Submitted:
16 July 2024
Posted:
17 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Patients’ Characteristics
2.2. Detection of cbDNA
3. Discussion
4. Materials and Methods
4.1. Patients Enrollment and Study Design
4.2. Ethics Approval
4.3. Blood Sampling and DNA Extraction
4.4. PCR Amplification of Microbial DNA
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A.; Kinzler, K.W. Cancer Genome Landscapes. Science 2013, 339, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Terzić, J.; Grivennikov, S.; Karin, E.; Karin, M. Inflammation and Colon Cancer. Gastroenterology 2010, 138, 2101–2114.e5. [Google Scholar] [CrossRef] [PubMed]
- Compton, C.; Fenoglio-Preiser, C.M.; Pettigrew, N.; Fielding, L.P. American Joint Committee on Cancer Prognostic Factors Consensus Conference: Colorectal Working Group. Cancer 2000, 88, 1739–1757. [Google Scholar] [CrossRef]
- Ilyas, M.I.M. Epidemiology of Stage IV Colorectal Cancer: Trends in the Incidence, Prevalence, Age Distribution, and Impact on Life Span. Clin Colon Rectal Surg 2023, 37, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Bien, J.; Palagani, V.; Bozko, P. The Intestinal Microbiota Dysbiosis and Clostridium Difficile Infection: Is There a Relationship with Inflammatory Bowel Disease? Therap Adv Gastroenterol 2013, 6, 53–68. [Google Scholar] [CrossRef] [PubMed]
- Abreu, M.T.; Peek, R.M. Gastrointestinal Malignancy and the Microbiome. Gastroenterology 2014, 146, 1534–1546.e3. [Google Scholar] [CrossRef] [PubMed]
- Irrazábal, T.; Belcheva, A.; Girardin, S.E.; Martin, A.; Philpott, D.J. The Multifaceted Role of the Intestinal Microbiota in Colon Cancer. Mol Cell 2014, 54, 309–320. [Google Scholar] [CrossRef]
- Shimizu, K.; Ogura, H.; Goto, M.; Asahara, T.; Nomoto, K.; Morotomi, M.; Yoshiya, K.; Matsushima, A.; Sumi, Y.; Kuwagata, Y.; et al. Altered Gut Flora and Environment in Patients with Severe SIRS. J Trauma 2006, 60, 126–133. [Google Scholar] [CrossRef]
- Koliarakis, I.; Athanasakis, E.; Sgantzos, M.; Mariolis-Sapsakos, T.; Xynos, E.; Chrysos, E.; Souglakos, J.; Tsiaoussis, J. Intestinal Microbiota in Colorectal Cancer Surgery. Cancers (Basel) 2020, 12, 3011. [Google Scholar] [CrossRef]
- Signat, B.; Roques, C.; Poulet, P.; Duffaut, D. Fusobacterium Nucleatum in Periodontal Health and Disease. Curr Issues Mol Biol 2011, 13, 25–36. [Google Scholar]
- Shang, F.-M.; Liu, H.-L. Fusobacterium Nucleatum and Colorectal Cancer: A Review. World J Gastrointest Oncol 2018, 10, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium Nucleatum in Colorectal Carcinoma Tissue and Patient Prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef]
