1. Introduction
Despite the critical importance of images in biomedicine, there are relatively few image databases that provide structured, annotated, and standardized collections of images using FAIR guidelines, i.e., they are findable, accessible, interoperable, and reusable. For images related to biodiversity and taxonomy, two such databases are Morphobank (O’Leary & Kaufman 2011) and Morphosource (
www.morphosource.org). These repositories focus on collecting images and traits in a phylogenetic context (Morphobank) and on 3D images such as CT scans (Morphosource), respectively.
There is no shortage of animal photos on the web and several attempts have been made to survey them in a more systematic way, including reptiles (Durso et al. 2021; Marshall et al. 2020). Some databases such as iNaturalist have huge image collections counting in the millions but their main purpose is to document observations in nature. Although iNaturalist is not a taxonomic project, it has recently started to add images of museum specimens. Many taxon-specific databases, such as the Reptile Database, focus on taxonomy and but not specifically on image collection. We believe that taxonomists need specialized databases that provide standardized images, e.g., in order to permit direct comparison of specimens in high resolution so that diagnostic features can be easily recognized. While many natural history collections offer images on their websites, this is only true for a subset of collections or a subset of their specimens. Importantly, the distributed nature of having images spread over numerous websites makes those images hard to find and even harder to use (Ruthsatz et al. 2021).
The purpose of this project is to provide a starting point for a systematic collection of reptile images, taken from preserved specimens that can be revisited in those collections, with the pertinent meta-data such as localities and other information. We started by taking pictures of more than 1,000 reptile species in 20 collections around the world, resulting in more than 14,000 images that have been tagged with meta-data, so that they can be sorted by specific features such as particular body parts and cross-referenced with other databases such as VertNet (Guralnick et al. 2016) or the Reptile Database. We are making this collection accessible through Morphobank, where each image can be traced back to a specific specimen based on its collection and catalogue number.
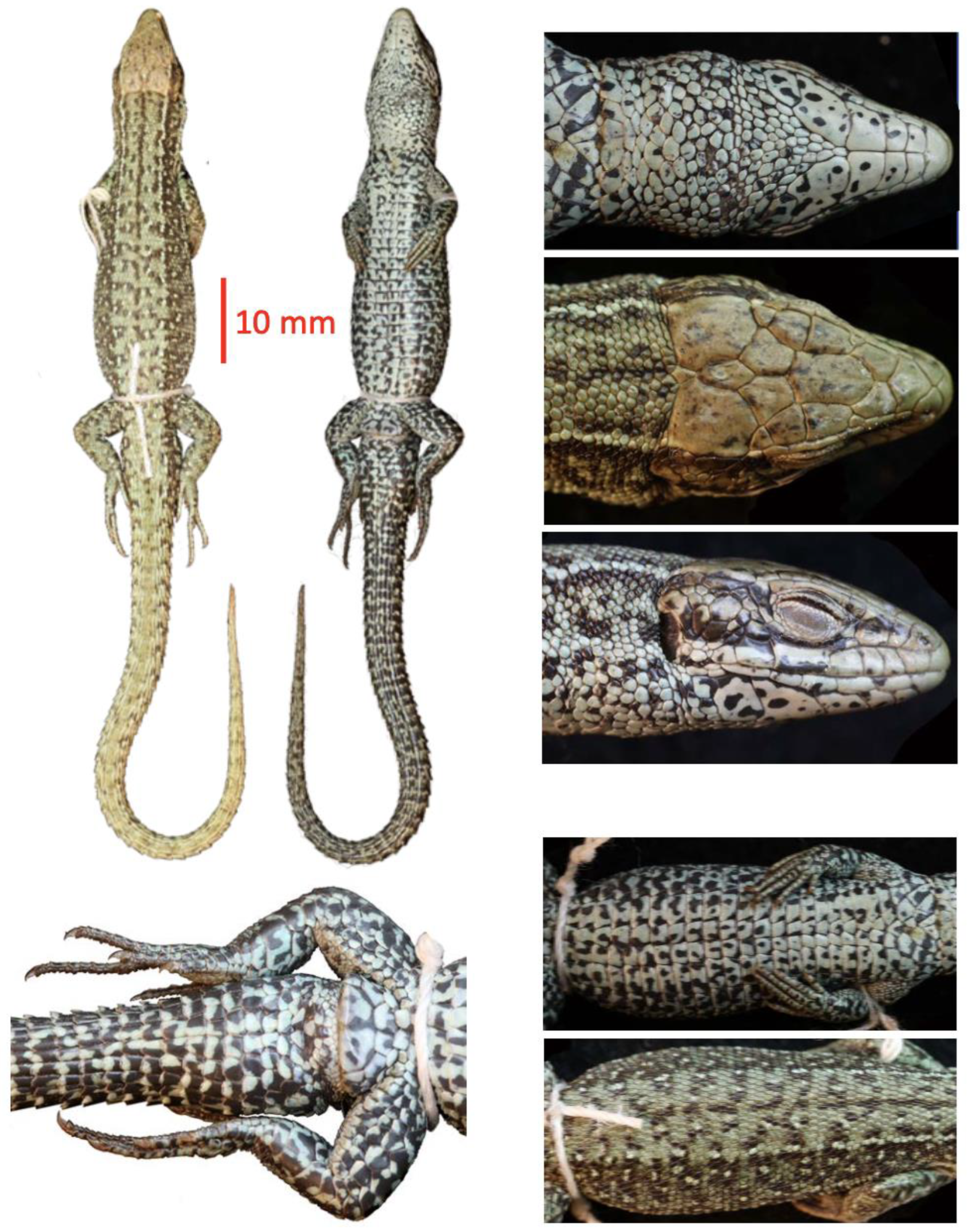
As an example and case study, we present photos of 8 species of the subfamily Lacertinae (currently identical with the tribus Lacertini), an Old World group containing 19 genera and 139 species of lizards to show the utility of such image collections for comparative studies (
