Submitted:
16 July 2024
Posted:
16 July 2024
You are already at the latest version
Abstract
Keywords:
I. Introduction
- Ranks among the top-performing models in the European region, indicating its potential to effectively address regional medical challenges.
- Achieves competitive results with our custom model based on EfficientNetB3, demonstrating efficient utilization of limited training data.
- Enhances practical feasibility and cost-effectiveness of deployment due to its modest computational requirements.
- It shows robust performance with fewer images compared to models that achieve similar or better results with larger datasets.
II. Methodology of Research
2.1. Description of the Training Dataset
2.2. Data Pre-Processing
- Resizing: Images were resized to 300x300, ensuring uniformity in input sizes.
- Normalization: Pixel values were scaled to the [0, 1] interval to reduce data discrepancies and aid training convergence.
- Mean Subtraction and Standardization: Each pixel’s value had the dataset’s mean subtracted and was then divided by the standard deviation to further normalize the data, enhancing model convergence.
- Data Augmentation: This technique creates new images by modifying existing ones. We employ mirroring, translation, rotation, scaling, brightness adjustment, and noise addition to augment the existing pictures [15]. These augmented images are then added to the categories with less data, thereby balancing the training dataset.
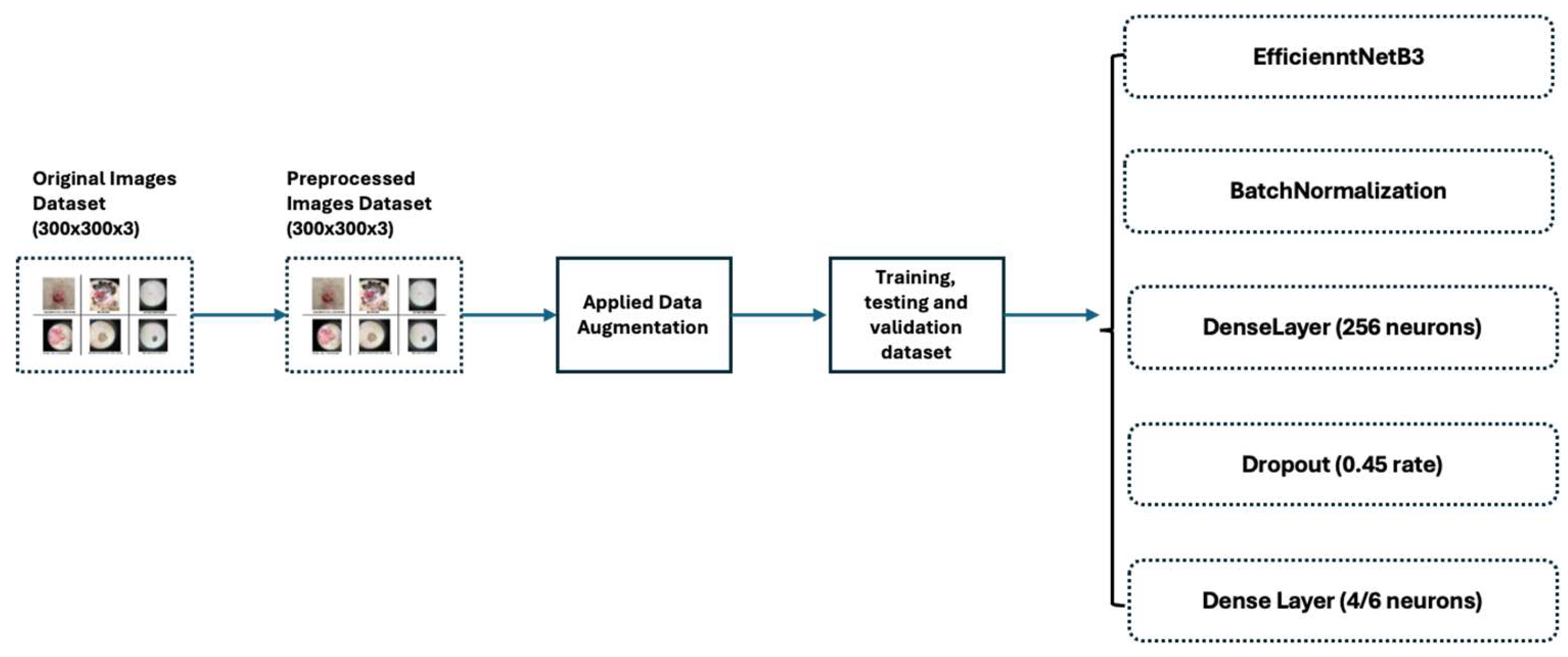
2.3. The Usage and the Architecture of the Model
- On top of the EfficientNetB3 model, adding a Batch Normalization [21] layer improved accuracy by enhancing convergence and helped reduce overfitting. Batch Normalization contributed to smoother training and improved generalization on unseen data by stabilizing and normalizing activations throughout the network.
