1. Introduction
Exosomes are small (average diameter 30-150 nm) extracellular vesicles produced by almost all cell types in the human body and they serve to promote intercellular communication and immunomodulatory functions. In most cases, cancer cells contain higher concentrations of exosomes than healthy cells [
1,
2]. Current researches on the development of exosome isolation technology have been reported using size exclusion chromatography, ultracentrifugation, density gradient centrifugation, and immunoaffinity exosome capture [
3,
4,
5,
6,
7,
8]. The ultracentrifugation method, the most common of these methods and considered the gold standard for exosome isolation, is simple but requires large amounts of sample and expensive, high-end equipment, such as a high-speed centrifuge. Therefore, it is important to obtain more efficient exosome isolation technologies that exhibit high sensitivity and high throughput. In that respect, immunoaffinity exosome capture is evaluated as an ideal method for isolating pure exosomes based on the principle of immunoaffinity interaction between exosome surface proteins (antigens) and target antibodies. It can isolate specific types of exosomes according to their surface markers [
9]. Exosome contents include unique mRNAs, microRNAs (miRNAs), DNA fragments, lipids, and proteins, which genetically specify the cell of origin [
3]. The most notable fact is that miRNAs exist in exosomes and, moreover, exosomal miRNAs are considered as biomarkers for many serious diseases [
10]. Exosomes from diseased people contain miRNAs not found in healthy individuals [
11]. MicroRNAs are small (21–25 nucleotides in length) endogenous non-coding naturally occurring single-stranded RNA molecules. Previous studies have reported that abnormal expression of specific miRNAs is closely associated with serious human diseases, such as cancer and cardiovascular diseases. Therefore, miRNAs can be used as biomarkers for early detection of several diseases and are also valuable therapeutic targets for the treatment of these diseases [
12].
The overexpression of miRNA-106b (miR-106b) has been noted in several tumor types. Abnormal miR-106b expression is associated with breast cancer, lung cancer, gastric cancer, prostate cancer, colorectal cancer, hepatocellular carcinoma, and esophageal squamous cell carcinoma [
13,
14,
15]. However, detecting miRNAs including miR-106b is technically difficult because they are small in size, low in abundance, and easily degradable. Therefore, accurate and sensitive miR-106b detection is important not only for understanding biological roles of miRNAs, but also for early and rigorous diagnosis of various human cancers. Among various approaches to detect miRNAs, RT-qPCR is preferred as the most convenient and practical method to detect miRNAs with high sensitivity and accuracy, and several technologies have been commercialized [
16,
17]. Recently, miRNA detection using various types of biosensor technology (fluorescence, electrochemistry, SPR, QCM, etc.) has been reported [
18,
19,
20,
21,
22,
23,
24,
25,
26]. Although some DNA analysis studies have been conducted using surface acoustic wave (SAW) biosensors [
27], only our previous study has reported studies on miRNA detection using SAW biosensors [
28].
A SAW biosensor is a piezoelectric biosensor based on proprietary surface acoustic waves to measure biomolecular interactions on the sensor surface in real time [
29,
30,
31,
32]. Specifically, this sensor measures changes in mass as a result of biomolecular interactions that occur on the sensor surface. The SAW sensor consists of a piezoelectric substrate as the base material and an input- and output-interdigitized transducer (IDT) on both sides of the substrate. When used as a biosensor, a waveguide layer is deposited on an IDT-patterned substrate for operation in aqueous conditions. Silicon dioxide (SiO
2) has been commonly used as a waveguide layer due to its high chemical and mechanical resistance, low acoustic loss, and ease of functionalization of a variety of biomaterials by self-assembled monolayer techniques of silane compounds.
This study demonstrates a SAW biosensor for detection of miR-106b isolated from cancer cells based on immunoaffinity separation technique using our unique paddle screw type devices. To improve detection reproducibility, an internal reference sensor was introduced in our SAW biosensor. This enables differential or normalized data acquisition from the working sensor signal and reference sensor signal, which can compensate unexpected signal drift or noise and discriminate against non-specific binding. Normalization (working sensor response divided by reference sensor response) can also be used to suppress disturbances known to similarly influence signals of both working and reference sensors.
