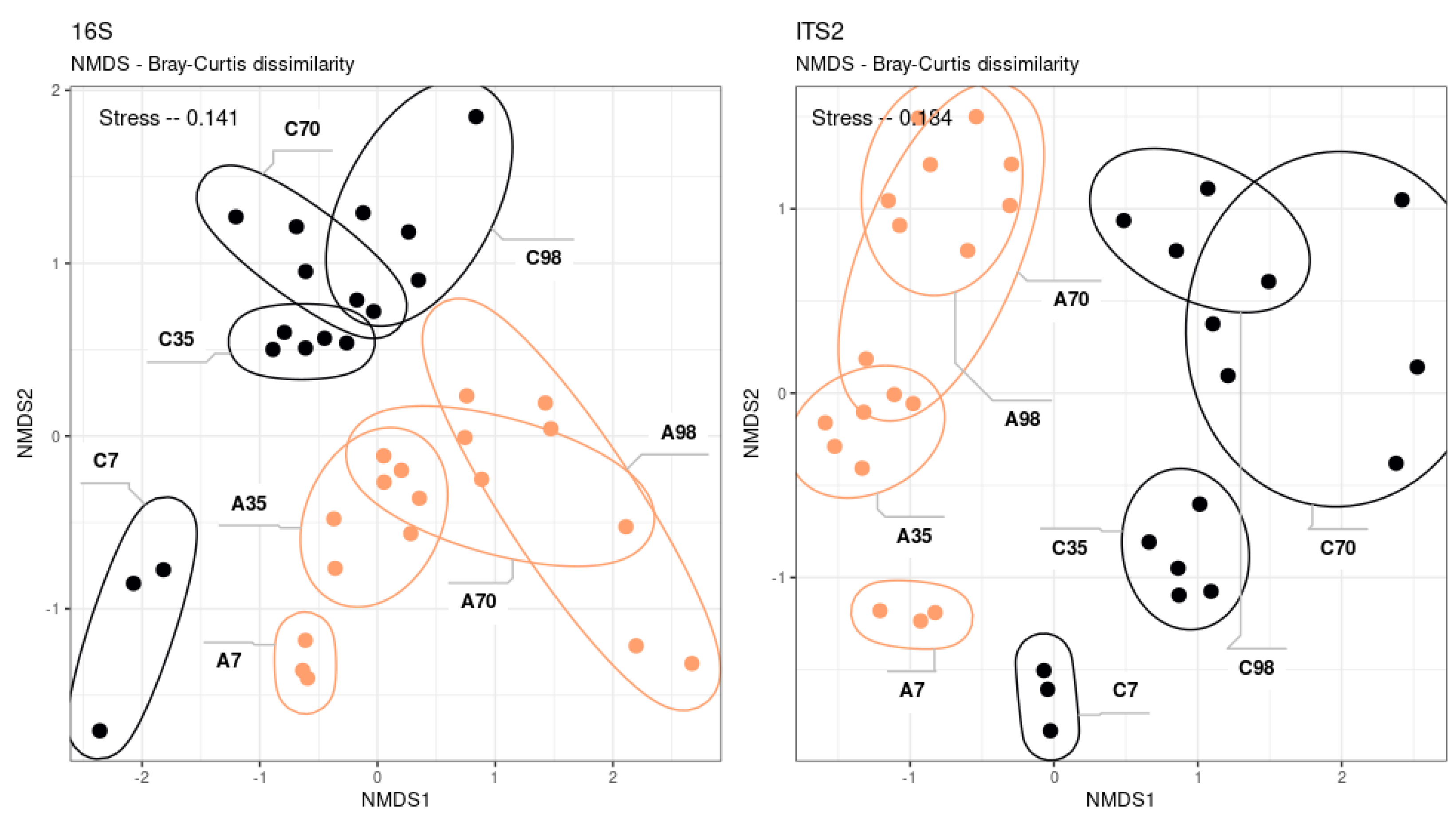
The resulting communities exhibit significant differences from one another (adonis p-value < 0.001) (
Figure 2). The development of the prokaryotic and eukaryotic communities is strictly linear from the early to the late stages. The partitioning of the community by substrate and the correspondence between the eukaryotic and prokaryotic successions were observed. The data demonstrate that alterations in community dynamics occurred up to day 35, after which the taxonomic composition of the community reached a state of stability.