- Two additional dense layers significantly enhanced classification performance by introducing non-linear features, extracting higher-level features, reducing parameter count and dimensionality of input images, and serving as a regularization technique.
- Finally, one dropout layer [22] randomly deactivated the neurons during training, which helped prevent overfitting by encouraging the model to generalize better. This technique improves the robustness and performance of the neural network on unseen data.
2.4. Training and Validating the Model
2.4.1. Hyperparameters
2.4.2. Techniques Used to Combat Overfitting
- Dropout: Dropout selectively deactivates neurons in neural network layers during training, simulating smaller networks within the model. This approach encourages the network to diversify its learning strategies, enhancing generalization and mitigating overfitting by preventing reliance on individual neurons. [31].
- Batch Normalization: Normalization adjusts data to a mean of zero and a standard deviation of one, aligning and scaling inputs. Batch Normalization speeds up training by preventing gradients from becoming too small, facilitating faster convergence with higher learning rates. It also acts as a regularizer, reducing overfitting and improving model generalization on new data. This stability reduces sensitivity to initial weight choices and simplifies experimenting with different architectures. [32].
- Regularization: We used the regularization techniques to reduce overfitting: L2 regularization with a strength of 0.016 for the kernel and L1 regularization at a strength of 0.006 for both activity and bias regularization. These methods were chosen to mitigate overfitting by penalizing large parameter values in the model, thereby promoting more straightforward and more generalized outcomes across varying datasets and scenarios.
III. Results
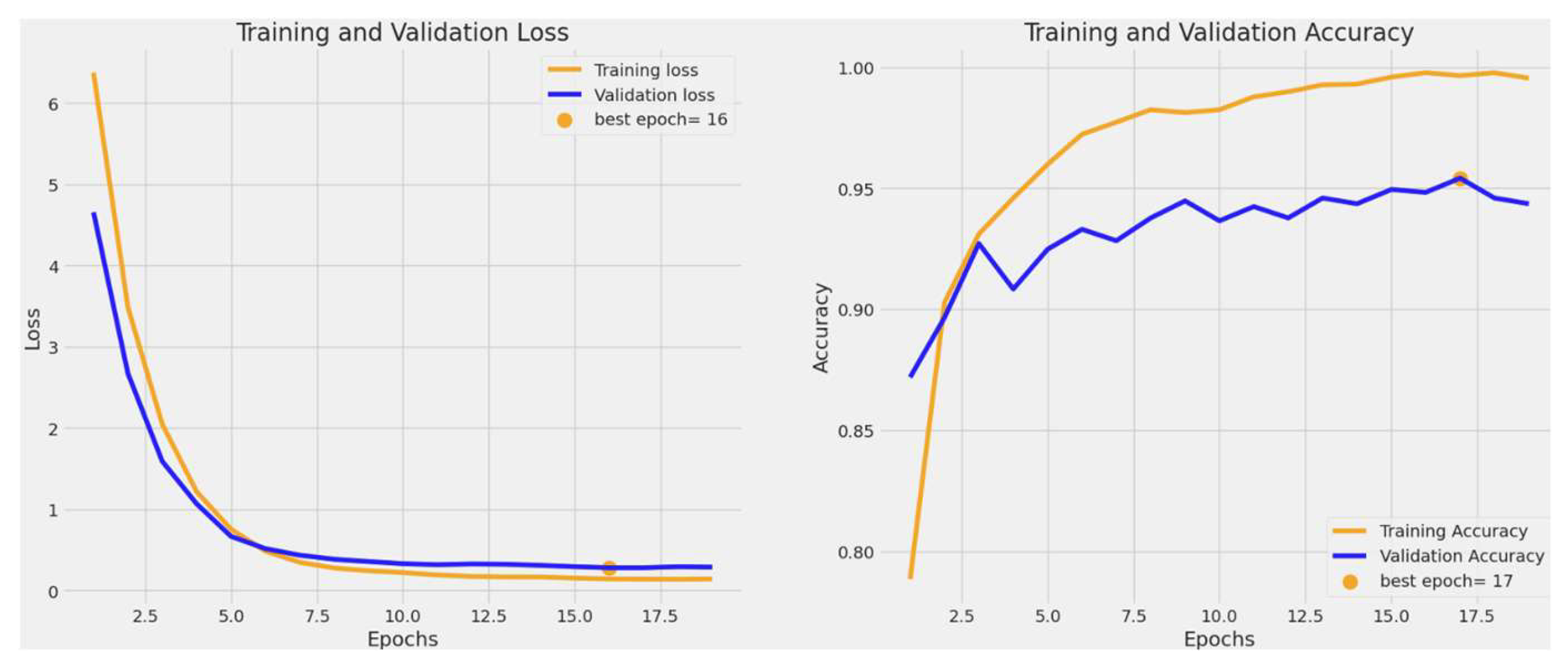
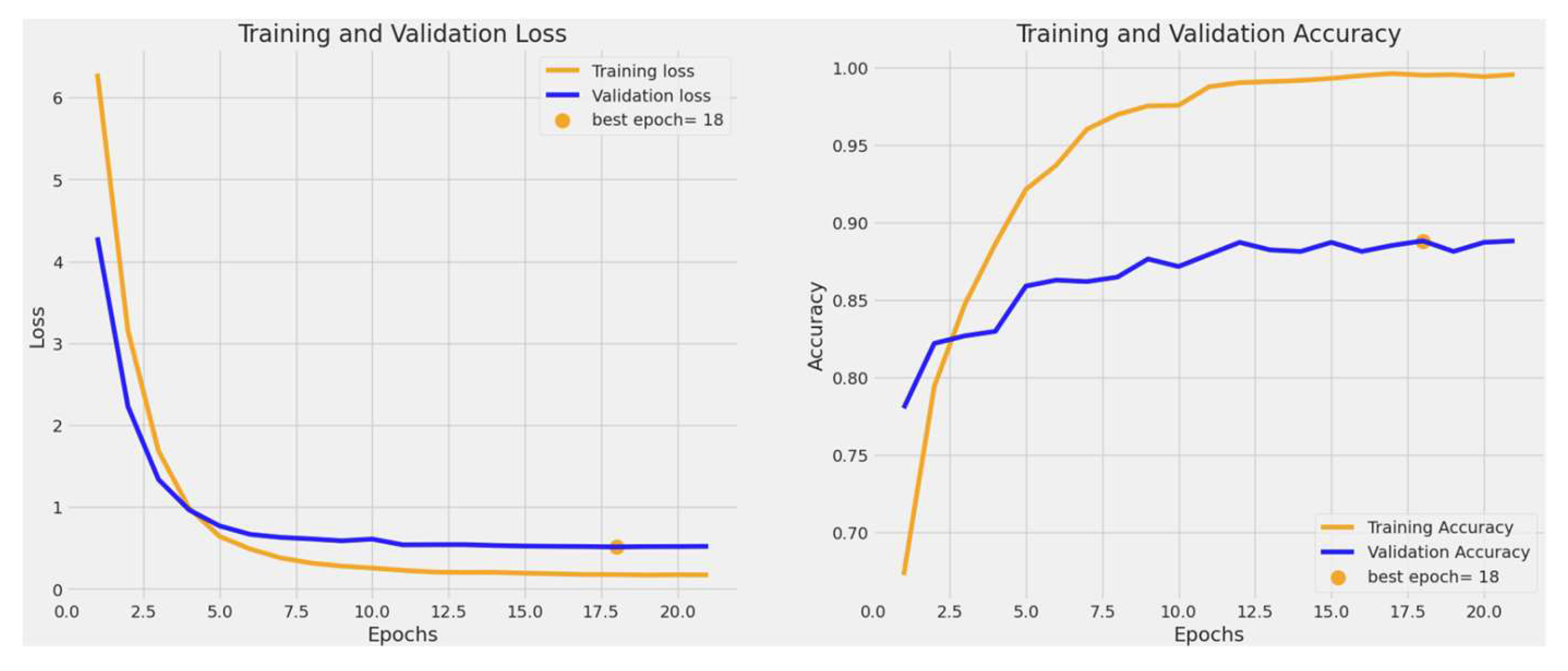
3.1. Training and Validation Accuracy and Loss
3.2. Classification Performance
3.3. Receiver Operating Characteristic (ROC) Curve
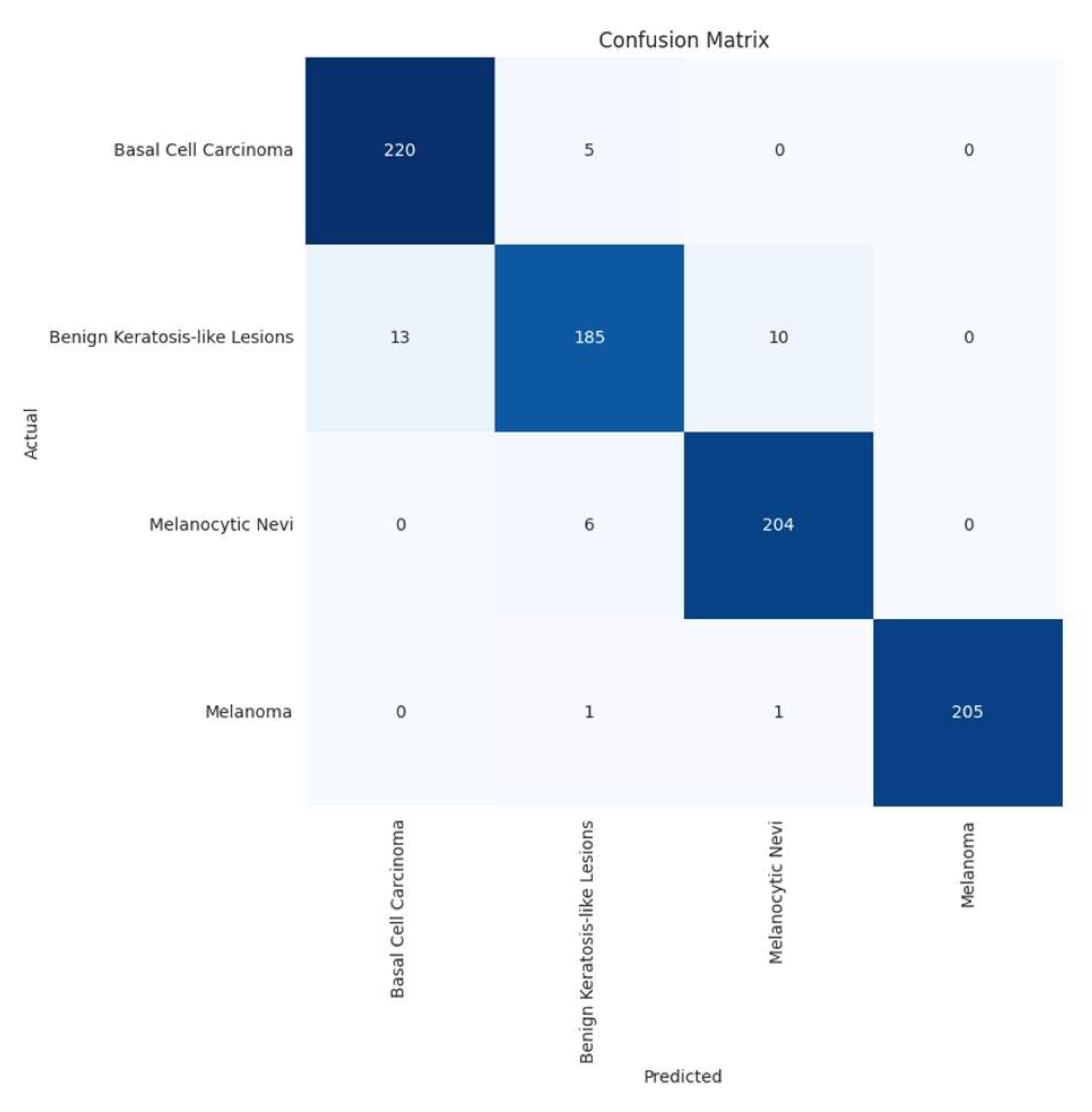
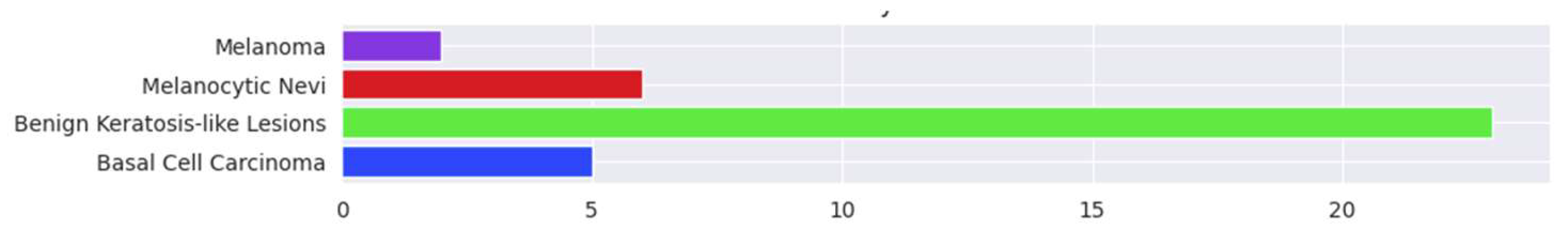
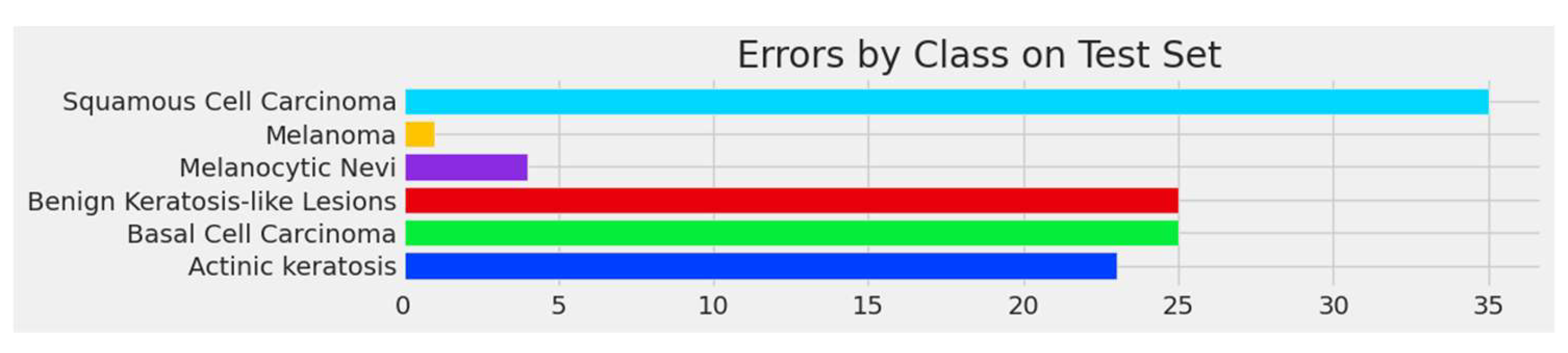
3.4. Confusion Matrix and Errors by Class
IV. Discussion
Limitations of Current Research
V. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- https://www.skincancer.org/skin-cancer-information/skin-cancer-facts/ (Last accessed 9June2024).
- Reichrath J, Leiter U, Eigentler T, Garbe C. Epidemiology of skin cancer. Sunlight, vitamin D and skin cancer. 2014;120-140. [CrossRef]
