Introduction
Evidence of a Role for a lncRNA in Leukemic Cell Differentiation
1. Long Non-Coding RNAs
Over the past few decades, lncRNAs have been discovered to function as regulators of gene expression and numerous additional biological processes. They are transcribed from loci throughout the genomes of most eukaryotes (Engreitz et al., 2016; Kopp and Mendell, 2018; Herman et al., 2022), and reports indicate that many lncRNAs are involved in the regulation of pluripotency and differentiation (Zibitt et al., 2021). Although the mechanisms by which these lncRNAs function are still being explored (Nuñez-Martinez and Recillas-Targa, 2022), one model that has emerged is that nuclear lncRNAs modulate gene expression through interactions with histone modifying proteins and/or transcription factors (Kopp and Mendell, 2018; Statello et al., 2020; Mangiavacchi et al., 2023).
Other lncRNAs, as does lncRNAp53int1, function in the cytoplasm (Atianand et al., 2016). Lnc-MD1, for example, is a muscle-specific lncRNA that regulates muscle differentiation by binding to microRNAs (miRNA) and limiting their availability (Fernandes et al., 2021). Another lncRNA, TINCR, is induced during epidermal differentiation and interacts with a number of differentiation-specific mRNAs to regulate their stability (Kretz et al., 2013). These, and other results, demonstrate that lncRNAs can act to regulate genes required for cell differentiation through multiple mechanisms (Farzaneh et al., 2022).
2. A p53-Associated lncRNA Is Linked to the Differentiation of Human Myeloid Leukemia Cells
A number of years ago, we identified a transcription unit located in the 1st intron of the human p53 tumor suppressor gene that encodes a lncRNA, lncRNAp53int1 (Reisman, 1997; NCBI Accession U58658). This transcript is classified as a lncRNA and is listed as GC17M015273 in the GeneCard Human Gene Database (
http://www.genecards.org/genecarna) and NONHSAG020729 in NONCODE v6 (
http://www.noncode.org). The lncRNAp53int1 transcript is approximately 1125 nucleotides in length, is polyadenylated, and contains no introns. While there appears to be no functional or regulatory relationship to p53 itself, the abundance of this lncRNA is significantly reduced during differentiation of human myeloid leukemia cells (Reisman et al., 2016). We have hypothesized that lncRNAp53Int1 plays a crucial role in maintenance of the undifferentiated proliferative state of myeloid leukemia cells, thus contributing to the genesis and maintenance of the disease. That lncRNAp53Int1 is generally expressed in immature cell types is supported by lncRNA expression data collated in various publicly available databases. The RNA is expressed in a variety of human tissues at elevated levels including those that contain proliferative cell types such as lymph node, foreskin fibroblasts and umbilical endothelial cells. Interestingly, a number of recent studies have identified lncRNAs as playing a role in myeloid leukemias and other malignancies (McCabe and Rasmussen, 2021; Farzaneh , et al., 2022; Jiang et al., 2024; Lobo-Alves et al., 2024).
3. lncRNAp53Int1 Is Expressed in Myeloid Leukemia
lncRNAp53int1 is a 1125 nucleotide lncRNA, transcribed from the first intron of the human p53 gene (Reisman et al., 1997) and the transcriptional regulation of this gene demonstrated differentiation-specific expression in myeloid leukemia cells (Reisman and Rotter, 1989). Elevated levels of this transcript, primarily located in the cytoplasm, are observed in the human leukemic cell lines K562, HEL, HL-60, and U937. The RNA is at low or undetectable levels in the colon tumor-derived cell lines COLO320 and SW837, lymphoma-derived cell lines IM-9, 7666 and Namalwa, as well as HeLa cells. White blood cell fractions from unidentified leukemic patient samples (University of South Carolina Center for Colon Cancer Research Tissue Biorepository) as well as a small panel of Acute Myeloid Leukemia samples (Discovery Life Sciences; Huntsville, Al) were assayed, and the lncRNA transcript was detected in all samples.
