Submitted:
16 July 2024
Posted:
17 July 2024
You are already at the latest version
Abstract
Keywords:
Procedures
- Tissue dissociation.
- Tissue sample fixation:
- add fixative (ethanol: chloroform: glacial acetic acid = 6:3:1), change new fixative every 2 or 3 hours, for larger tissues, please change more times, then fix the samples overnight.
- Dehydration:
- rinse with 70% ethanol 2 times with 30min each, then 96% ethanol 2 times with 30min each, at last 100% ethanol 2 times with 30min each.
- Transparency:
- samples were transparent with xylene 2 times with 30min each.
- Embedding:
- First the paraffin was heated at 65 °C, then the samples were embedded in paraffin overnight. Put them on bench to cool down, store in 4 °C for long term use.
- slicing, spreading and preservation:
- Trim paraffin blocks as needed; next the blocks were cut into 5 μm thickness with a microtome machine. All slices are put on the glass slides with gelatin, then put them on slide warmer for 15min to stretch the paraffin samples, then cool down at room temperature, wipe the gelatin, and warm the slides for 24h.
- deparaffinization:
- samples were rinsed with xylene 2 times with 10min each.
- Rinse with 95% ethanol 2 times with 5min each.
- Rinse with 80% ethanol 2 times with 5min each.
- Rinse with 70% ethanol 2 times with 5min each.
- Rinse with water 2 times with 3min each.
- Rinse with PBS one time.
- Stain with hematoxylin for 5 to 10min.
- Discard the hematoxylin and rinse with water for 10min.
- Rinse with 95% ethanol for 5s.
- Stain with eosin for 30s to 2min.
- Rinse with 70% ethanol 2 times for 3min each.
- Rinse with PBS 2 times.
- Stain with DAPI for 10min.
- Discard the extra DAPI with PBS.
- Put on the coverslip.
- Microscopy detection.
- 11.
- Low pH sodium citrate buffer for 15min.
- 12.
- PBS wash 2 times, 1min each.
- 13.
- First antibody for 1h.
- 14.
- PBS wash 3 times, 1min each.
- 15.
- Fluorescence labelled second antibody for 1h.
- 16.
- PBS wash 3 times, 1min each
- 17.
- Stain with DAPI for 10min.
- 18.
- Discard the extra DAPI with PBS.
- 19.
- Put on the coverslip.
- 20.
- Microscopy detection.
-
Lavel morphology

- 2.
-
HE staining
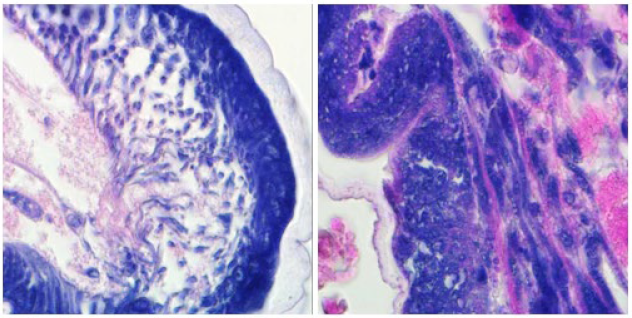
- 3.
-
TUNEL staining
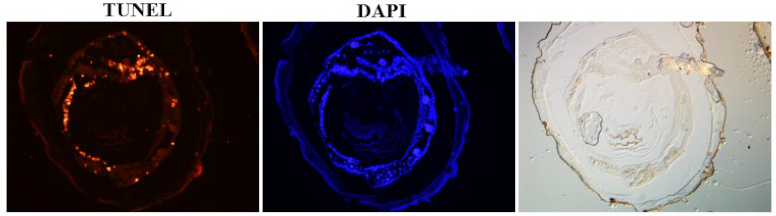
References
- Huang, W., Zhang, J., Yang, B., Beerntsen, B.T., Song, H. and Ling, E. (2016) DNA duplication is essential for the repair of gastrointestinal perforation in the insect midgut. Scientific reports, 6, 19142. [CrossRef]
- Zhang, H., Cheng, C., Li, F., Gu, S., Zhou, Z. and Cheng, L. (2013) Cloning and characterization of ubiquitin ribosome fusion gene RpS27a, a deltamethrin-resistance-associated gene from diamondback moth (Plutella xylostella L.). Turkish Journal of Zoology, 37, 506-513. [CrossRef]
- Zhang, H., Feng-Liang, L., Chen, C., Liu, B.-D. and Luo-Gen, C. (2013) cDNA Representational Difference Analysis of Deltamethrin-Resistant and-Susceptible Strains of Diamondback Moth. Pakistan Journal of Zoology, 45.
- Zhang NaNa, Z.N., Zhang Hong, Z.H., Cheng Chen, C.C., Li FengLiang, L.F., Gao ShaoQi, G.S. and Cheng LuoGen, C.L. (2013) A comparative profiling of protein expression in the deltamethrin-sensitive and resistant strains of the diamondback moth (Plutella xylostella).
- Zhang, H., Li, F., Cheng, C., Jiao, D., Zhou, Z. and Cheng, L. (2013) The identification and characterisation of a new deltamethrin resistance-associated gene, UBL40, in the diamondback moth, Plutella xylostella (L.). Gene, 530, 51-56. [CrossRef]
- Huang, W., Tang, R., Li, S., Zhang, Y., Chen, R., Gong, L., Wei, X., Tang, Y., Liu, Q. and Geng, L. (2021) Involvement of epidermis cell proliferation in defense against Beauveria bassiana infection. Frontiers in Immunology, 12, 741797. [CrossRef]
- Migliore, S., Condorelli, L., Galluzzo, P., Galuppo, L., Corrente, A., Lepri, E., Ridley, A., Loria, G.R. and Puleio, R. (2024) First Description of Mycoplasma agalactiae Anatomical Localization in Naturally Infected Hard Ticks (Rhipicephalus bursa). Microorganisms, 12, 1390. [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).