- https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2022.html (Last accessed 9June2024).
- Argenziano G, Soyer HP, Chimenti S, Talamini R, Corona R, Sera F, et al. Dermoscopy of pigmented skin lesions: results of a consensus meeting via the Internet. J Am Acad Dermatol. 2003;48(5):679-693. [CrossRef]
- https://www.iarc.who.int/cancer-type/skin-cancer/ (Last accessed 9June2024).
- Tschandl P, Codella N, Akay BN, Argenziano G, Braun RP, Cabo H, et al. Comparison of the accuracy of human readers versus machine-learning algorithms for pigmented skin lesion classification: an open, web-based, international, diagnostic study. Lancet Oncol. 2019;20(7):938-947. [CrossRef]
- Marchetti MA, Liopyris K, Dusza SW, Codella NC, Gutman DA, Helba B, et al. Computer algorithms show potential for improving dermatologists’ accuracy to diagnose cutaneous melanoma: Results of the International Skin Imaging Collaboration 2017. J Am Acad Dermatol. 2020;82(3):622-627. [CrossRef]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542(7639):115-118. [CrossRef]
- Codella NC, Nguyen QB, Pankanti S, Gutman DA, Helba B, Halpern AC, et al. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J Res Dev. 2017;61(4/5):5-1. [CrossRef]