4. lncRNAp53Int1 Levels Are Reduced during Terminal Differentiation of Myeloid Cells
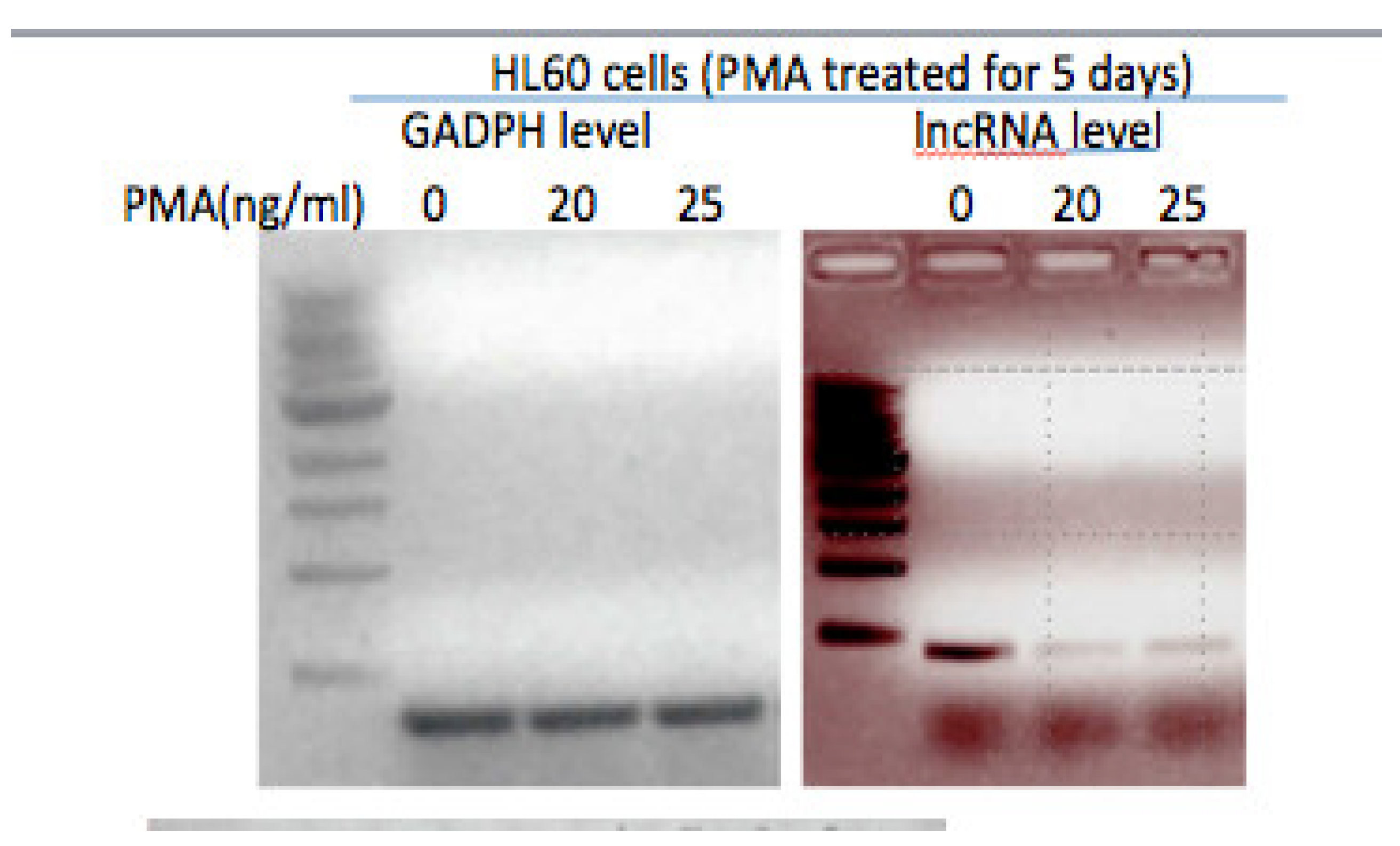
Many human acute myeloid leukemia-derived cell lines represent proliferative stem-cell like populations that can be induced to differentiate into mature monocytes or macrophages (Suzuki et al., 2009). These cells have been used as model systems for particular stages of monocyte differentiation (FANTOM Consortium, Suzuki et al., 2009; Tasseff et al., 2017). For example, when HL-60 or U937 cells are treated with 20ng/ml Phorbol myristate acetate (PMA), the cells undergo terminal differentiation and cease proliferation, take on macrophage-like morphology, carry out phagocytosis, and display the cell surface macrophage marker CD11b. Interestingly, as determined by qRT-PCR, while the lncRNA transcript is detected in HL-60 promyelocytes, its level is significantly reduced in cells induced to differentiate into macrophage-like cells by PMA (
Figure 1).