- Lelouvier, B.; Servant, F.; Païssé, S.; Brunet, A.-C.; Benyahya, S.; Serino, M.; Valle, C.; Ortiz, M.R.; Puig, J.; Courtney, M.; et al. Changes in Blood Microbiota Profiles Associated with Liver Fibrosis in Obese Patients: A Pilot Analysis. Hepatology 2016, 64, 2015–2027. [Google Scholar] [CrossRef]
- Bruns, T.; Reuken, P.A.; Stengel, S.; Gerber, L.; Appenrodt, B.; Schade, J.H.; Lammert, F.; Zeuzem, S.; Stallmach, A. The Prognostic Significance of Bacterial DNA in Patients with Decompensated Cirrhosis and Suspected Infection. Liver Int 2016, 36, 1133–1142. [Google Scholar] [CrossRef]
- Merlini, E.; Bellistri, G.M.; Tincati, C.; d’Arminio Monforte, A.; Marchetti, G. Sequencing of Bacterial Microflora in Peripheral Blood: Our Experience with HIV-Infected Patients. J Vis Exp 2011, 2830. [Google Scholar] [CrossRef]
- Messaritakis, I.; Vogiatzoglou, K.; Tsantaki, K.; Ntretaki, A.; Sfakianaki, M.; Koulouridi, A.; Tsiaoussis, J.; Mavroudis, D.; Souglakos, J. The Prognostic Value of the Detection of Microbial Translocation in the Blood of Colorectal Cancer Patients. Cancers (Basel) 2020, 12, 1058. [Google Scholar] [CrossRef]
- Koulouridi, A.; Messaritakis, I.; Theodorakis, E.; Chondrozoumaki, M.; Sfakianaki, M.; Gouvas, N.; Tsiaoussis, J.; Mavroudis, D.; Tzardi, M.; Souglakos, J. Detection of Circulating Tumor Cells and Microbial DNA Fragments in Stage III Colorectal Cancer Patients under Three versus Six Months of Adjuvant Treatment. Cancers (Basel) 2021, 13, 3552. [Google Scholar] [CrossRef] [PubMed]
- Lescut, D.; Colombel, J.F.; Vincent, P.; Cortot, A.; Fournier, L.; Quandalle, P.; Vankemmel, M.; Triboulet, J.P.; Wurtz, A.; Paris, J.C. Bacterial Translocation in Colorectal Cancers. Gastroenterol Clin Biol 1990, 14, 811–814. [Google Scholar]
- Ono, S.; Tsujimoto, H.; Yamauchi, A.; Hiraki, S.; Takayama, E.; Mochizuki, H. Detection of Microbial DNA in the Blood of Surgical Patients for Diagnosing Bacterial Translocation. World J Surg 2005, 29, 535–539. [Google Scholar] [CrossRef]
- Giacconi, R.; Donghia, R.; Arborea, G.; Savino, M.T.; Provinciali, M.; Lattanzio, F.; Caponio, G.R.; Coletta, S.; Bianco, A.; Notarnicola, M.; et al. Plasma Bacterial DNA Load as a Potential Biomarker for the Early Detection of Colorectal Cancer: A Case-Control Study. Microorganisms 2023, 11, 2360. [Google Scholar] [CrossRef]
- Messaritakis, I.; Koulouris, A.; Boukla, E.; Vogiatzoglou, K.; Lagkouvardos, I.; Intze, E.; Sfakianaki, M.; Chondrozoumaki, M.; Karagianni, M.; Athanasakis, E.; et al. Exploring Gut Microbiome Composition and Circulating Microbial DNA Fragments in Patients with Stage II/III Colorectal Cancer: A Comprehensive Analysis. Cancers (Basel) 2024, 16, 1923. [Google Scholar] [CrossRef]