Community differences manifest at the highest taxonomic level. Bacillota, Chloroflexota, and Bdellovibrionota were more characteristic of sod-podzolic soil. Chernozem was distinguished by the initial presence of Archaea and a considerably higher proportion of
Streptomyces relative to other organisms. It is distinctive that certain microorganisms are identified in two distinct soil types, although at varying stages. This phenomenon is particularly prevalent among Acidobacteriota. Concurrently, at the low taxonomic level (ASV, genus), soil samples exhibit notable differences, particularly among representatives of the Bacteroidota phylum. This observation can be attributed to the high functional diversity observed among members of this phylum [
18].
The identified eukaryotic community displayed a considerably lower level of diversity than the prokaryotic community (334 vs. 2427 ASVs), but its dynamics exhibited a correspondence to the prokaryotic community. In contrast to sod-podzolic soil, the data for chernozem indicate differences at day 35 from later phases. At day 35, the community included basidiomycetes
Coprinellus, ascomycetes
Chaetomium and
Parachaetomium (previously identified as cellulolytic [
19]), and
Schizothecium and
Dictyosporium. In the late stages, a high proportion of nematodes was observed in the community, with the main fungus represented by predatory fungi of the genus
Arthrobotrys [
20]. The cellulolytic component of the community is represented by members of the genus
Stachybotrys, which emerge at the second point of the chronoseries [
21].
In both soil types, by the late stages, there was an increase in the relative representation of nematodes in comparison to the remainder of the eukaryotic community. In sod-podzolic soil, the proportion of nematodes among the total number of eukaryotes increases from the second to the third stage, from 29.5 to 41.5 percent. In chernozem soil, the same trend is observed, but with a lower range, from 8.1 to 16.2 percent. Concurrently, nematodes from disparate ecological niches were represented in the two soil types. For example, in sod-podzolic soil,
Acrobeloides,
Pseudacrobeles, and
Aphelenchoides [
22] were present. For the sod-podzolic soil, the nematodes observed were