- Brinker TJ, Hekler A, Enk AH, Klode J, Hauschild A, Berking C, et al. Deep neural networks are superior to dermatologists in melanoma image classification. Eur J Cancer. 2019;119:11-17. [CrossRef]
- Liu Y, Jain A, Eng C, Way DH, Lee K, Bui P, et al. A deep learning system for differential diagnosis of skin diseases. Nat Med. 2020;26(6):900-908. [CrossRef]
- Tan M, Le Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In: International conference on machine learning. PMLR; 2019. p. 6105-6114. [CrossRef]
- The International Skin Imaging Collaboration: https://gallery.isic-archive.com/.
- ImageNet Website and Dataset - https://www.image-net.org/.
- Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data. 2019;6(1):1-48. [CrossRef]
- Sharma N, Jain V, Mishra A. An analysis of convolutional neural networks for image classification. Procedia Comput Sci. 2018;132:377-384. [CrossRef]
- Pan SJ, Yang Q. A survey on transfer learning. IEEE Trans Knowl Data Eng. 2009;22(10):1345-1359. [CrossRef]
- Jain S, Singhania U, Tripathy B, Nasr EA, Aboudaif MK, Kamrani AK. Deep learning-based transfer learning for classification of skin cancer. Sensors. 2021;21(23):8142. [CrossRef]
- An end-to-end platform for machine learning - www.tensorflow.org.
- Keras, a deep learning API written in Python - https://keras.io/about/.
- https://keras.io/api/layers/normalization_layers/batch_normalization/.