- Xiao, Q.; Lu, W.; Kong, X.; Shao, Y.W.; Hu, Y.; Wang, A.; Bao, H.; Cao, R.; Liu, K.; Wang, X.; et al. Alterations of Circulating Bacterial DNA in Colorectal Cancer and Adenoma: A Proof-of-Concept Study. Cancer Lett 2021, 499, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Mutignani, M.; Penagini, R.; Gargari, G.; Guglielmetti, S.; Cintolo, M.; Airoldi, A.; Leone, P.; Carnevali, P.; Ciafardini, C.; Petrocelli, G.; et al. Blood Bacterial DNA Load and Profiling Differ in Colorectal Cancer Patients Compared to Tumor-Free Controls. Cancers (Basel) 2021, 13, 6363. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.C.S.; Ko, K.K.K.; Chen, H.; Liu, J.; Loh, M.; SG10K_Health Consortium; Chia, M; Nagarajan, N. No Evidence for a Common Blood Microbiome Based on a Population Study of 9,770 Healthy Humans. Nat Microbiol 2023, 8, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, V.; Moreau, R.; Kamath, P.S.; Jalan, R.; Ginès, P.; Nevens, F.; Fernández, J.; To, U.; García-Tsao, G.; Schnabl, B. Acute-on-Chronic Liver Failure in Cirrhosis. Nat Rev Dis Primers 2016, 2, 16041. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Kouzu, K.; Tsujimoto, H.; Kishi, Y.; Ueno, H.; Shinomiya, N. Bacterial Translocation in Gastrointestinal Cancers and Cancer Treatment. Biomedicines 2022, 10, 380. [Google Scholar] [CrossRef]
- Potgieter, M.; Bester, J.; Kell, D.B.; Pretorius, E. The Dormant Blood Microbiome in Chronic, Inflammatory Diseases. FEMS Microbiol Rev 2015, 39, 567–591. [Google Scholar] [CrossRef]
- Komiya, Y.; Shimomura, Y.; Higurashi, T.; Sugi, Y.; Arimoto, J.; Umezawa, S.; Uchiyama, S.; Matsumoto, M.; Nakajima, A. Patients with Colorectal Cancer Have Identical Strains of Fusobacterium Nucleatum in Their Colorectal Cancer and Oral Cavity. Gut 2019, 68, 1335–1337. [Google Scholar] [CrossRef] [PubMed]
- Koliarakis, I.; Messaritakis, I.; Nikolouzakis, T.K.; Hamilos, G.; Souglakos, J.; Tsiaoussis, J. Oral Bacteria and Intestinal Dysbiosis in Colorectal Cancer. Int J Mol Sci 2019, 20, 4146. [Google Scholar] [CrossRef] [PubMed]
- Chelakkot, C.; Ghim, J.; Ryu, S.H. Mechanisms Regulating Intestinal Barrier Integrity and Its Pathological Implications. Exp Mol Med 2018, 50, 1–9. [Google Scholar] [CrossRef]
- Wasinger, V.C.; Lu, K.; Yau, Y.Y.; Nash, J.; Lee, J.; Chang, J.; Paramsothy, S.; Kaakoush, N.O.; Mitchell, H.M.; Leong, R.W.L. Spp24 Is Associated with Endocytic Signalling, Lipid Metabolism, and Discrimination of Tissue Integrity for “leaky-Gut” in Inflammatory Bowel Disease. Sci Rep 2020, 10, 12932. [Google Scholar] [CrossRef]