Acrobeloides,
Pseudacrobeles,
Aphelenchoides (only at the second stage),
Ditylenchus medicaginis, and
Pseudacrobeles curvatus (only at the third stage). For the chernozem soil, the nematodes observed were
Acrobeloides,
Pseudacrobeles,
Aphelenchoides (only at the second stage),
Ditylenchus medicaginis, and
Pseudacrobeles curvatus (only at the third stage) [
21].
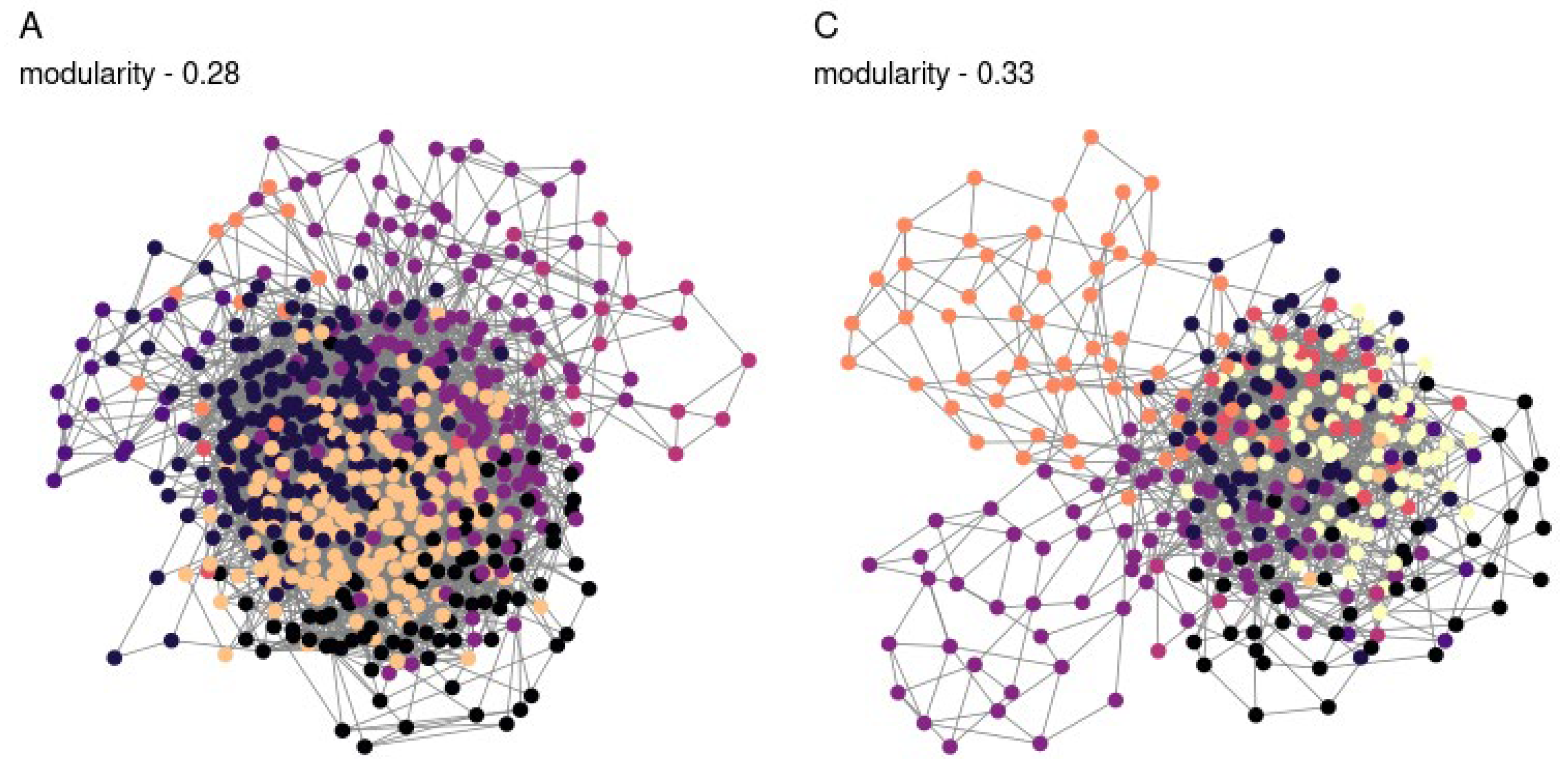
We applied a compositional approach to construct co-correlation networks (
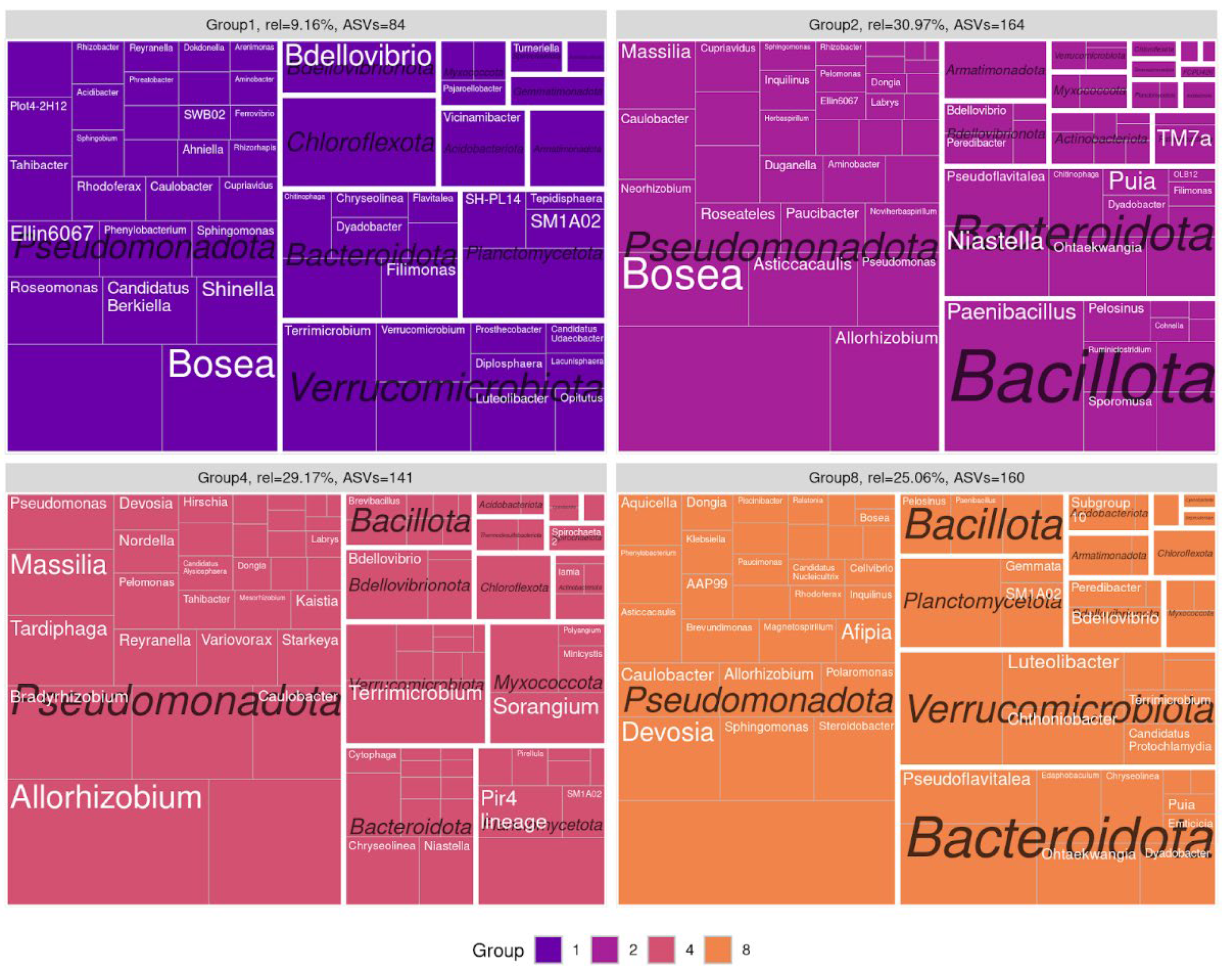
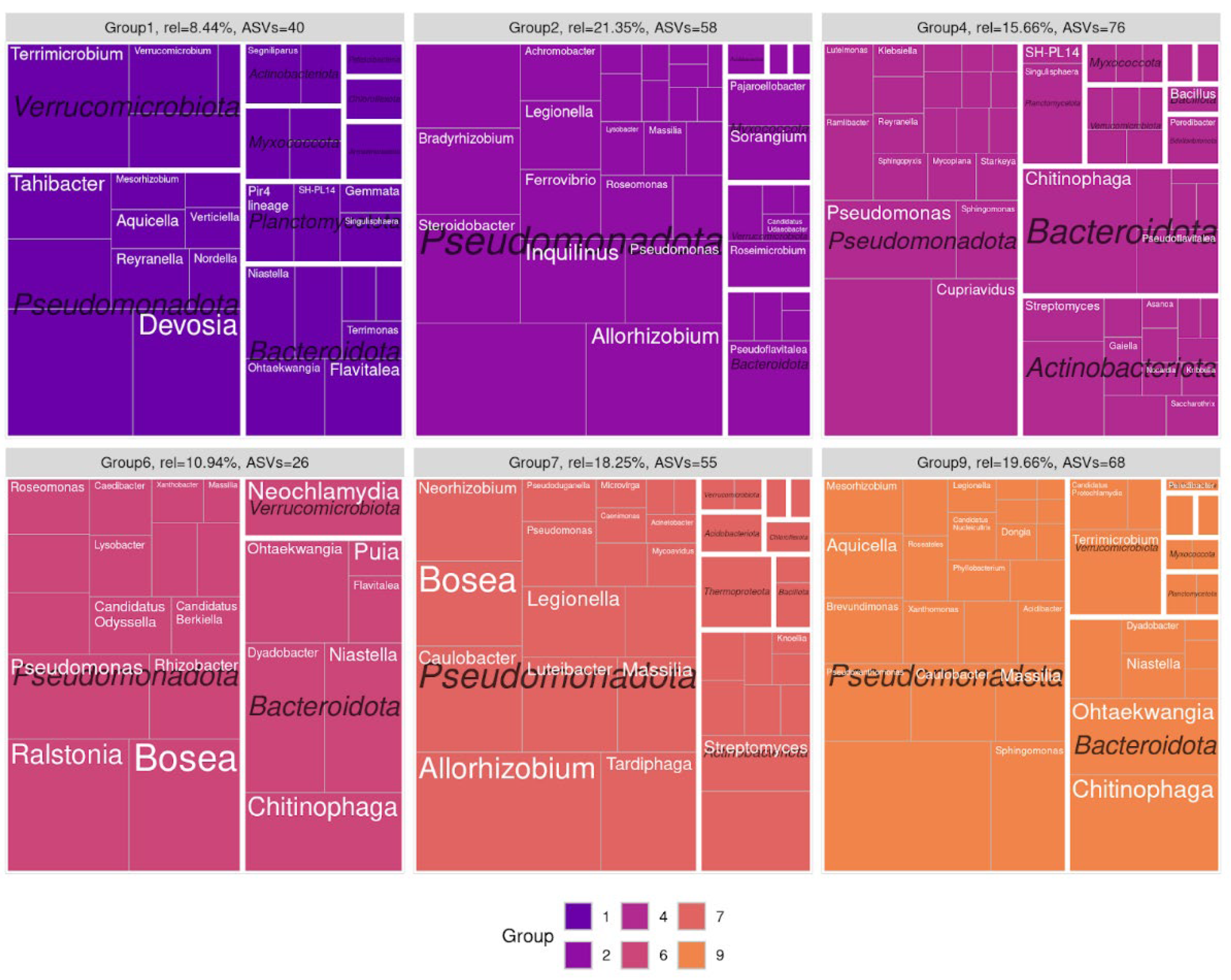
Figure 3), which enabled us to normalize the varying amplicon library sizes while avoiding the distortions associated with rarefaction or normalization. It was demonstrated that the resulting network structure varied by the soil type. The communities obtained from sod-podzolic soil displayed greater structural homogeneity compared to those from chernozem. The chernozem community exhibited lower modularity and network connectivity (network modularity index for sod-podzolic soil: 0.28; for chernozem: 0.32). Clusters identified using a random walk algorithm showed analogous alterations in relative representation. In the case of sod-podzolic soil, eight groups of microorganisms were identified, four of which were classified as major (i.e., comprising more than 5% of the total community) (
Figure 4). In the case of the chernozem soil, nine groups were identified, of which four were classified as major (see
Figure 5). The taxonomic composition of the sod-podzolic soil was relatively homogeneous, in contrast to the chernozem, which was characterized by the presence of small taxon-specific clusters. Cluster 6, which is characterized by a high prevalence of major ASVs belonging to the Bacteroidetes phylum (specifically,
Niastella and
Dyadobacter), is worthy of closer examination. The most abundant bacteria in this cluster are specifically associated with the second point in the chronoseries. Additionally, сluster 4, dominated by actinobacteria (
Streptomyces), is also noteworthy, with representatives most abundant at the first point in the chronoseries.
4. Discussion