- https://www.tensorflow.org/api_docs/python/tf/keras/layers/Dropout.
- https://www.tensorflow.org/guide/mixed_precision.
- https://www.tensorflow.org/api_docs/python/tf/keras/callbacks/EarlyStopping.
- https://keras.io/api/callbacks/reduce_lr_on_plateau/.
- https://www.tensorflow.org/tutorials/images/transfer_learning.
- https://keras.io/api/callbacks/model_checkpoint/.
- https://www.kaggle.com/code/residentmario/full-batch-mini-batch-and-online-learning.
- Ruder S. An overview of gradient descent optimization algorithms. arXiv preprint arXiv:1609.04747. 2016. 10.48550/arXiv.1609.04747.
- Goodfellow I, Bengio Y, Courville A. Deep learning. MIT Press; 2016.
- Srivastava N, Hinton G, Krizhevsky A, Sutskever I, Salakhutdinov R. Dropout: a simple way to prevent neural networks from overfitting. J Mach Learn Res. 2014;15(1):1929-1958. [CrossRef]
- Ioffe S, Szegedy C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In: International conference on machine learning. PMLR; 2015. p. 448-456. [CrossRef]
- Karthik R, Vaichole TS, Kulkarni SK, Yadav O, Khan F. Eff2Net: An efficient channel attention-based convolutional neural network for skin disease classification. Biomed Signal Process Control. 2022;73:103406. [CrossRef]
- Ali K, Shaikh ZA, Khan AA, Laghari AA. Multiclass skin cancer classification using EfficientNets–a first step towards preventing skin cancer. Neurosci Inform. 2022;2(4):100034. [CrossRef]
- Rafay A, Hussain W. EfficientSkinDis: An EfficientNet-based classification model for a large manually curated dataset of 31 skin diseases. Biomed Signal Process Control. 2023;85:104869. [CrossRef]
- Venugopal V, Raj NI, Nath MK, Stephen N. A deep neural network using modified EfficientNet for skin cancer detection in dermoscopic images. Decis Anal J. 2023;8:100278. [CrossRef]
- Harahap M, Husein AM, Kwok SC, Wizley V, Leonardi J, Ong DK, et al. Skin cancer classification using EfficientNet architecture. Bull Electr Eng Inform. 2024;13(4):2716-2728. [CrossRef]
- Bazgir E, Haque E, Maniruzzaman M, Hoque R. Skin cancer classification using Inception Network. World J Adv Res Rev. 2024;21(2):839-849. [CrossRef]
- Rahman MA, Bazgir E, Hossain SS, Maniruzzaman M. Skin cancer classification using NASNet. Int J Sci Res Arch. 2024;11(1):775-785. [CrossRef]
- Anand V, Gupta S, Altameem A, Nayak SR, Poonia RC, Saudagar AKJ. An enhanced transfer learning based classification for diagnosis of skin cancer. Diagnostics. 2022;12(7):1628. [CrossRef]
- Singh RK, Gorantla R, Allada SGR, Narra P. SkiNet: A deep learning framework for skin lesion diagnosis with uncertainty estimation and explainability. PLoS One. 2022;17(10). [CrossRef]