- Païssé, S.; Valle, C.; Servant, F.; Courtney, M.; Burcelin, R.; Amar, J.; Lelouvier, B. Comprehensive Description of Blood Microbiome from Healthy Donors Assessed by 16S Targeted Metagenomic Sequencing. Transfusion 2016, 56, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Martinson, J.N.V.; Walk, S.T. Escherichia Coli Residency in the Gut of Healthy Human Adults. EcoSal Plus 2020, 9, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Wassenaar, T.M. E. Coli and Colorectal Cancer: A Complex Relationship That Deserves a Critical Mindset. Crit Rev Microbiol 2018, 44, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Genua, F.; Raghunathan, V.; Jenab, M.; Gallagher, W.M.; Hughes, D.J. The Role of Gut Barrier Dysfunction and Microbiome Dysbiosis in Colorectal Cancer Development. Front Oncol 2021, 11, 626349. [Google Scholar] [CrossRef]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium Nucleatum Infection Is Prevalent in Human Colorectal Carcinoma. Genome Res 2012, 22, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic Analysis Identifies Association of Fusobacterium with Colorectal Carcinoma. Genome Res 2012, 22, 292–298. [Google Scholar] [CrossRef]
- Flanagan, L.; Schmid, J.; Ebert, M.; Soucek, P.; Kunicka, T.; Liska, V.; Bruha, J.; Neary, P.; Dezeeuw, N.; Tommasino, M.; et al. Fusobacterium Nucleatum Associates with Stages of Colorectal Neoplasia Development, Colorectal Cancer and Disease Outcome. Eur J Clin Microbiol Infect Dis 2014, 33, 1381–1390. [Google Scholar] [CrossRef]
- Joo, J.E.; Chu, Y.L.; Georgeson, P.; Walker, R.; Mahmood, K.; Clendenning, M.; Meyers, A.L.; Como, J.; Joseland, S.; Preston, S.G.; et al. Intratumoral Presence of the Genotoxic Gut Bacteria Pks+ E. Coli, Enterotoxigenic Bacteroides Fragilis, and Fusobacterium Nucleatum and Their Association with Clinicopathological and Molecular Features of Colorectal Cancer. Br J Cancer 2024, 130, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Liu, L.; Li, H.; Qin, H.; Sun, Z. Clinical Significance of Fusobacterium Nucleatum, Epithelial-Mesenchymal Transition, and Cancer Stem Cell Markers in Stage III/IV Colorectal Cancer Patients. Onco Targets Ther 2017, 10, 5031–5046. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Coker, O.O.; Nakatsu, G.; Wu, W.K.K.; Zhao, L.; Chen, Z.; Chan, F.K.L.; Kristiansen, K.; Sung, J.J.Y.; Wong, S.H.; et al. Multi-Cohort Analysis of Colorectal Cancer Metagenome Identified Altered Bacteria across Populations and Universal Bacterial Markers. Microbiome 2018, 6, 70. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.-H.; Jiang, J.-K.; Luo, J.-C.; Lin, C.-C.; Ting, P.-H.; Yang, U.-C.; Lan, Y.-T.; Huang, Y.-H.; Hou, M.-C.; Lee, F.-Y. The Long Term Microbiota and Metabolic Status in Patients with Colorectal Cancer after Curative Colon Surgery. PLoS One 2019, 14, e0218436. [Google Scholar] [CrossRef] [PubMed]