Figure 1).
2. Methods and Materials
A total of 62.6 GB of 15,675 photographs encompassing 1,045 reptile species were taken across 20 collections around the world (
Table 1). 14,239 of these photos show specimens, the remaining 1436 show labels which may or may not contain additional information such as localities. The photos depict 1,342 unique specimens, with the explicit intention to take pictures from the same perspective, namely the dorsal and ventral side of the whole body, plus close-ups of the head (dorsal, lateral, and ventral sides), as well as details of the trunk (lateral view) and the cloacal region. In some cases, close-ups of other body parts were taken too (such as the toe pads in geckos), depending on preservation state and other factors.
Since many photos were similar and created some redundancy, we removed some of this redundancy using Duplicate Photo Cleaner version 7.5.0.12 (
https://www.easyduplicatefinder. com/). This removed photos that exhibited at least an 85% similarity threshold to one or more other photos.
Metadata was added to the photos using a custom-made tool built in Claris FileMaker Pro (https://www.claris.com/filemaker/pro/). The view and visible body parts were marked for each image. Keywords for view included dorsal, ventral, left lateral, right lateral, and lateral, while keywords for body parts consisted of the whole body, head, trunk, forelimb, hindlimb, tail, hand, foot, and cloaca. After all the tagging was completed, the metadata was exported from FileMaker in the form of Excel sheets showing file names, body parts, and views for each image file.
3. Results
We have taken more than 14,239 standardized photos representing 1,045 species of reptiles and made them available in Morphobank. These photos can serve as a reference dataset for comparative morphology, species identification, or other purposes. While only 81 of the 1,342 specimens are primary types, images of other type specimen can be added as they become available.
Taxonomically, our initial goal was to obtain photos of all lizard genera, that is, at least one species of each genus (ideally the type species). While we have not reached that goal yet, our collection has photos of 490 lizard genera (out of 603), that is, 81% of all lizard genera. However, representing all genera is a moving target: since the turn of the century (2000), 122 new lizard genera have been described (Uetz et al. 2024), including 16 genera in the years 2021 and 2022, not counting those from taxonomic vandalism (Wüster et al. 2021). Most of these were not covered by this project, given that our current set of photos were mostly taken from 2019 to 2020 (before Covid made museum visits rather difficult).
