Submitted:
16 July 2024
Posted:
17 July 2024
You are already at the latest version
Abstract
Keywords:
Introduction
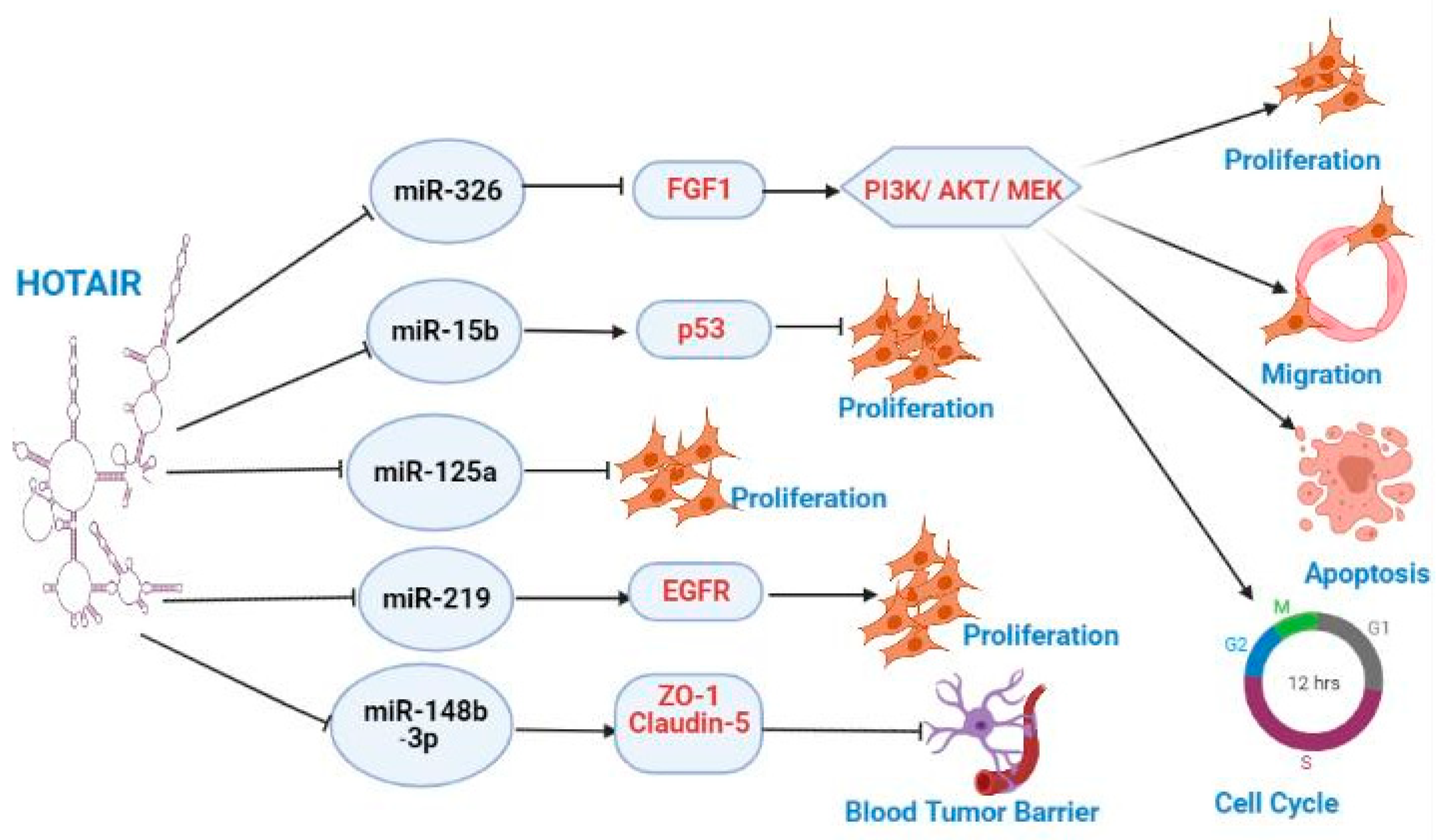
| Role of HOTAIR | Reference |
|---|---|
|
- HOTAIR inhibits the transcription of NLK in U87 GBM cells, regulate Wnt/β-catenin pathway, inhibit cell cycle arrest and promote cell migration. |
[70] |
| - HOTAIR mRNA levels are increased in A172 glioma cells compared to normal astrocytes. . |
[71] |
| - miR-141 directly binds to the 3UTR of HOTAIR in U251 and U87 glioma cells, inhibiting its expression. |
[72] |
| - miR-148b-3p downregulates the expression of tight junction-related proteins including ZO-1, clauidin-5, and occludin |
[69] |
| - HOTAIR rs920778 and rs12826786 frequencies do not differ between glioma patients and controls |
[73] |
| - HOTAIR levels positively correlate with MMP-7, MMP-9, and VEGF levels in human glioma |
[74] |
| - HOTAIR upregulates the expression of hexokinase 2 by downregulating miR-125 |
[75] |
| - HOTAIR is upregulated in temozolomide-resistant GBM cells - Serum exosome HOTAIR levels are higher in GBM patients’ resistant to temozolomide compared with responders. |
[68] |
Conclusions
Credit Author Statement
Conflict of Interest
Funding
References
- Bush, N.A.O.; Chang, S.M.; Berger, M.S. Current and future strategies for treatment of glioma. Neurosurg. Rev. 2016, 40, 1–14. [Google Scholar] [CrossRef]