- Kong, C.; Gao, R.; Yan, X.; Huang, L.; He, J.; Li, H.; You, J.; Qin, H. Alterations in Intestinal Microbiota of Colorectal Cancer Patients Receiving Radical Surgery Combined with Adjuvant CapeOx Therapy. Sci China Life Sci 2019, 62, 1178–1193. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota Disbiosis Is Associated with Colorectal Cancer. Front Microbiol 2015, 6, 20. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-J.; Zhang, Y.-L.; Shang, Y.; Wu, B.; Yang, E.; Luo, Y.-Y.; Li, X.-R. Intestinal Bacteria Detected in Cancer and Adjacent Tissue from Patients with Colorectal Cancer. Oncol Lett 2019, 17, 1115–1127. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural Segregation of Gut Microbiota between Colorectal Cancer Patients and Healthy Volunteers. ISME J 2012, 6, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Cass, S.; White, M.G. The Influence of the Microbiome on Metastatic Colorectal Cancer. Clin Colon Rectal Surg 2023, 36, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, P.; Wang, J.; Wei, X.; Wang, M. Unveiling Intratumoral Microbiota: An Emerging Force for Colorectal Cancer Diagnosis and Therapy. Pharmacological Research 2024, 203, 107185. [Google Scholar] [CrossRef]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.-J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal Inflammation Targets Cancer-Inducing Activity of the Microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef] [PubMed]
- Glebov, O.K.; Rodriguez, L.M.; Nakahara, K.; Jenkins, J.; Cliatt, J.; Humbyrd, C.-J.; DeNobile, J.; Soballe, P.; Simon, R.; Wright, G.; et al. Distinguishing Right from Left Colon by the Pattern of Gene Expression. Cancer Epidemiol Biomarkers Prev 2003, 12, 755–762. [Google Scholar]
- Tamas, K.; Walenkamp, A.M.E.; de Vries, E.G.E.; van Vugt, M. a. T.M.; Beets-Tan, R.G.; van Etten, B.; de Groot, D.J.A.; Hospers, G. a. P. Rectal and Colon Cancer: Not Just a Different Anatomic Site. Cancer Treat Rev 2015, 41, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Wu, J.; Shi, L.; Zhou, B.; Shang, F.; Chang, X.; Dong, X.; Deng, S.; Liu, L.; Cai, K.; et al. Gut Microbiota Distinct between Colorectal Cancers with Deficient and Proficient Mismatch Repair: A Study of 230 CRC Patients. Front Microbiol 2022, 13, 993285. [Google Scholar] [CrossRef]
- Qi, Z.; Zhibo, Z.; Jing, Z.; Zhanbo, Q.; Shugao, H.; Weili, J.; Jiang, L.; Shuwen, H. Prediction Model of Poorly Differentiated Colorectal Cancer (CRC) Based on Gut Bacteria. BMC Microbiol 2022, 22, 312. [Google Scholar] [CrossRef] [PubMed]