Overall, we collected photos of 53 families, 29 of which having all genera covered (
Table 3). While most of these families are rather small, two of them have 10 or more genera, namely Anguidae and Phyllodactylidae. Of 41 families or nearly half of all families, we have captured 80% or more of all genera.
Given that our focus was on lizard genera, we attempted to obtain photos of the type species of each genus. This was achieved for 530 genera, including 478 lizard genera and for 52 snake genera (Supplementary Table S1).
In order to make the images more accessible, we removed about 20% of images as duplicates using a 85% similarity threshold (see methods). The remaining 14,239 image files were renamed and marked with metadata, so that they can be searched for features such as “head” or “ventral” sides. The tagged photos can be searched on or downloaded from the Morphobank and Figshare web sites (see Materials).
4. Discussion
This project aims to fill the need for a reference database of specimen photos that are highly standardized in terms of what and how they show it. The image collection presented here is also designed to be FAIR, that is, it is findable, accessible, interoperable, and reusable. The latter two criteria are fulfilled by their creative commons licenses but also by their connection to other databases such as the Reptile Database (especially for names) and Vertnet (for specimen information, including localities). The Reptile Database also provides and/or directly links to the original description and other relevant literature so that access to the primary sources is provided.
4.1. Morphological Features.
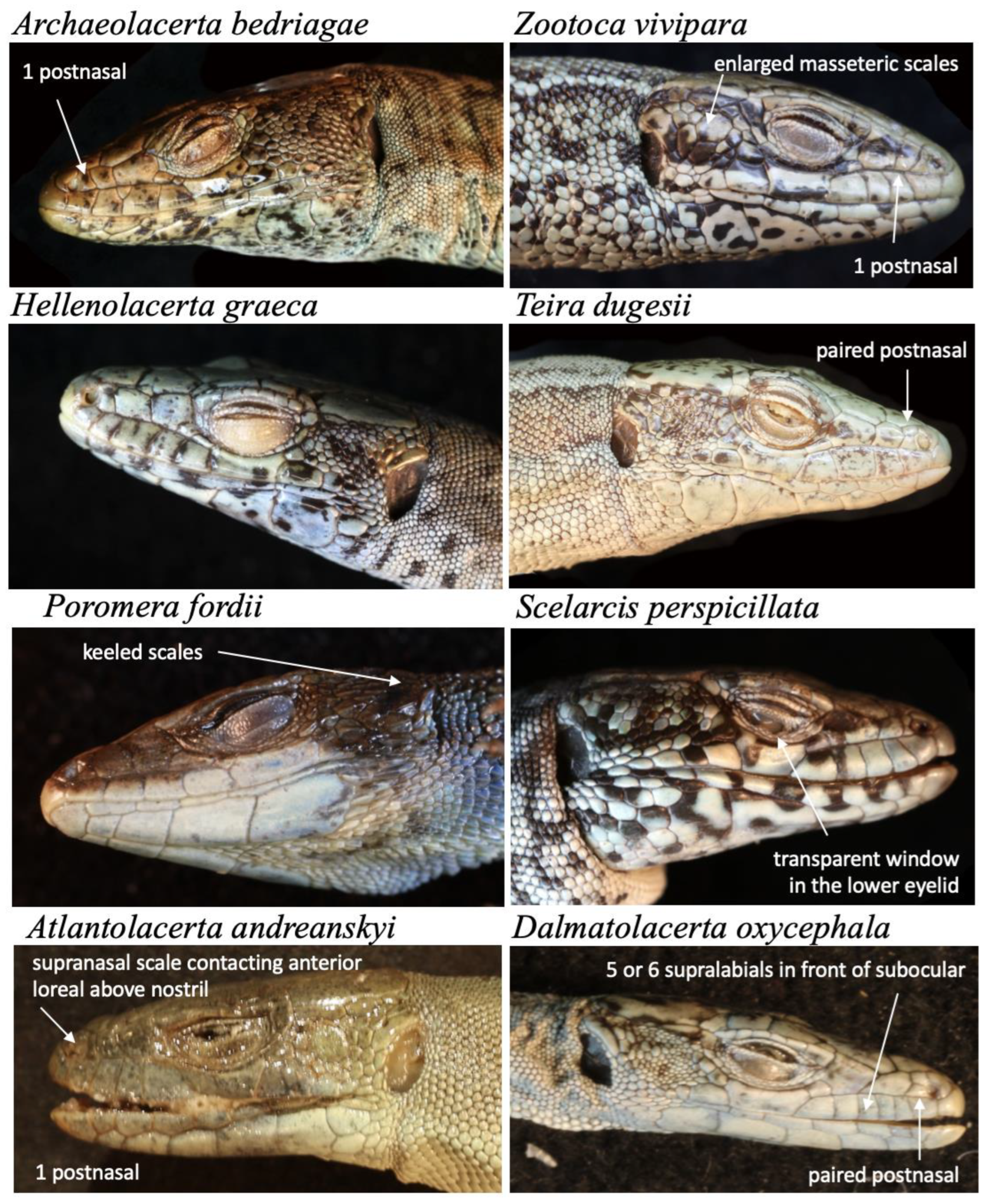
Each of our photos shows dozens or hundreds of individual scales and their features (size, shape, relationships, pattern, surface features etc), hence we cautiously estimate that we have documented at least a million, but probably millions of individual characters with our project. This database will become especially valuable when used in combination with other data sources such as the Reptile Database which now has descriptions of about 8000 species, mostly from the primary literature (Uetz et al. 2023). Most of these descriptions are of little use without images and many of the images in the primary literature are hard to find or behind paywalls (or there are no images, as in the majority of older descriptions). However, many historical descriptions have detailed descriptions of scalation features, and these will become available in conjunction with our image database. For instance, some lacertid genera have a single or paired postnasal and this character is easy to recognize, even for non-experts (