- Sherrod, B.A.; Gamboa, N.T.; Wilkerson, C.; Wilde, H.; Azab, M.A.; Karsy, M.; Jensen, R.L.; Menacho, S.T. Effect of patient age on glioblastoma perioperative treatment costs: a value driven outcome database analysis. J. Neuro-Oncology 2019, 143, 465–473. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Kapranov, P.; Cheng, J.; Dike, S.; Nix, D.A.; Duttagupta, R.; Willingham, A.T.; Stadler, P.F.; Hertel, J.; Hackermüller, J.; Hofacker, I.L.; et al. RNA Maps Reveal New RNA Classes and a Possible Function for Pervasive Transcription. Science 2007, 316, 1484–1488. [Google Scholar] [CrossRef]
- Wahlestedt, C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat. Rev. Drug Discov. 2013, 12, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef]
- Gomes, A.Q.; Nolasco, S.; Soares, H. Non-Coding RNAs: Multi-Tasking Molecules in the Cell. Int. J. Mol. Sci. 2013, 14, 16010–16039. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Nowakowski, T.J.; Pollen, A.A.; Lui, J.H.; Horlbeck, M.A.; Attenello, F.J.; He, D.; Weissman, J.S.; Kriegstein, A.R.; Diaz, A.A.; et al. Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 2016, 17, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Dong, X. , et al., Knockdown of long non-coding RNA ANRIL inhibits proliferation, migration, and invasion but promotes apoptosis of human glioma cells by upregulation of miR-34a. J Cell Biochem, 2018. 119(3): p. 2708-2718.