- Carelli, L.L.; D’Aquila, P.; Rango, F.D.; Incorvaia, A.; Sena, G.; Passarino, G.; Bellizzi, D. Modulation of Gut Microbiota through Low-Calorie and Two-Phase Diets in Obese Individuals. Nutrients 2023, 15, 1841. [Google Scholar] [CrossRef]
- Wilkins, L.J.; Monga, M.; Miller, A.W. Defining Dysbiosis for a Cluster of Chronic Diseases. Sci Rep 2019, 9, 12918. [Google Scholar] [CrossRef]
- Amar, J.; Lelouvier, B.; Servant, F.; Lluch, J.; Burcelin, R.; Bongard, V.; Elbaz, M. Blood Microbiota Modification After Myocardial Infarction Depends Upon Low-Density Lipoprotein Cholesterol Levels. J Am Heart Assoc 2019, 8, e011797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Chu, J.; Hao, W.; Zhang, J.; Li, H.; Yang, C.; Yang, J.; Chen, X.; Wang, H. Gut Microbiota and Type 2 Diabetes Mellitus: Association, Mechanism, and Translational Applications. Mediators Inflamm 2021, 2021, 5110276. [Google Scholar] [CrossRef] [PubMed]
- Proença, M.A.; Biselli, J.M.; Succi, M.; Severino, F.E.; Berardinelli, G.N.; Caetano, A.; Reis, R.M.; Hughes, D.J.; Silva, A.E. Relationship between Fusobacterium Nucleatum, Inflammatory Mediators and microRNAs in Colorectal Carcinogenesis. World J Gastroenterol 2018, 24, 5351–5365. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | Group 1 (n = 25) | Group 2 (n = 25) | Group 3 (n = 25) | Group 4 (n = 25) |
|---|---|---|---|---|
| Mean (range) | ||||
| Age | 20.5 (20-21) | 61.5 (25-86) | 62.9 (39-80) | 57.8 (26-78) |
| BMI (kg/m2) | 23.1 (20.3-24.8) | 28.4 (22.1-46.9) | 27.7 (20.2-37.2) | 27.9 (17.1-52.5) |
| Frequency (percentage) | ||||
| Sex | ||||
| Male | 18 (72%) | 11 (44%) | 18 (72%) | 16 (64%) |
| Female | 7 (28%) | 14 (56%) | 7 (28%) | 9 (36%) |
| Surgery | ||||
| Yes | n/a * | 25 (100%) | 25 (100%) | 0 (0%) |
| No | n/a | 0 (0%) | 0 (0%) | 25 (100%) |
| Metastasis | ||||
| Yes | n/a | 0 (0%) | 25 (100%) | 25 (100%) |
| No | n/a | 25 (100%) | 0 (0%) | 0 (0%) |
| Tumor Location | ||||
| Right Colon | n/a | 10 (40%) | 10 (40%) | 5 (20%) |
| Left Colon | n/a | 9 (36%) | 8 (32%) | 10 (40%) |
| Rectum | n/a | 6 (24%) | 7 (28%) | 10 (40%) |
| Stage | ||||
| II | n/a | 5 (20%) | 0 (0%) | 0 (0%) |
| III | n/a | 20 (80%) | 0 (0 %) | 0 (0%) |
| IV | n/a | 0 (0%) | 25 (100%) | 25 (100%) |
| Grade | ||||
| Low | n/a | 6 (24%) | 2 (8%) | 2 (8%) |
| Intermediate | n/a | 13 (52%) | 15 (60%) | 16 (64%) |
| High | n/a | 6 (24%) | 8 (32%) | 7 (28%) |
| Mismatch Repair (MMR) status | ||||
| Microsatellite Stability (MSS) | n/a | 19 (76%) | 23 (92%) | 21 (84%) |
| Microsatellite Instability (MSI) | n/a | 3 (12%) | 2 (8%) | 2 (8%) |
| Unreported | n/a | 3 (12%) | 0 (0%) | 2 (8%) |
| Smoking | ||||
| Yes | 0 (0%) | 7 (28%) | 12 (48%) | 8 (32%) |
| No | 25 (100%) | 18 (72%) | 13 (52%) | 17 (68%) |