The application of metagenomic methodologies in the search for effective biodegrades is a common practice in the scientific community. However, the utilization of the obtained strains in biopreparations frequently demonstrates considerable instability of active microorganisms when applied to soil. An alternative approach involves the use of complex biopreparations [
23,
24]. This strategy requires a comprehensive understanding of both the decomposition processes and the ecological impacts specific to the soil type where the biopreparation is applied [
25]. This study focuses on the temporal dynamics of sterile crystalline cellulose decomposition, with a particular interest in the ecological implications of cellulolytic activity among soil microorganisms, particularly concerning the soil type in which the substrate is applied.
Previously, we conducted a similar study using a more biologically accessible substrate (oat straw) in a single soil type (sod-podzolic). The results demonstrated a clear correlation between the shift in prokaryotic community composition from simpler substrates (simple sugars) to more complex ones (polysaccharides) and the change in total soil respiration. In this study, cellulose was selected as the substrate to eliminate the influence of the source material and to focus on the role of soil microorganisms in the degradation of complex polysaccharides. The chronoseries on crystalline cellulose demonstrated a relatively stable diversity over time. Additionally, the inverse correlation between alpha-diversity indices and respiration, previously observed in other studies [
10], was not evident in this experiment.
In contrast to the minimal alteration in alpha diversity indices, the prokaryotic community structure undergoes a significant transformation throughout the study period. Our findings demonstrate the presence of cellulolytic microorganisms in both the early and late stages. Early on, we identified cellulolytic microorganisms described in the literature, such as representatives of the genera Streptomyces for chernozem and
Paenibacillus, and
Ochtaekwangia for sod-podzolic soil [
26]. By the later stages, the proportion of Proteobacteria had decreased, while the proportion of minor Bacillota representatives and the total proportion of Bacteroidota and minor phyla had increased.