- Shi, Y.; Wang, Y.; Luan, W.; Wang, P.; Tao, T.; Zhang, J.; Qian, J.; Liu, N.; You, Y. Long Non-Coding RNA H19 Promotes Glioma Cell Invasion by Deriving miR-675. PLOS ONE 2014, 9, e86295. [Google Scholar] [CrossRef]
- Shang, C.; Guo, Y.; Hong, Y.; Xue, Y.-X. Long Non-coding RNA TUSC7, a Target of miR-23b, Plays Tumor-Suppressing Roles in Human Gliomas. Front. Cell. Neurosci. 2016, 10, 235. [Google Scholar] [CrossRef]
- Bhan, A.; Mandal, S.S. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim. et Biophys. Acta (BBA) - Rev. Cancer 2015, 1856, 151–164. [Google Scholar] [CrossRef]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.-C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Morales, D.R.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. 2009, 106, 11667–11672. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Mandal, S.S. Long Noncoding RNAs: Emerging Stars in Gene Regulation, Epigenetics and Human Disease. ChemMedChem 2014, 9, 1932–1956. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.-C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science 2010, 329, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Mozdarani, H.; Ezzatizadeh, V.; Parvaneh, R.R. The emerging role of the long non-coding RNA HOTAIR in breast cancer development and treatment. J. Transl. Med. 2020, 18, 1–15. [Google Scholar] [CrossRef]
- Shi, J.; Dong, B.; Cao, J.; Mao, Y.; Guan, W.; Peng, Y.; Wang, S. Long non-coding RNA in glioma: signaling pathways. Oncotarget 2017, 8, 27582–27592. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Qin, Y.; Zhi, Q.; Wang, J.; Qin, C. Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3K/Akt and Wnt/β-catenin signaling pathways by up-regulating miR-34a. Int. J. Biol. Macromol. 2017, 107, 2620–2629. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lin, C.; Yong, W.; Ye, Y.; Huang, Z. Calycosin and Genistein Induce Apoptosis by Inactivation of HOTAIR/p-Akt Signaling Pathway in Human Breast Cancer MCF-7 Cells. Cell. Physiol. Biochem. 2015, 35, 722–728. [Google Scholar] [CrossRef]
- Liang, H.; Huang, W.; Wang, Y.; Ding, L.; Zeng, L. Overexpression of MiR-146a-5p Upregulates lncRNA HOTAIR in Triple-Negative Breast Cancer Cells and Predicts Poor Prognosis. Technol. Cancer Res. Treat. 2019, 18. [Google Scholar] [CrossRef]
- Wu, Y.; Liu, J.; Zheng, Y.; You, L.; Kuang, D.; Liu, T. Suppressed expression of long non-coding RNA HOTAIR inhibits proliferation and tumourigenicity of renal carcinoma cells. Tumor Biol. 2014, 35, 11887–11894. [Google Scholar] [CrossRef]
- Xavier-Magalhães, A. , et al., The long non-coding RNA HOTAIR is transcriptionally activated by HOXA9 and is an independent prognostic marker in patients with malignant glioma. Oncotarget, 2018. 9(21): p. 15740-15756.
- Huang, K.; Sun, J.; Yang, C.; Wang, Y.; Zhou, B.; Kang, C.; Han, L.; Wang, Q. HOTAIR upregulates an 18-gene cell cycle-related mRNA network in glioma. Int. J. Oncol. 2017, 50, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.X. , et al., HOTAIR, a cell cycle-associated long noncoding RNA and a strong predictor of survival, is preferentially expressed in classical and mesenchymal glioma. Neuro Oncol, 2013. 15(12): p. 1595-603.
- Pojo, M.; Gonçalves, C.S.; Xavier-Magalhães, A.; Oliveira, A.I.; Gonçalves, T.; Correia, S.; Rodrigues, A.J.; Costa, S.; Pinto, L.; Pinto, A.A.; et al. A transcriptomic signature mediated by HOXA9 promotes human glioblastoma initiation, aggressiveness and resistance to temozolomide. Oncotarget 2015, 6, 7657–7674. [Google Scholar] [CrossRef] [PubMed]
- Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature, 2008. 455(7216): p. 1061-8.