| Alcohol | ||||
| Yes | 0 (0%) | 1 (4%) | 3 (12%) | 4 (16%) |
| No | 25 (100%) | 24 (96%) | 22 (88%) | 21 (84%) |
| Diabetes | ||||
| Yes | 0 (0%) | 5 (20%) | 11 (44%) | 3 (12%) |
| No | 25 (100%) | 20 (80%) | 14 (56%) | 22 (88%) |
| Dyslipidemia | ||||
| Yes | 0 (0%) | 9 (36%) | 5 (20%) | 6 (24%) |
| No | 25 (100%) | 16 (64%) | 20 (80%) | 19 (76%) |
| Cardiovascular Disease (CVD) | ||||
| Yes | 0 (0%) | 12 (48%) | 13 (52%) | 9 (36%) |
| No | 25 (100%) | 13 (52%) | 12 (48%) | 16 (64%) |
| Gene Target | Detection | Group 1 (n = 25) |
Group 2 (n = 25) |
Group3 (n = 25) |
Group 4 (n = 25) |
|---|---|---|---|---|---|
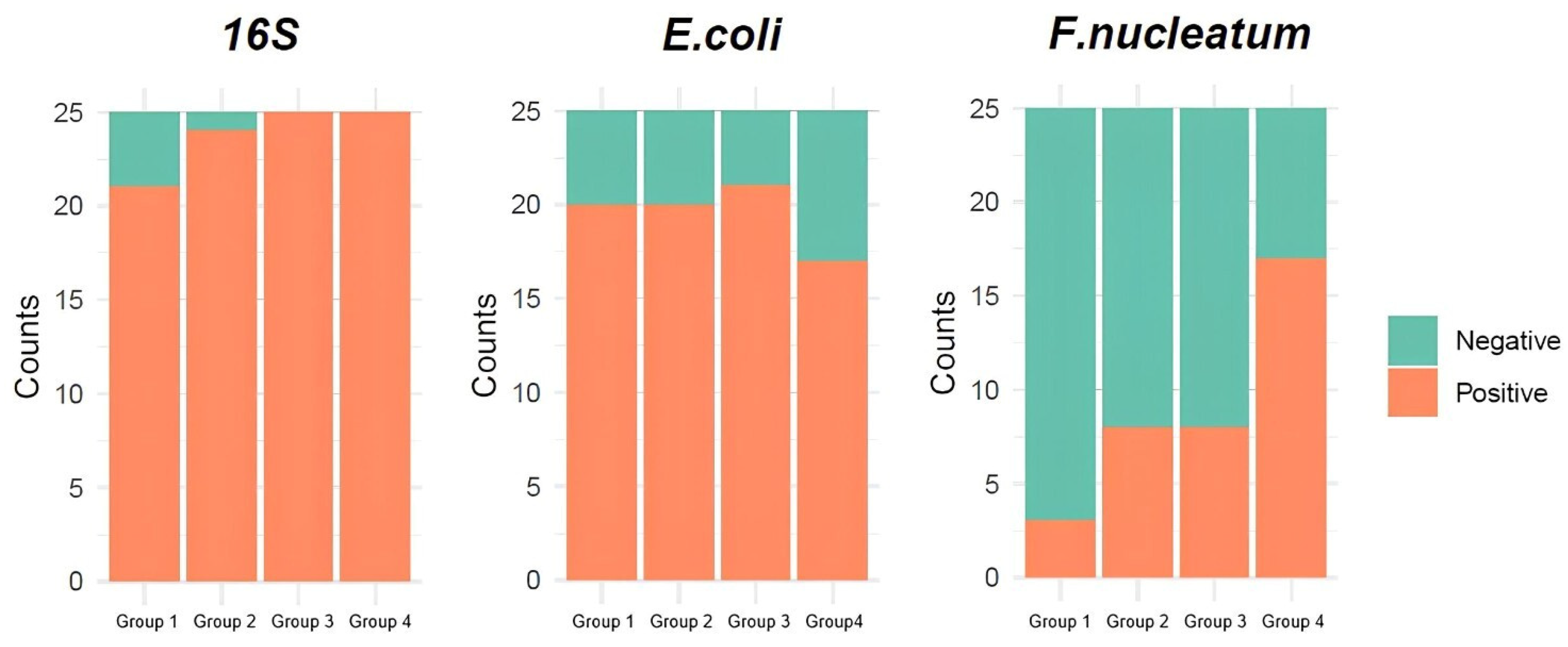
| 16S rRNA | Positive | 21 (84%) | 24 (96%) | 25 (100%) | 25 (100%) |
| Negative | 4 (16%) | 1 (4%) | 0 (0%) | 0 (0%) | |
| β-galactosidase gene of Escherichia coli | Positive | 20 (80%) | 20 (80%) | 21 (84%) | 17 (68%) |
| Negative | 5 (20%) | 5 (20%) | 4 (16%) | 8 (32%) | |
| NusG gene of Fusobacterium nucleatum | Positive | 3 (12%) | 8 (32%) | 8 (32%) | 17 (68%) |
| Negative | 22 (88%) | 17 (68%) | 17 (68%) | 8 (32%) |
| Pairwise comparison | 16S rRNA | E. coli | F. nucleatum | |||
|---|---|---|---|---|---|---|
| p-value | adj. p-value | p-value | adj. p-value | p-value | adj. p-value | |
| Group 1 – Group 2 | 0.349 | 0.697 | - * | - | 0.171 | 0.205 |
| Group 1 – Group 3 | 0.11 | 0.33 | - | - | 0.171 | 0.205 |
| Group 1 – Group 4 | 0.11 | 0.33 | 0.52 | - | <0.001 | <0.001 |
| Group 2 – Group 3 | - | - | - | - | - | - |
| Group 2 – Group 4 | - | - | 0.52 | - | 0.023 | 0.045 |
| Group 3 – Group 4 | - | - | 0.321 | - | 0.023 | 0.045 |
| Parameter / Pairwise comparison | E. coli | F. nucleatum | ||
|---|---|---|---|---|
| p-value | adj. p-value | p-value | adj. p-value | |
| Sex | ||||
| Female – Male | 0.403 | n/a 1 | 0.348 | n/a |
| Surgery | ||||
| No – Yes | 0.242 | n/a | 0.006 | n/a |
| Metastasis | ||||
| No (stage II/III) – Yes (stage IV) | 0.777 | n/a | 0.217 | n/a |
| Tumor Location | ||||
| Left Colon – Rectum | 0.308 | 0.462 | 0.252 | 0.596 |
| Right Colon – Rectum | 0.292 | 0.462 | - 2 | - |
| Left Colon – Right Colon | - | - | 0.397 | 0.596 |
| Grade | ||||
| High – Low | 0.634 | 0.951 | 0.701 | - |
| Intermediate – Low | 0.427 | 0.951 | - | - |
| High – Intermediate | - | - | 0.605 | - |
| MMR status | ||||
| MSI – MSS | - | n/a | - | n/a |
| Smoking | ||||
| No – Yes | 0.15 | n/a | 0.634 | n/a |
| Alcohol | ||||
| No – Yes | - | n/a | 0.725 | n/a |
| Diabetes | ||||
| No – Yes | 0.004 | n/a | 0.286 | n/a |
| Dyslipidemia | ||||
| No – Yes | - | n/a | - | n/a |
| CVD | ||||
| No – Yes | 0.414 | n/a | 0.816 | n/a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