These observations are largely aligned with previous reports. For instance, the initial colonization of the site by members of the
Burkholderia,
Pseudomonas, and
Streptomyces genera [
27] has been previously documented. Notably, a subset of Bacteroides (
Niastella) was present in the initial stages but was subsequently displaced from the community. For each substrate species, two distinct stages of microbial community development can be identified: an early stage (7 days) and a late stage (35-98 days). In the case of the eukaryotic amplicon libraries of chernozem, an intermediate stage at day 35 is also distinguished, which is absent in the sod-podzolic soil community. At this stage, a decline in soil respiration, a reduction in the diversity of prokaryotes, and a shift in the taxonomic profile of eukaryotes were observed.
The changes in the prokaryotic profile, as revealed by network analysis, appear to be indirectly associated with modifications in the eukaryotic component. It is hypothesized that the key drivers of community formation are the presence of specific nematodes in the soil and their subsequent interactions with fungal and prokaryotic components. Despite the relatively low diversity (only 11 phylotypes), nematodes -- account for 41.5% of all eukaryotic reads in sod-podzolic soil. The predominant group of reads belonged to the predatory fungus
Arthrobotrys, which prey on nematodes. This fungus was present in sod-podzolic soil at the initial stage and in chernozem soil at the third stage, with a representation of 64% in the latter. Notably, the predominant nematodes in different soil types belong to disparate trophic groups. For example,
Acrobeloides and
Pseudacrobeles, prevalent in sod-podzolic soil, are bacteria feeders [
22], whereas
Ditylenchus and
Aphelenchoides are mycotrophs or plant parasites [
28]. A specific response of the prokaryotic component to the overrepresentation of nematodes in the second stage of chernozem has been demonstrated. The presence of
Vampiriovibrio was observed in the second stage of chernozem, in contrast to sod-podzolic soil, where
Chitinofaga was the predominant species. These microorganisms have been associated with the decomposition of fungal mycelium residues [
29] and predation [
30].