- Chen, Y.; Bian, Y.; Zhao, S.; Kong, F.; Li, X. Suppression of PDCD4 mediated by the long non-coding RNA HOTAIR inhibits the proliferation and invasion of glioma cells. Oncol. Lett. 2016, 12, 5170–5176. [Google Scholar] [CrossRef] [PubMed]
- Xia, S.; Ji, R.; Zhan, W. Long noncoding RNA papillary thyroid carcinoma susceptibility candidate 3 (PTCSC3) inhibits proliferation and invasion of glioma cells by suppressing the Wnt/β-catenin signaling pathway. BMC Neurol. 2017, 17, 1–11. [Google Scholar] [CrossRef]
- Xiao, D.; Cui, X.; Wang, X. LncRNA PTCSC3 inhibits cell proliferation in laryngeal squamous cell carcinoma by down-regulating lncRNA HOTAIR. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef]
- Visconti, R.; Della Monica, R.; Grieco, D. Cell cycle checkpoint in cancer: A therapeutically targetable double-edged sword. J. Exp. Clin. Cancer Res. 2016, 35, 153. [Google Scholar] [CrossRef] [PubMed]
- Bucher, N.; Britten, C.D. G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. Br. J. Cancer 2008, 98, 523–528. [Google Scholar] [CrossRef]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef]
- Tripathi, V.; Shen, Z.; Chakraborty, A.; Giri, S.; Freier, S.M.; Wu, X.; Zhang, Y.; Gorospe, M.; Prasanth, S.G.; Lal, A.; et al. Long Noncoding RNA MALAT1 Controls Cell Cycle Progression by Regulating the Expression of Oncogenic Transcription Factor B-MYB. PLOS Genet. 2013, 9, e1003368. [Google Scholar] [CrossRef]
- Ke, J.; Yao, Y.-L.; Zheng, J.; Wang, P.; Liu, Y.-H.; Ma, J.; Li, Z.; Liu, X.-B.; Li, Z.-Q.; Wang, Z.-H.; et al. Knockdown of long non-coding RNA HOTAIR inhibits malignant biological behaviors of human glioma cells via modulation of miR-326. Oncotarget 2015, 6, 21934–21949. [Google Scholar] [CrossRef]
- Stangeland, B.; Mughal, A.A.; Grieg, Z.; Sandberg, C.J.; Joel, M.; Nygård, S.; Meling, T.; Murrell, W.; Mo, E.O.V.; Langmoen, I.A. Combined expressional analysis, bioinformatics and targeted proteomics identify new potential therapeutic targets in glioblastoma stem cells. Oncotarget 2015, 6, 26192–26215. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.-L.; Hsieh, T.-H.; Ng, K.-H.; Tsai, Y.-N.; Tsai, C.-F.; Chao, M.-E.; Liu, D.-J.; Chu, S.-S.; Chen, W.; Liu, Y.-R.; et al. Downregulation of miR-137 and miR-6500-3p promotes cell proliferation in pediatric high-grade gliomas. Oncotarget 2016, 7, 19723–19737. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Dai, Y.; Grant, S.; Dent, P. Enhancing CHK1 inhibitor lethality in glioblastoma. Cancer Biol. Ther. 2012, 13, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar-Sinha, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.J.; Sewalt, R.G.A.B.; Otte, A.P.; et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R. , et al., Downregulation of Ezh2 expression by RNA interference induces cell cycle arrest in the G0/G1 phase and apoptosis in U87 human glioma cells. Oncol Rep, 2012. 28(6): p. 2278-84.
- Jalali, S. , et al., Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One, 2013. 8(2): p. e53823.