5. Expression of lncRNAp53Int1 from a Lentivirus Vector Leads to a Partial Block in Differentiation
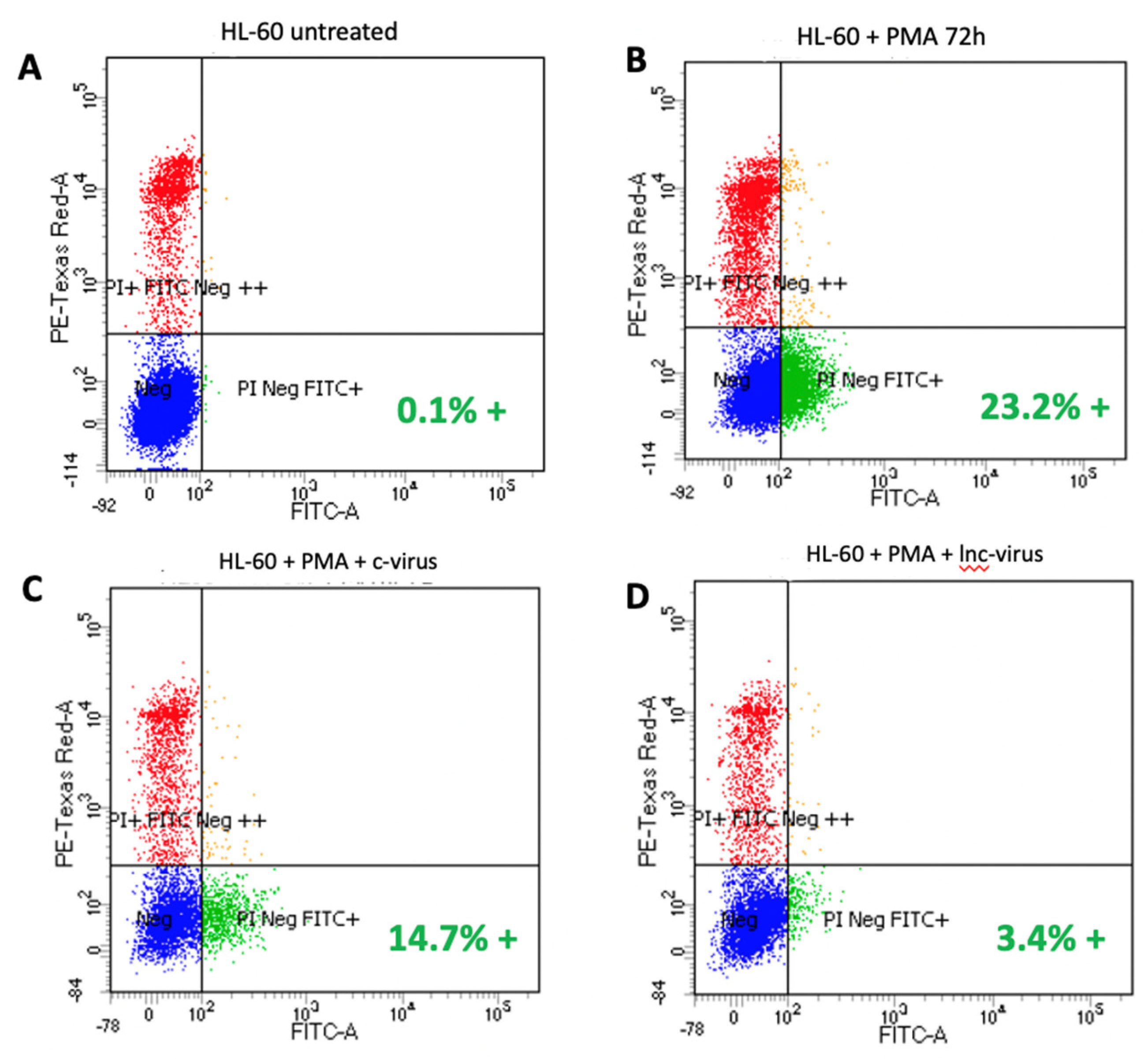
Lentivirus vectors were generated expressing high levels of lncRNAp53int1 and introduced at a multiplicity of infection of 5, into HL-60 cells. Measurement of lncRNAp53int1 levels by qPCR revealed an average of a
29-fold increase in the level of lncRNAp53int. As predicted, treatment of uninfected HL-60 with PMA induced differentiation in over 23% of the cells identified by the presence of cell surface CD-11b. However, when lncRNAp53Int1 levels were highly elevated by virtue of lentivirus expression, terminal differentiation was significantly inhibited resulting in only 3.4% of the cells expressing CD-11b: a reduction of 85% (
Figure 2).