- Ahmed T, Mou FS, Hossain A. SCCNet: An Improved Multi-Class Skin Cancer Classification Network using Deep Learning. In: 2024 3rd International Conference on Advancement in Electrical and Electronic Engineering (ICAEEE); 2024 Apr; IEEE. p. 1-5. [CrossRef]
- Al-Rasheed A, Ksibi A, Ayadi M, Alzahrani AI, Zakariah M, Hakami NA. An ensemble of transfer learning models for the prediction of skin cancers with conditional generative adversarial networks. Diagnostics. 2022;12(12):3145. [CrossRef]
- Naeem A, Anees T, Khalil M, Zahra K, Naqvi RA, Lee SW. SNC_Net: Skin Cancer Detection by Integrating Handcrafted and Deep Learning-Based Features Using Dermoscopy Images. Mathematics. 2024;12(7):1030. [CrossRef]
- Naeem A, Anees T. DVFNet: A deep feature fusion-based model for the multiclassification of skin cancer utilizing dermoscopy images. PLoS One. 2024;19(3). [CrossRef]
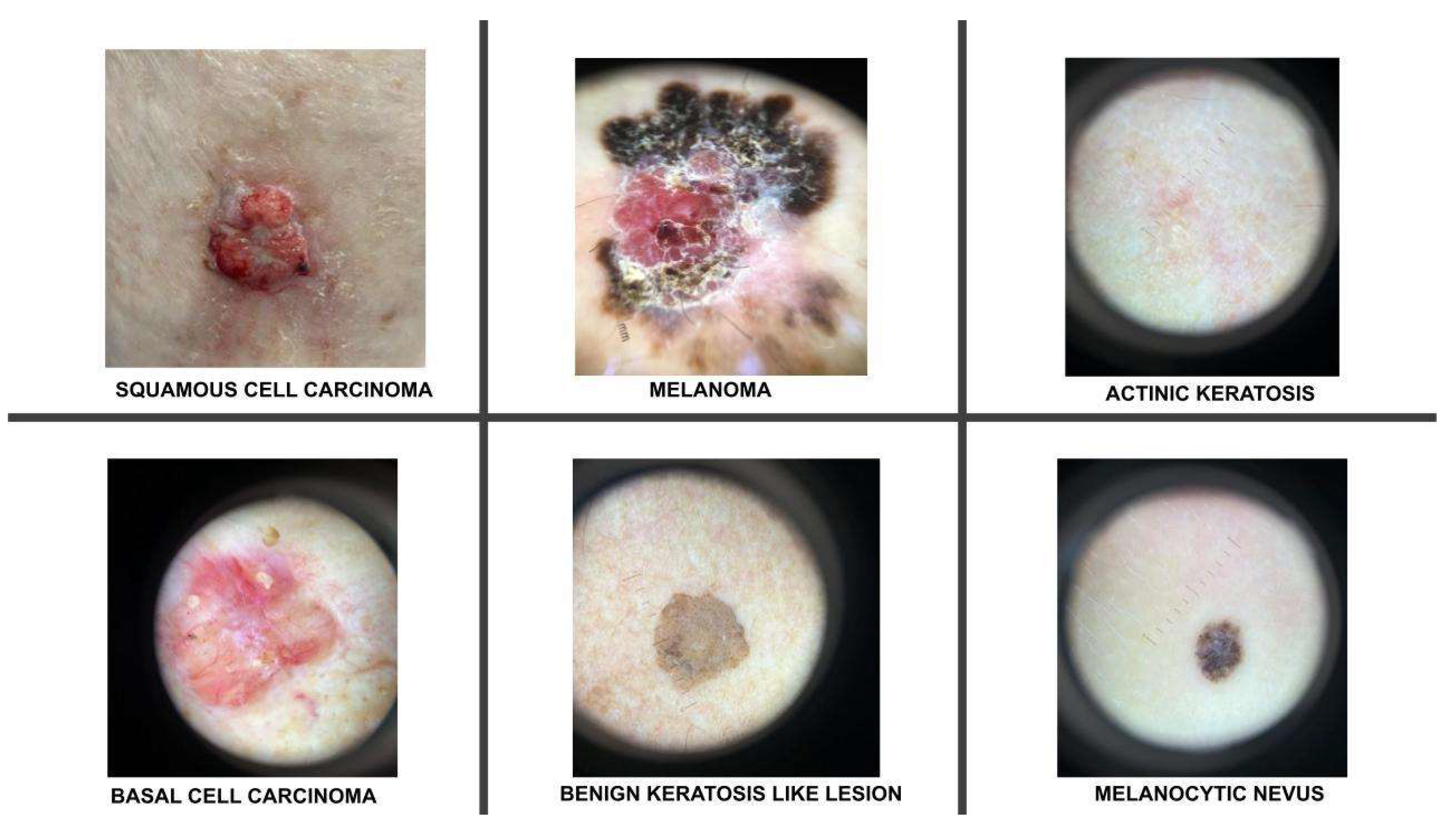



| Classes | No of Images | No of Augmented Images | Total |
|---|---|---|---|
| Melanoma | 1655 | 489 | 2144 |
| BCC | 1811 | 333 | 2144 |
| Benign Keratosis-like lesions | 1663 | 481 | 2144 |
| Melanocytic Nevi | 1686 | 458 | 2144 |
| SCC | 606 | 1538 | 2144 |
| AK | 801 | 1343 | 2144 |
| Total | 8222 | 4642 | 12864 |
| Hyperparameters | Values |
|---|---|
| Learning Rate | 0.001 |
| Batch size | 32 |
| Number of Epochs | 19 |
| Optimizer | Adamax |
| Dropout Rate | 0.45 |
| Activation Functions | Relu, Softmax |
| Regularization Parameters | Kernel Regularizer: L2 regularization with strength 0.016 Activity Regularizer: L1 regularization with strength 0.006 Bias Regularizer: L1 regularization with strength 0.006 |
| Loss Function | Categorical Cross Entropy |
| Augmentation techniques | Rotate, Scale, Flip, Zoom |
| Precision | Recall | F1-score | Support | |
|---|---|---|---|---|
| Basal cell carcinoma | 0.94 | 0.98 | 0.96 | 225 |
| Benign keratosis-like lesions | 0.94 | 0.89 | 0.91 | 208 |
| Melanocytic nevi | 0.95 | 0.97 | 0.96 | 210 |
| Melanoma | 1.00 | 0.99 | 1.00 | 207 |
| Accuracy | 0.96 | 850 | ||
| Macro Avg | 0.96 | 0.96 | 0.96 | 850 |
| Weighted Avg | 0.96 | 0.96 | 0.96 | 850 |
| Precision | Recall | F1-score | Support | |
|---|---|---|---|---|
| Actinic keratosis | 0.74 | 0.77 | 0.75 | 100 |
| Basal cell carcinoma | 0.87 | 0.84 | 0.85 | 227 |
| Benign keratosis-like lesions | 0.85 | 0.85 | 0.85 | 208 |
| Melanocytic nevi | 0.94 | 0.97 | 0.96 | 210 |
| Melanoma | 1.00 | 1.00 | 1.00 | 207 |
| Squamous cell carcinoma | 0.69 | 0.54 | 0.61 | 76 |
| Accuracy | 0.89 | 1028 | ||
| Macro Avg | 0.85 | 0.84 | 0.85 | 1028 |
| Weighted Avg | 0.89 | 0.89 | 0.89 | 1028 |
| Model | Year | Dataset | Model Used | Scope | Accuracy |
|---|---|---|---|---|---|