- Liu, X.-H.; Sun, M.; Nie, F.-Q.; Ge, Y.-B.; Zhang, E.-B.; Yin, D.-D.; Kong, R.; Xia, R.; Lu, K.-H.; Li, J.-H.; et al. Lnc RNA HOTAIR functions as a competing endogenous RNA to regulate HER2 expression by sponging miR-331-3p in gastric cancer. Mol. Cancer 2014, 13, 92–92. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cai, K.; Wang, J.; Wang, X.; Cheng, K.; Shi, F.; Jiang, L.; Zhang, Y.; Dou, J. MiR-7, Inhibited Indirectly by LincRNA HOTAIR, Directly Inhibits SETDB1 and Reverses the EMT of Breast Cancer Stem Cells by Downregulating the STAT3 Pathway. STEM CELLS 2014, 32, 2858–2868. [Google Scholar] [CrossRef]
- Sun, G.; Wang, Y.; Zhang, J.; Lin, N.; You, Y. MiR-15b/HOTAIR/p53 form a regulatory loop that affects the growth of glioma cells. J. Cell. Biochem. 2018, 119, 4540–4547. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Xiao, G.; Peng, G.; Liu, D.; Wang, Z.; Liao, Y.; Liu, Q.; Wu, M.; Yuan, X. MiRNA-125a-5p inhibits glioblastoma cell proliferation and promotes cell differentiation by targeting TAZ. Biochem. Biophys. Res. Commun. 2015, 457, 171–176. [Google Scholar] [CrossRef]
- Tang, L.; Shen, H.; Li, X.; Li, Z.; Liu, Z.; Xu, J.; Ma, S.; Zhao, X.; Bai, X.; Li, M.; et al. MiR-125a-5p decreases after long non-coding RNA HOTAIR knockdown to promote cancer cell apoptosis by releasing caspase 2. Cell Death Dis. 2016, 7, e2137–e2137. [Google Scholar] [CrossRef]
- Jiang, Y. , et al., Schisandrin B inhibits the proliferation and invasion of glioma cells by regulating the HOTAIR-micoRNA-125a-mTOR pathway. Neuroreport, 2017. 28(2): p. 93-100.
- Rao, S.A.M.; Arimappamagan, A.; Pandey, P.; Santosh, V.; Hegde, A.S.; Chandramouli, B.A.; Somasundaram, K. miR-219-5p Inhibits Receptor Tyrosine Kinase Pathway by Targeting EGFR in Glioblastoma. PLOS ONE 2013, 8, e63164. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Guan, C. HOTAIR inhibits the proliferation of glioblastoma cells by targeting miR-219. Cancer Biomarkers 2020, 28, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Plate, K.H.; Breier, G.; Weich, H.A.; Mennel, H.D.; Risau, W. Vascular endothelial growth factor and glioma angiogenesis: Coordinate induction of VEGF receptors, distribution of VEGF protein and possible In vivo regulatory mechanisms. Int. J. Cancer 1994, 59, 520–529. [Google Scholar] [CrossRef]
- A Azab, M.; Alomari, A.; Azzam, A.Y. Featuring how calcium channels and calmodulin affect glioblastoma behavior. A review article. Cancer Treat. Res. Commun. 2020, 25, 100255. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.-M.; Lu, Y.-F.; Hu, B.-G.; Liang, W.-C.; Zhu, X.; Yang, H.-D.; Li, G.; Zhang, J.-F. Long noncoding RNA hotair mediated angiogenesis in nasopharyngeal carcinoma by direct and indirect signaling pathways. Oncotarget 2015, 7, 4712–4723. [Google Scholar] [CrossRef]
- Subramaniam, S.R.; Federoff, H.J. Targeting Microglial Activation States as a Therapeutic Avenue in Parkinson’s Disease. Front. Aging Neurosci. 2017, 9, 176–176. [Google Scholar] [CrossRef] [PubMed]
- Cata, J.; Bhavsar, S.; Hagan, K.; Arunkumar, R.; Grasu, R.; Dang, A.; Carlson, R.; Arnold, B.; Popat, K.; Rao, G.; et al. Intraoperative serum lactate is not a predictor of survival after glioblastoma surgery. J. Clin. Neurosci. 2017, 43, 224–228. [Google Scholar] [CrossRef]
- Jin, Z.; Jin, R.-H.; Ma, C.; Li, H.-S.; Xu, H.-Y. Serum expression level of miR-504 can differentiate between glioblastoma multiforme and solitary brain metastasis of non-small cell lung carcinoma. . 2017, 22, 474–480. [Google Scholar]