Figure 1). However, some characters are relative, such as having “enlarged masseteric scales”: without at least two photos to compare the sizes of scale in different specimens it is practically impossible to know what “enlarged” means, given that most lacertids have more or less enlarged masseteric scales (
Figure 1).
4.2. Limitations
Obviously, this project is only a first step towards a complete database of reference images. Even if we had images of all 12,000 reptile species, that would still not reflect the diversity and variation within species, as geographic and individual variation needs to be represented by additional images. Similarly, even those specimens that are represented often lack critical meta-data, such as precise localities or biological data such as sex or age (the latter can be estimated from size information, but that is usually not included in collection data).
A final concern is the notorious problem of taxonomic uncertainty: during the process of taking pictures, we found several mis-identified specimens which may have been simply mis-identified but it is increasingly common that species change names because of splitting. That is, if a species gets split into multiple other species, a subset of specimens needs to be re-allocated to the newly split off species which rarely happens in collections. This problem can only be solved by taxonomists and collection managers who closely follow the literature (or online resources such as the Reptile Database) and relabel those specimens as needed. However, our online resource will also allow the community to re-examine specimens and thus correct such misidentifications.
While the reference image database could also incorporate images of live animals, these specimens are likely not available for follow-up studies.
4.3. Linking Data and Databases
A critical component of this project was the linking of specimen data to both images but also collection databases such as VertNet. However, numerous other links are needed and possible, e.g., links to observation data (iNaturalist, GBIF), conservation data (IUCN), or DNA sequence databases which increasingly reference specimen information. DNA sequences will become increasingly important once phenotypes can be linked to genome data.