2. Materials and Methods
2.1. Reagent and Materials
HPLC purified synthetic miRNAs (miR-21, miR-124, miR-106b, and miR-155), 5′-amine modified oligonucleotide probes, and 3′-thiol modified detecting oligonucleotide probes were obtained from Bioneer (Daejeon, South Korea). Their sequences are listed in
Table 1. Analytical grade gold(III) chloride, sodium citrate, and hydroxylamine hydrochloride for AuNP synthesis and gold staining reaction were obtained from Sigma-Aldrich Chemical Co. (St. Louis, MO, USA). (3-Glycidoxypropyl)triethoxysilane (3-GPTES), 3-amino-1-propanol, mercury(II) chloride and dithiothreitol (DTT) were purchased from Sigma-Aldrich. Human breast total RNA (AM6952) was obtained from Thermo Fisher Scientific (Waltham, MA, USA). MCF-7 human breast cancer cell line (ATCC
® HTB-22™) was obtained from American Type Culture Collection (ATCC, Rockville, MD, USA). An RNeasy Mini Kit for extracting total RNA was brought from Qiagen (Valencia, CA, USA). An HB miR Multi Assay kit™ for RT-qPCR assay to detect miRNAs was purchased from HeimBiotek, Inc. (Pangyo, South Korea).
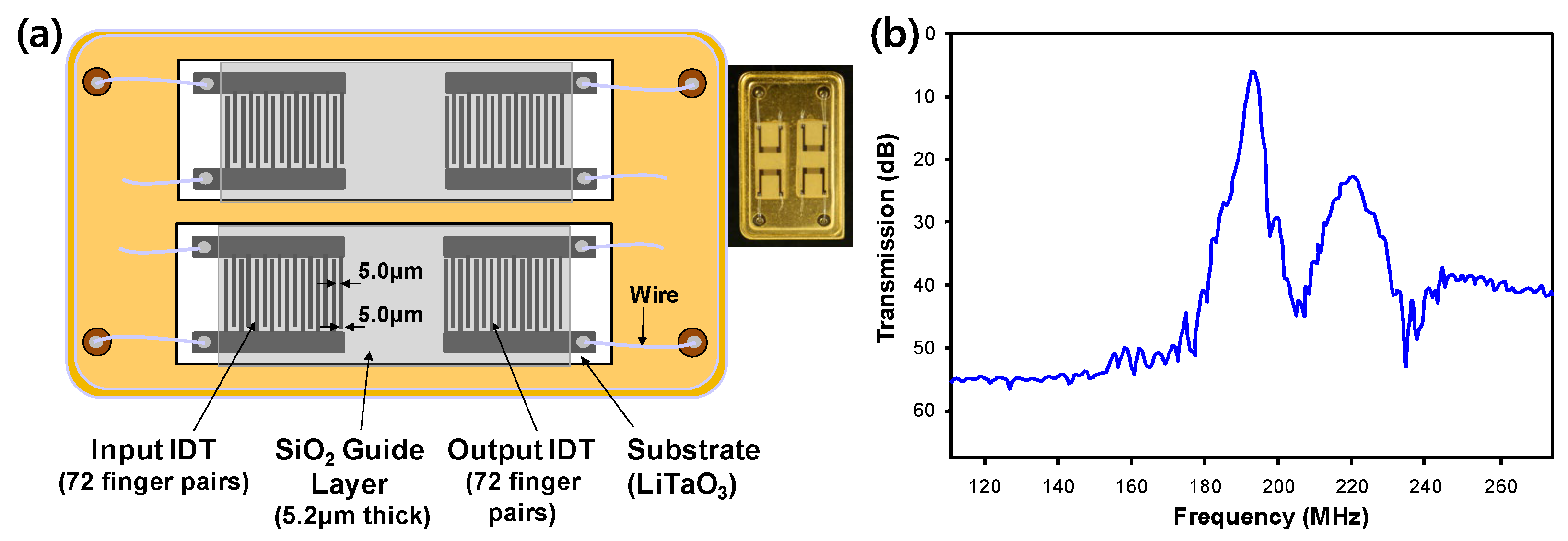
2.2. Design and Fabrication of the SAW Sensor
Two pairs of IDT electrodes forming a 2-port SAW delay line were patterned onto a 36
◦YX-LiTaO
3 piezoelectric substrate (Yamaju Ceramics Co. Ltd., Anada-Cho Seto City, Japan), which has been widely used due to its large electromechanical coupling factor (K
2) with low propagation and insertion loss [
33]. Higher center frequency and low insertion loss will lead to a higher sensitivity of the SAW sensor. Aluminium input and output IDT electrodes consisted of 72 finger electrode pairs with a width of 5.0 μm and a center-to-center separation of 10.0 μm. The spacing between delay lines was 100 λ. The area of the SAW sensor was 3.0 mm × 9.0 mm and the aperture of the IDT electrodes was 1.6 mm. This configuration allowed transmission (S
21) and reflection (S
11) measurements. The SAW sensor response was measured in terms of insertion loss of the S
21 transmission parameter. In order to confine the acoustic energy near the surface and protect the IDT electrodes from the test buffer, a 5.2 μm thick SiO
2 guide layer determined through simulation was deposited on the sensor surface [
30]. The fabricated SAW sensor could operate at a canter frequency of approximately 200 MHz. After dicing the fabricated wafer, SAW sensors were mounted in individual packages to form a dual-type SAW sensor, followed by aluminium wire bonding to establish an electric connection.
Figure 1a presents a detailed configuration of the dual-type SAW sensor.