- Vietheer, J.-M.; Rieger, J.; Wagner, M.; Senft, C.; Tichy, J.; Foerch, C. Serum concentrations of glial fibrillary acidic protein (GFAP) do not indicate tumor recurrence in patients with glioblastoma. J. Neuro-Oncology 2017, 135, 193–199. [Google Scholar] [CrossRef]
- Tan, S.K.; Pastori, C.; Penas, C.; Komotar, R.J.; Ivan, M.E.; Wahlestedt, C.; Ayad, N.G. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol. Cancer 2018, 17, 1–7. [Google Scholar] [CrossRef]
- Cantile, M.; Scognamiglio, G.; Marra, L.; Aquino, G.; Botti, C.; Falcone, M.R.; Malzone, M.G.; Liguori, G.; Di Bonito, M.; Franco, R.; et al. HOTAIR role in melanoma progression and its identification in the blood of patients with advanced disease. J. Cell. Physiol. 2017, 232, 3422–3432. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; He, X.; Zheng, Z.; Ma, X.; Hu, X.; Wu, D.; Wang, M. Serum HOTAIR as a novel diagnostic biomarker for esophageal squamous cell carcinoma. Mol. Cancer 2017, 16, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ren, Y.; Wang, Y.; Tan, Y.; Wang, Q.; Cai, J.; Zhou, J.; Yang, C.; Zhao, K.; Yi, K.; et al. A Compound AC1Q3QWB Selectively Disrupts HOTAIR-Mediated Recruitment of PRC2 and Enhances Cancer Therapy of DZNep. Theranostics 2019, 9, 4608–4623. [Google Scholar] [CrossRef] [PubMed]
- Shi, J. , et al., HOTAIR-EZH2 inhibitor AC1Q3QWB upregulates CWF19L1 and enhances cell cycle inhibition of CDK4/6 inhibitor palbociclib in glioma. Clin Transl Med, 2020. 10(1): p. 182-198.
- Filippakopoulos, P.; Knapp, S. Targeting bromodomains: epigenetic readers of lysine acetylation. Nat. Rev. Drug Discov. 2014, 13, 337–356. [Google Scholar] [CrossRef] [PubMed]
- Pastori, C.; Kapranov, P.; Penas, C.; Peschansky, V.; Volmar, C.-H.; Sarkaria, J.N.; Bregy, A.; Komotar, R.; St Laurent, G.; Ayad, N.G.; et al. The Bromodomain protein BRD4 controls HOTAIR, a long noncoding RNA essential for glioblastoma proliferation. Proc. Natl. Acad. Sci. USA 2015, 112, 8326–8331. [Google Scholar] [CrossRef] [PubMed]
- Li, C.H.; Chen, Y. Targeting long non-coding RNAs in cancers: Progress and prospects. Int. J. Biochem. Cell Biol. 2013, 45, 1895–1910. [Google Scholar] [CrossRef] [PubMed]
- Bruniaux, J.; Allard-Vannier, E.; Aubrey, N.; Lakhrif, Z.; Ben Djemaa, S.; Eljack, S.; Marchais, H.; Hervé-Aubert, K.; Chourpa, I.; David, S. Magnetic nanocarriers for the specific delivery of siRNA: Contribution of breast cancer cells active targeting for down-regulation efficiency. Int. J. Pharm. 2019, 569, 118572. [Google Scholar] [CrossRef] [PubMed]
- Fang, K.; Liu, P.; Dong, S.; Guo, Y.; Cui, X.; Zhu, X.; Li, X.; Jiang, L.; Liu, T.; Wu, Y. Magnetofection based on superparamagnetic iron oxide nanoparticle-mediated low lncRNA HOTAIR expression decreases the proliferation and invasion of glioma stem cells. Int. J. Oncol. 2016, 49, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; He, A.; Chen, B.; Bi, J.; Chen, J.; Guo, D.; Qian, Y.; Wang, W.; Shi, T.; Zhao, Z.; et al. A HOTAIR regulatory element modulates glioma cell sensitivity to temozolomide through long-range regulation of multiple target genes. Genome Res. 2020, 30, 155–163. [Google Scholar] [CrossRef]
- Yuan, Z. , et al., Exosome-Mediated Transfer of Long Noncoding RNA HOTAIR Regulates Temozolomide Resistance by miR-519a-3p/RRM1 Axis in Glioblastoma. Cancer Biother Radiopharm, 2020.