4.4. Usage of Reference Images
The reference library has several possible uses. The first and most obvious is to use it in taxonomic studies for comparison and for the identification of species. This includes the use by amateurs, e.g., in iNaturalist where many photos do not show sufficient details for exact species identification. Second, reference images will allow researchers to investigate traits much more easily and systematically, especially when they are combined with ontologies of terms. For instance, lacertids such as the species shown in
Figure 1 and
Figure 2 have essentially all the same types of scales. As long as a user knows which characters to look for, all images with that character can be easily found, simply by searching for all images that show, for instance, the dorsal side of a head (which has prefrontal, frontal, parietal, etc. scales). This could also facilitate the analysis of geographic variation. Third, we also envision possible uses for image analysis and automated analysis of the dataset. Reptiles are particularly suited for image analysis as the scales allow for easy demarcation and quantification, in contrast to the skin of amphibians or the fur of mammals which has much fewer landmarks. There are many more uses possible, and we invite the scientific community to explore such uses.
4.5. Future Improvements and Outlook
Most obviously, we need to expand the image collection to all reptile species, and eventually to all species in the animal and plant kingdoms. It remains to be seen to what extent this is possible, but with the inclusion of variation, the task as well as the possibilities are nearly endless. While it may be desirable to have 3D scans of all species, including photogrammetric images, the added benefit appears to be limited at this time, given the huge investment for 3D imaging, in addition to the storage and time requirements.
It will remain a challenge to maintain databases and the links to numerous other resources, but the increasing use of standardized identifiers (such as NCBI taxonIDs) should facilitate that in the future.
Acknowledgments
We thank the collection managers and curators who provided access to the specimens in their care, namely Dane Trembath (AMS), Stephanie Tessier (CMNAR), Andreas Schmitz (MHNG), Nicolas Vidal (MNHN), Bryan Stuart (NCSM), Eduard Stöckli (NMBA), Jane Melville (NMV), Silke Schweiger and Georg Gassner (NMW), Andrew Amey (QM), Garin Cael (RMCA), Ralph Foster (SAMA), Gunther Köhler (SMF), Alexander Kupfer (SMNS), Shai Meiri (TAU), Coleman Sheehy (UF), Paul Doughty (WAM), Flecks Morris (ZFMK), Frank Tillack (ZMB), Chan Kin Onn (ZRC), and Michael Franzen (ZSM). Kenzley Adolphe and Tanya Berardini generously helped with this project’s implementation at Morphobank.
References
- Durso, A.M. , et al. Citizen Science and Online Data: Opportunities and Challenges for Snake Ecology and Action against Snakebite. Toxicon 2021, 9–10, 100071. [Google Scholar] [CrossRef] [PubMed]
- Guralnick, R.P. , et al. The Importance of Digitized Biocollections as a Source of Trait Data and a New VertNet Resource. Database 2016, 2016, baw158. [Google Scholar] [CrossRef] [PubMed]
- Marshall, B.M. , et al. An Inventory of Online Reptile Images. Zootaxa 2020, 4896, 251–264. [Google Scholar] [CrossRef]
- Matthews, P.C. FAIRness in Scientific Publishing. F1000Research 2017, 5, 2816. [Google Scholar] [CrossRef]
- O’Leary, M.A.; Kaufman, S. MorphoBank: Phylophenomics in the ‘Cloud. ’ Cladistics 2011, 27, 529–537. [Google Scholar] [CrossRef]
- Ruthsatz, K.; Scherz, M.D.; Vences, M. Dissecting the tree of life: the prospect of open-access digital resources in morphology, anatomy and taxonomy in training the next generation of zoologists. Zootaxa 2021, 5016, 448–450. [Google Scholar] [CrossRef] [PubMed]
- Uetz, P.; Darko, Y.A.; Voss, O. Towards digital descriptions of all extant reptile species. Megataxa 2023, 10, 027–042. [Google Scholar] [CrossRef]
- Uetz, P. et al. (eds.) (2024) The Reptile Database, http://www.reptile-database.org , accessed 8 March 2024.
- Wüster, W. , Thomson, S.A., O’shea, M., & Kaiser, H. (2021) Confronting taxonomic vandalism in biology: conscientious community self-organization can preserve nomenclatural stability. Biol. J. of the Linnean Society 2021, 133, 645–670. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).