The observed differences in the substrate occupation by soil cellulolytic organisms between the two soil types can be linked to the fact that the sod-podzolic soil is distinguished by a higher prevalence of slow-growing microorganisms with high catalytic potential, in contrast to the more nutrient-rich chernozem [
31]. Additionally, the dissimilarity between the two soil types can be explained by the fact that the chernozem was a less diverse source of microorganisms (as indicated by lower alpha diversity indices of the original soil [
32]), which resulted in a longer formation of a stable community. These findings are consistent with the previously described phenomenon, wherein the microbial community’s response to a eukaryotic component serves as a site-specific marker [
33]. Additionally, this response is closely correlated with the available soil carbon levels [
34].
5. Conclusions
This study demonstrates the distinction between primary colonization by soil cellulolytic originating from two contrasting soil types: chernozem and sod-podzolic soil. The community structure of microorganisms colonizing the sod-podzolic soil substrate exhibited a higher degree of taxonomic homogeneity. Additionally, a gradual increase in the specificity of the prokaryotic component of the communities was observed, contingent upon the dynamics of the eukaryotic component.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
Conceptualization, G.V.G., E.E.A. and O.V.O.; Data curation, G.V.G., A.K.K. and A.D.B.; Formal analysis, G.V.G., A.D.B., and O.V.O.; Funding acquisition, E.E.A.; Investigation, G.V.G. and O.V.O.; Methodology, G.V.G., O.V.O., T.O.L. and E.E.A; Project administration, E.E.A.; Resources, A.A.K. and T.O.L..; Software, G.V.G. and A.D.B.; Supervision, E.E.A.; Validation, G.V.G., and A.K.K.; Visualization, G.V.G.; Writing—original draft, G.V.G.; Writing—review & editing, G.V.G., A.D.B., A.K.K. and E.E.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Russian Science Foundation, grant number 23-16-00147.
Data Availability Statement
Data is available at the NCBI BioProject ID number PRJNA841641.
Acknowledgments
We thank the Centre for Genomic Technologies, Proteomics and Cell Biology (ARRIAM, Russia) for performing the preparation and sequencing of gene and genomic libraries.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Drula, E.; Garron, M.-L.; Dogan, S.; Lombard, V.; Henrissat, B.; Terrapon, N. The Carbohydrate-Active Enzyme Database: Functions and Literature. Nucleic Acids Res. 2022, 50, D571–D577. [Google Scholar] [CrossRef]
- Hayat, R.; Sheirdil, R.A.; Iftikhar-ul-Hassan, M.; Ahmed, I. Characterization and Identification of Compost Bacteria Based on 16S rRNA Gene Sequencing. Ann. Microbiol. 2013, 63, 905–912. [Google Scholar] [CrossRef]
- Soares, F.L.; Melo, I.S.; Dias, A.C.F.; Andreote, F.D. Cellulolytic Bacteria from Soils in Harsh Environments. World J. Microbiol. Biotechnol. 2012, 28, 2195–2203. [Google Scholar] [CrossRef] [PubMed]
- Cantarel, B.L.; Coutinho, P.M.; Rancurel, C.; Bernard, T.; Lombard, V.; Henrissat, B. The Carbohydrate-Active EnZymes Database (CAZy): An Expert Resource for Glycogenomics. Nucleic Acids Res. 2009, 37, D233–D238. [Google Scholar] [CrossRef]
- Terrapon, N.; Lombard, V.; Drula, É.; Lapébie, P.; Al-Masaudi, S.; Gilbert, H.J.; Henrissat, B. PULDB: The Expanded Database of Polysaccharide Utilization Loci. Nucleic Acids Res. 2018, 46, D677–D683. [Google Scholar] [CrossRef] [PubMed]