6. Inhibited Expression of lncRNAp53int1 after Treatment with siRNA Leads to Spontaneous Cell Differentiation
Another prediction of our model is that if expression of the lncRNA were to be inhibited, then the cells would be predicted to undergo spontaneous differentiation even in the absence of an inducer such as PMA. In order to test this, we had three inhibitory siRNA molecules synthesized based on algorithms employed by Invitrogen Corp (Waltham, MA). siRNA molecules were introduced into HL-60 cells and after 48hrs, the level of lncRNAp53int1 was found to be reduced greater than 10,000-fold, effectively eliminating the transcript to background levels. When the cells were then assayed for spontaneous differentiation by cell surface expression of CD-11a, the number of terminally differentiated and non-proliferative cells in the population increased by 39-fold after 48hrs.
Strategies to Interrogate lncRNA Function in Leukemic Cells
A. Identification of Protein and/or RNA Partners
To identify proteins that associate with lncRNAp53Int1, a biotinylated derivative of lncRNAp53int1 has been generated to capture either proteins or RNA molecules that interact with the RNA. Total cell lysates that are prepared from untreated HL-60 cells in an RNA binding buffer can be incubated with biotinylated RNA and RNA-protein complexes isolated using streptavidin magnetic beads. By subjecting these products to SDS gel electrophoresis, prominent and specific bands are isolated, digested with trypsin, and subjected to Mass Spectrometry. Alternatively, to capture low abundance proteins, the entire lane of the gel can be also isolated, cut into small segments, subjected to trypsinization and mass spectrometry.
Once a candidate protein or proteins are detected, validation studies to determine whether these interactions occur in cells. Antibodies directed against the identified protein or proteins are then used for immunoprecipitations followed by RT-PCR to detect the lncRNAp53Int1 transcript in the complex. Alternatively, the introduction of a FLAG-tag onto a cDNA of the protein identified as part of an expression vector such as pcDNA (CMV promoter) can be introduced into cells and the protein precipitated with anti-FLAG antibody. RT-PCR will detect lncRNAp53Int1in the complex.
Since some lncRNAs, designated competing endogenous RNAs (ceRNA), function through interactions with specific miRNAs, miRNA sequences expressed in myeloid cells can be obtained from the miRBase database (version 22.1; 2018; Kozomara et al. 2019), and the likelihood of binding of a specific miRNA to lncRNAp53Int1 estimated using TargetScan (
http://www.targetscan.org/ version 8.0, 2021).
Once validated miRNAs that complex with lncRNAp53Int1 are identified, strategies to understand their role in differentiation can be developed. For example, if lncRNAp53int1 acts as a “sponge” or ceRNA for any of these miRNAs then we expect that the abundance of the particular miRNA would increase as lncRNAp53int1 levels decrease either during induced differentiation or in response to inhibition by siRNA, whereas overexpression of lncRNAp53int1 by lentivirus expression should prevent the increase in miRNA levels.
B. Identification of Genetic Program Requiring lncRNAp53Int1
RNAseq technology and transcriptome profiling will ultimately be a powerful strategy to investigate differentially expressed genes in response to a variety of stimuli (Wang et al., 2009; Chung et. al., 2021). By identifying the genes regulated in response to the expression of this lncRNA, the biology and regulatory pathways influenced by this lncRNA will become more apparent. This information will provide insight not only into how this lncRNA functions, but also ways that these pathways lead to leukemia when defective or inappropriately regulated. The results and detailed analysis of these experiments will allow the ability to focus in on the genes that this lncRNA participates in regulating and provide further insight into both the process of terminal differentiation.
Future Prospects
In that differentiation therapy for myeloid leukemias continues to hold promise as new targets that control differentiation are defined (Nowak et al., 2009; Su et al., 2014), the observation that a lncRNA, lncRNAp53int1, plays an important role in the proliferative to terminal differentiation transition in acute myeloid leukemia-derived cells, additional studies will shed light on regulatory role(s) for lncRNAp53int1 in leukemic cell differentiation. This may then lead to new potential therapeutic target(s) that will induce differentiation. The identification of protein(s) and or RNA partners and initiated studies into their functions will provide inroads into the pathways through which lncRNAp53int1 functions. As this research progresses, in vivo studies in mouse models of myeloid leukemia will test the consequences of targeting lncRNAp53int1 or it’s binding partners as potential therapies for the disease.
Acknowledgments
This work was supported by funding from the Children’s Leukemia Research Foundation.