- Sa, L.; Li, Y.; Zhao, L.; Liu, Y.; Wang, P.; Liu, L.; Li, Z.; Ma, J.; Cai, H.; Xue, Y. RETRACTED: The Role of HOTAIR/miR-148b-3p/USF1 on Regulating the Permeability of BTB. Front. Mol. Neurosci. 2017, 10, 194. [Google Scholar] [CrossRef]
- Zhou, X.; Ren, Y.; Zhang, J.; Zhang, C.; Zhang, K.; Han, L.; Kong, L.; Wei, J.; Chen, L.; Yang, J.; et al. HOTAIR is a therapeutic target in glioblastoma. Oncotarget 2015, 6, 8353–8365. [Google Scholar] [CrossRef]
- Wang, G.; Li, Z.; Tian, N.; Han, L.; Fu, Y.; Guo, Z.; Tian, Y. miR-148b-3p inhibits malignant biological behaviors of human glioma cells induced by high HOTAIR expression. Oncol. Lett. 2016, 12, 879–886. [Google Scholar] [CrossRef]
- Bian, E.-B.; Ma, C.-C.; He, X.-J.; Wang, C.; Zong, G.; Wang, H.-L.; Zhao, B. Epigenetic modification of miR-141 regulates SKA2 by an endogenous ‘sponge’ HOTAIR in glioma. Oncotarget 2016, 7, 30610–30625. [Google Scholar] [CrossRef]
- Xavier-Magalhães, A.; Oliveira, A.I.; de Castro, J.V.; Pojo, M.; Gonçalves, C.S.; Lourenço, T.; Viana-Pereira, M.; Costa, S.; Linhares, P.; Vaz, R.; et al. Effects of the functional HOTAIR rs920778 and rs12826786 genetic variants in glioma susceptibility and patient prognosis. J. Neuro-Oncology 2017, 132, 27–34. [Google Scholar] [CrossRef]
- Zhao, W.-H.; Yuan, H.-Y.; Ren, X.-Y.; Huang, K.; Guo, Z.-Y. Association between expression of HOTAIR and invasiveness of gliomas, and its predictive value. Adv. Clin. Exp. Med. 2019, 28, 1179–1183. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, G.; Gao, Y.; Liang, H. HOTAIR/miR-125 axis-mediated Hexokinase 2 expression promotes chemoresistance in human glioblastoma. J. Cell. Mol. Med. 2020, 24, 5707–5717. [Google Scholar] [CrossRef]
- Hanisch, A.; Silljé, H.H.W.; A Nigg, E. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 2006, 25, 5504–5515. [Google Scholar] [CrossRef]
- Wang, G.; Li, Z.; Tian, N.; Han, L.; Fu, Y.; Guo, Z.; Tian, Y. miR-148b-3p inhibits malignant biological behaviors of human glioma cells induced by high HOTAIR expression. Oncol. Lett. 2016, 12, 879–886. [Google Scholar] [CrossRef]
- Chen, L.; Han, L.; Wei, J.; Zhang, K.; Shi, Z.; Duan, R.; Li, S.; Zhou, X.; Pu, P.; Zhang, J.; et al. SNORD76, a box C/D snoRNA, acts as a tumor suppressor in glioblastoma. Sci. Rep. 2015, 5, 8588. [Google Scholar] [CrossRef]
- Chen, L.; Han, L.; Wei, J.; Zhang, K.; Shi, Z.; Duan, R.; Li, S.; Zhou, X.; Pu, P.; Zhang, J.; et al. SNORD76, a box C/D snoRNA, acts as a tumor suppressor in glioblastoma. Sci. Rep. 2015, 5, 8588. [Google Scholar] [CrossRef]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).