- Aakko, J.; Pietilä, S.; Toivonen, R.; Rokka, A.; Mokkala, K.; Laitinen, K.; Elo, L.; Hänninen, A. A Carbohydrate-Active Enzyme (CAZy) Profile Links Successful Metabolic Specialization of Prevotella to Its Abundance in Gut Microbiota. Sci. Rep. 2020, 10, 12411. [Google Scholar] [CrossRef]
- Štursová, M.; Žifčáková, L.; Leigh, M.B.; Burgess, R.; Baldrian, P. Cellulose Utilization in Forest Litter and Soil: Identification of Bacterial and Fungal Decomposers. FEMS Microbiol. Ecol. 2012, 80, 735–746. [Google Scholar] [CrossRef]
- Li, S.; Lian, W.-H.; Han, J.-R.; Ali, M.; Lin, Z.-L.; Liu, Y.-H.; Li, L.; Zhang, D.-Y.; Jiang, X.-Z.; Li, W.-J.; et al. Capturing the Microbial Dark Matter in Desert Soils Using Culturomics-Based Metagenomics and High-Resolution Analysis. Npj Biofilms Microbiomes 2023, 9, 67. [Google Scholar] [CrossRef]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil Bacterial and Fungal Communities across a pH Gradient in an Arable Soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Kimeklis, A.K.; Gladkov, G.V.; Orlova, O.V.; Afonin, A.M.; Gribchenko, E.S.; Aksenova, T.S.; Kichko, A.A.; Pinaev, A.G.; Andronov, E.E. The Succession of the Cellulolytic Microbial Community from the Soil during Oat Straw Decomposition. Int. J. Mol. Sci. 2023, 24, 6342. [Google Scholar] [CrossRef]
- Ábrego-Gacía, A.; Poggi-Varaldo, H.M.; Mendoza-Vargas, A.; Mercado-Valle, F.G.; Ríos-Leal, E.; Ponce-Noyola, T.; Calva-Calva, G. Effects of Fermented Oat Straw as a Lovastatin Carrier on in Vitro Methane Production and Rumen Microbiota. Front. Energy Res. 2021, 9, 630701. [Google Scholar] [CrossRef]
- Hobbie, S.E. Interactions between Litter Lignin and Nitrogenitter Lignin and Soil Nitrogen Availability during Leaf Litter Decomposition in a Hawaiian Montane Forest. Ecosystems 2000, 3, 484–494. [Google Scholar] [CrossRef]
- Orlova, O.V.; Kichko, A.A.; Pershina, E.V.; Pinaev, A.G.; Andronov, E.E. Succession of Bacterial Communities in the Decomposition of Oats Straw in Two Soils with Contrasting Properties. Eurasian Soil Sci. 2020, 53, 1620–1628. [Google Scholar] [CrossRef]
- Gladkov, G.; Kimeklis, A.; Zverev, A.; Pershina, E.; Ivanova, E.; Kichko, A.; Andronov, E.; Abakumov, E. Soil Microbiome of the Postmining Areas in Polar Ecosystems in Surroundings of Nadym, Western Siberia, Russia. Open Agric. 2019, 4, 684–696. [Google Scholar] [CrossRef]
- Kurtz, Z.D.; Müller, C.L.; Miraldi, E.R.; Littman, D.R.; Blaser, M.J.; Bonneau, R.A. Sparse and Compositionally Robust Inference of Microbial Ecological Networks. PLOS Comput. Biol. 2015, 11, e1004226. [Google Scholar] [CrossRef]
- Meinshausen, N.; Bühlmann, P. High-Dimensional Graphs and Variable Selection with the Lasso. Ann. Stat. 2006, 34. [Google Scholar] [CrossRef]
- Csárdi, G.; Nepusz, T.; Müller, K.; Horvát, S.; Traag, V.; Zanini, F.; Noom, D. Igraph for R: R Interface of the Igraph Library for Graph Theory and Network Analysis 2024.
- Wolińska, A.; Kuźniar, A.; Zielenkiewicz, U.; Izak, D.; Szafranek-Nakonieczna, A.; Banach, A.; Błaszczyk, M. Bacteroidetes as a Sensitive Biological Indicator of Agricultural Soil Usage Revealed by a Culture-Independent Approach. Appl. Soil Ecol. 2017, 119, 128–137. [Google Scholar] [CrossRef]
- Moo-Young, M.; Chahal, D.S.; Swan, J.E.; Robinson, C.W. SCP Production by Chaetomium Cellulolyticum, a New Thermotolerant Cellulolytic Fungus. Biotechnol. Bioeng. 1977, 19, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Biodiversity of Fungi; Elsevier, 2004; ISBN 978-0-12-509551-8.