References
- Atianand MK, Hu W, Satpathy AT, et al. (2016). A long noncoding RNA lincRNA-EPS acts as a transcriptional brake to restrain inflammation. Cell 165, 1672-1685.
- Chung, M., Bruno, V.M., Rasko, D.A., et al. (2021) Best practices on the differential expression analysis on multi-species RNA-seq. Genome Biol 29, 121-136. [CrossRef]
- Engreitz JM, Haines JE, Perez EM, et al. (2016) Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539, 452-455.
- Farzaneh, M., Najafi, S., Sabzehpoush. M., et al., (2023) The stem cell-specific long non-cofding ZRNAs in leukemia. Clin Transl Oncol. 25, 345-351.
- Fernandes, M., Marques, H., Tiexeira,A.L., and Medeiros, R. (2021) Competitive endogenous RNA network involving MiRNA and lncRNA in non-Hodgkin lymphoma: current advances and clinical perspectives, Biomed. 17, 1934-1947. [CrossRef]
- Herman, AB., Tsitsipatis, D., and Gorospe, M. (2022) Integrated lncRNA function upon genomic and epigenomic regulation. Mol. Cell. 82, 2252-2266.
- Jiang, Z., Liu, t., Wang, Y. et al. (2024). Effect of lncRNA XIST on acute myeloid leukemia cells via miR-142-5p-PFKP axis. Hematology 29, 2306444. [CrossRef]
- Kopp, F. and Mendell JT. (2018) Functional classification and experimental dissection of long noncoding RNAs. Cell 172, 393-407. [CrossRef]
- Kozomara, A, Birgaoanu, M, and Griffiths-Jones, S. (2019). MiRBase: from microRNA sequences to function. Nuc. Acid Res. 47, D155-D162. [CrossRef]
- Kretz, M, Siprashvili, Z., Chu, C., et al. (2013) Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493, 231-235. [CrossRef]
- Lobo-Alves, S., de Oliviera, L., Kretzschmar, GC., et al. (2024) Long noncoding RNA expresión in acute lymphoblastic leukemia: a systematic review. Crit Rev Oncol Hematol 196, 104290.
- Mangiavacchi, A., Morelli, G., and Orlando, V. (2023) Behind the scenes: How RNA orchestrates the epigenetic regulation of gene expression. Front Cell Dev Biol. 11,1123975. [CrossRef]
- McCabe, E.M., and Rasmussen, T.P. (2021) lncRNA involvement in cancer stem cell function and epithelial-mesenchymal transitions. Semin Cancer Biol. 75, 38-48. [CrossRef]
- Nunez-Martinez., H.N and Recillas-Targas, F. (2022) Emerging functions of lncRNA loci beyond the transcript itself. Int J Mol. Sci. 23, 6258.
- Reisman, D., and Rotter, V. (1989). Two promoters that map to 5′-sequences of the human p53 gene are differentially regulated during terminal differentiation of myeloid cells. Oncogene 4:945-953.
- Reisman, D. and Balint, E, Loging, et al. (1997). A novel transcript encoded within the 10-Kb first intron of the human p53 tumor suppressor gene (D17S2179E) is induced during differentiation of myeloid leukemia cells. Genomics 15, 364-370. [CrossRef]
- Reisman, D., Gibson, A., Patel, M., and Wang, Y. (2016). Evidence for a role of a lncRNA encoded from the p53 tumor suppressor gene in maintaining the undifferentiated state of human myeloid leukemias. Gene Reports, 5, 45-50. [CrossRef]
- Statello, L., Guo, C.J., Chen, L.L., and Huarte M. (2021). Gene regulation by long non-coding RNAs and its biological functions. Nature Reviews|Cell Biology 22, 96-118. [CrossRef]
- Su., R., Lin, HS., Zhang, XH., et al. (2014) MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene, 34: 3226-3239. [CrossRef]
- Suzuki, H., Forrest, A.R., van Nimwegen, E., et al. (2009) The transcriptional network that controls growth arrest and differentiation in a myeloid leukemia cell line. Nature Genetics. 41, 553-562.
- Tasseff, R., Jensen, H.A., Congleton, J., et al. (2017) An Effective Model of the Retinoic Acid Induced HL-60 Differentiation Program. Scientific Reports 7, 14317. [CrossRef]
- Wang, Z., Gerstein, M. and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genetics 10, 57-63. [CrossRef]
- Zibitt, S.M., Harford, C.C.R. and Lai, A. (2021). Interrogating lncRNA functions via CRISPR/CAS systems. RNA Biol. 18, 2097-2106.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).