| Karthik et al. [33] | 2022 | DermNet NZ, Derm7Pt, DermatoWeb, Fitzpatrick17k | EfficientNetV2 in conjunction with the Efficient Channel Attention block | Classification of 4 skin diseases: acne, AK, melanoma, and psoriasis. | 84.7% |
| Ali et al. [34] | 2022 | HAM10000 dataset of dermatoscopic images | EfficientNet variants (results presented refer to EfficientNet B0) | Classification of 7 skin diseases | 87.9% |
| Rafay et al. [35] | 2023 | Manually curated from Atlas Dermatology & ISIC Dataset |
Fine-tuned EfficientNet-B2 |
Classification of 31 skin diseases |
87.15% |
| Venugopal et al. [36] | 2023 | ISIC2019 dataset | EfficientNetV2-M | Binary classification: malignant vs benign | 95.49% |
| Venugopal et al. [36] | 2023 | ISIC2019 dataset | EfficientNet-B4 | Binary classification: malignant vs benign | 93.17% |
| Harahap et al. [37] | 2024 | ISIC2019 dataset | EfficientNet-B0 to EfficientNet-B7 (results reported to EfficientNet-B3) | Classification of 3 skin diseases: BCC, SCC, melanoma | 77.6% |
| Harahap et al. [37] | 2024 | ISIC2019 dataset | EfficientNet-B0 to EfficientNet-B7 (results reported to EfficientNet-B4, the highest result obtained) | Classification of 3 skin diseases: BCC, SCC, melanoma | 79.69% |
| Proposed model | ISIC2019 & personal images collection | EfficientNetB3 | Classification of 4 skin diseases (benign& malign) | 95.4% |
|
| Proposed model | ISIC2019 & personal images collection | EfficientNetB3 | Classification of 6 skin diseases (benign& malign) |
88.8% |
| Model | Year | Dataset | Model used | Scope | Accuracy |
|---|---|---|---|---|---|
| Bazgir et al. [38] | 2024 | Kaggle/ISIC | Inception Network | Binary classification: malign vs benign | 85.94% |
| Rahman et al. [39] | 2024 | Kaggle/ISIC | NASNet | Binary classification: malign vs benign | 86.73% |
| Anand et al. [40] | 2022 | Kaggle/ISIC | Modified VGG16 architecture | Binary classification: malign vs benign | 89.9% |
| Singh et al. [41] | 2022 | ISIC2018 | Bayesian DenseNet-169 | Classification of 7 skin diseases | 73.65% |
| Ahmed et al. [42] | 2024 | ISIC2018 | SCCNet derived from Xpection architecture | Classification of 7 skin diseases | 95.2% |
| Al-Rasheed et al. [43] | 2022 | HAM10000 | Combination of VGG16, ResNet50, ResNet101 | Classification of 7 skin diseases | 93.5% |
| Naeem et al. [44] | 2024 | ISIC2019 | SNC_Net | Classification of 8 skin diseases | 97.81% |
| Naeem et al. [45] | 2024 | ISIC2019 | DVFNet | Classification of 8 skin diseases | 98.32% |
| Proposed model | ISIC2019 | EfficientNetB3 | Classification of 4 skin diseases | 95.4% | |
| Proposed model | ISIC2019 | EfficientNetB3 | Classification of 6 skin diseases | 88.8% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