- Andersen, B.; Poulsen, R.; Hansen, G.H. Cellulolytic and Xylanolytic Activities of Common Indoor Fungi. Int. Biodeterior. Biodegrad. 2016, 107, 111–116. [Google Scholar] [CrossRef]
- Shokoohi, E.; Moyo, N.; Gouveia, F. Relationship of Nematodes in Natural and Disturbed Land with Physicochemical Properties in Magoebaskloof, Limpopo Province, South Africa. Biologia (Bratisl.) 2023, 78, 3223–3233. [Google Scholar] [CrossRef]
- Hidalgo, D.; Corona, F.; Martín-Marroquín, J.M. Manure Biostabilization by Effective Microorganisms as a Way to Improve Its Agronomic Value. Biomass Convers. Biorefinery 2022, 12, 4649–4664. [Google Scholar] [CrossRef]
- Pylak, M.; Oszust, K.; Frąc, M. Review Report on the Role of Bioproducts, Biopreparations, Biostimulants and Microbial Inoculants in Organic Production of Fruit. Rev. Environ. Sci. Biotechnol. 2019, 18, 597–616. [Google Scholar] [CrossRef]
- Kyrychenko. Market analysis and microbial biopreparations creation for crop growing in ukraine. Biotechnol. Acta 2015, 8, 42–52. [Google Scholar] [CrossRef]
- Balla, A.; Silini, A.; Cherif-Silini, H.; Bouket, A.C.; Boudechicha, A.; Luptakova, L.; Alenezi, F.N.; Belbahri, L. Screening of Cellulolytic Bacteria from Various Ecosystems and Their Cellulases Production under Multi-Stress Conditions. Catalysts 2022, 12, 769. [Google Scholar] [CrossRef]
- Vieira, S.; Sikorski, J.; Gebala, A.; Boeddinghaus, R.S.; Marhan, S.; Rennert, T.; Kandeler, E.; Overmann, J. Bacterial Colonization of Minerals in Grassland Soils Is Selective and Highly Dynamic. Environ. Microbiol. 2020, 22, 917–933. [Google Scholar] [CrossRef] [PubMed]
- Sanwal, K.C. A KEY TO THE SPECIES OF THE NEMATODE GENUS APHELENCHOIDES FISCHER, 1894. Can. J. Zool. 1961, 39, 143–148. [Google Scholar] [CrossRef]
- Tran, T.L.Q.; Anani, H.; Trinh, H.T.; Pham, T.P.T.; Dang, V.K.; Ho, V.M.; Bui, N.H.L.; Nguyen, N.H.; Raoult, D.; Trinh, T.T.; et al. Chitinophaga Vietnamensis Sp. Nov., a Multi-Drug Resistant Bacterium Infecting Humans. Int. J. Syst. Evol. Microbiol. 2020, 70, 1758–1768. [Google Scholar] [CrossRef] [PubMed]
- Hovde, B.T.; Steichen, S.A.; Starkenburg, S.R.; Brown, J.K. Vampirovibrio Chlorellavorus Draft Genome Sequence, Annotation, and Preliminary Characterization of Pathogenicity Determinants. Phycol. Res. 2020, 68, 23–29. [Google Scholar] [CrossRef]
- Trifonova, T.; Kosmacheva, A.; Sprygin, A.; Chesnokova, S.; Byadovskaya, O. Enzymatic Activity and Microbial Diversity of Sod-Podzolic Soil Microbiota Using 16S rRNA Amplicon Sequencing Following Antibiotic Exposure. Antibiotics 2021, 10, 970. [Google Scholar] [CrossRef]
- Evdokimova, E.V.; Gladkov, G.V.; Kuzina, N.I.; Ivanova, E.A.; Kimeklis, A.K.; Zverev, A.O.; Kichko, A.A.; Aksenova, T.S.; Pinaev, A.G.; Andronov, E.E. The Difference between Cellulolytic ‘Culturomes’ and Microbiomes Inhabiting Two Contrasting Soil Types. PLOS ONE 2020, 15, e0242060. [Google Scholar] [CrossRef]
- Bao, Z.; Ikunaga, Y.; Matsushita, Y.; Morimoto, S.; Takada-Hoshino, Y.; Okada, H.; Oba, H.; Takemoto, S.; Niwa, S.; Ohigashi, K.; et al. Combined Analyses of Bacterial, Fungal and Nematode Communities in Andosolic Agricultural Soils in Japan. Microbes Environ. 2012, 27, 72–79. [Google Scholar] [CrossRef]
- Karpinska, A.; Ryan, D.; Germaine, K.; Dowling, D.; Forrestal, P.; Kakouli-Duarte, T. Soil Microbial and Nematode Community Response to the Field Application of Recycled Bio-Based Fertilisers in Irish Grassland. Sustainability 2021, 13, 12342. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).