Figure 1b shows frequency response of the SAW sensor. A resonant frequency of 196 MHz was obtained, which was close to the theoretical value of 200 MHz. Then, the 3-GPTES-modified SAW sensor chip was prepared by general silanization protocol described in supplementary materials (
Figure S1).
2.3. Sensing and Fluidic Blocks of the SAW Sensor
In the sensing block, the SAW sensor was in contact with a custom-made oscillator circuitry. The mass loading effects on the SAW sensor were monitored by tracking the frequency of SAWs at the center frequency of 200 MHz with a HP8753ES network analyzer (Hewlett-Packard Company, Palo Alto, CA, USA). The instrument was optimized to measure the S
21 frequency response of the SAW sensor. A temperature controller was installed below the chamber to keep the temperature at 25 °C. The sensing block, where the SAW sensor was to be mounted, possessed a two-part plastic fixture. The bottom piece supported the SAW sensor in a recessed cavity. It was in contact with the oscillator circuitry. The upper piece contained recessed areas for reaction chambers and silicone gaskets to prevent liquid leakage. The sample and the buffer solution that flew to the reaction chamber was actuated with a peristaltic pump (ISM597; ISMATEC, Glattbrugg, Switzerland). Through careful optimization, the flow rate was kept at 1.0 ml/min and the volume of each reaction chamber was 30 μl. Detailed configuration of the fluidic cell is provided in the supplementary materials (
Figure S2).
2.4. Detection of Synthetic miR-106b Using SAW Biosensor
For miR-106b detection using the SAW biosensor, a hairpin loop structure was first formed through the addition of Hg2+ ion (HgCl2, 10 μM) on the working sensing area. After 5 min, various concentrations (0.1 pM to 1.0 μM) of synthetic miR-106b in 1X SSC buffer (pH 7.0) were added to the sensor surface and allowed to stand at 25 °C for 10 min to induce the formation of a hybrid duplex between the miR-106b and the hairpin loop-oligonucleotide probe on the surface. A DNA detection probe conjugated with AuNPs was then introduced to the partially hybridized sensor surface and allowed to stand at 25 °C for 10 min to form a sandwich hybridization. After washing with 1X SSC buffer solution for 1 min, a gold staining solution consisting of gold(III) chloride (10 mM, 50 μL) and hydroxylamine hydrochloride (20 mM, 50 μL) was added to the sensor surface and incubated at 25 °C for 2 min. The staining reaction was then stopped by washing the sensing surface with 1X SSC buffer. Identical experiments were also performed using control miRNAs (miR-21, m-R-124, miR-155). Meanwhile, in the reference sensing area, the same process as in the working sensing area was applied using a reference sensing probe (poly G10) conjugated with AuNPs.
2.5. Preparation of Antibodies-Conjugated Paddle Screw Type Devices for Exosome Isolation
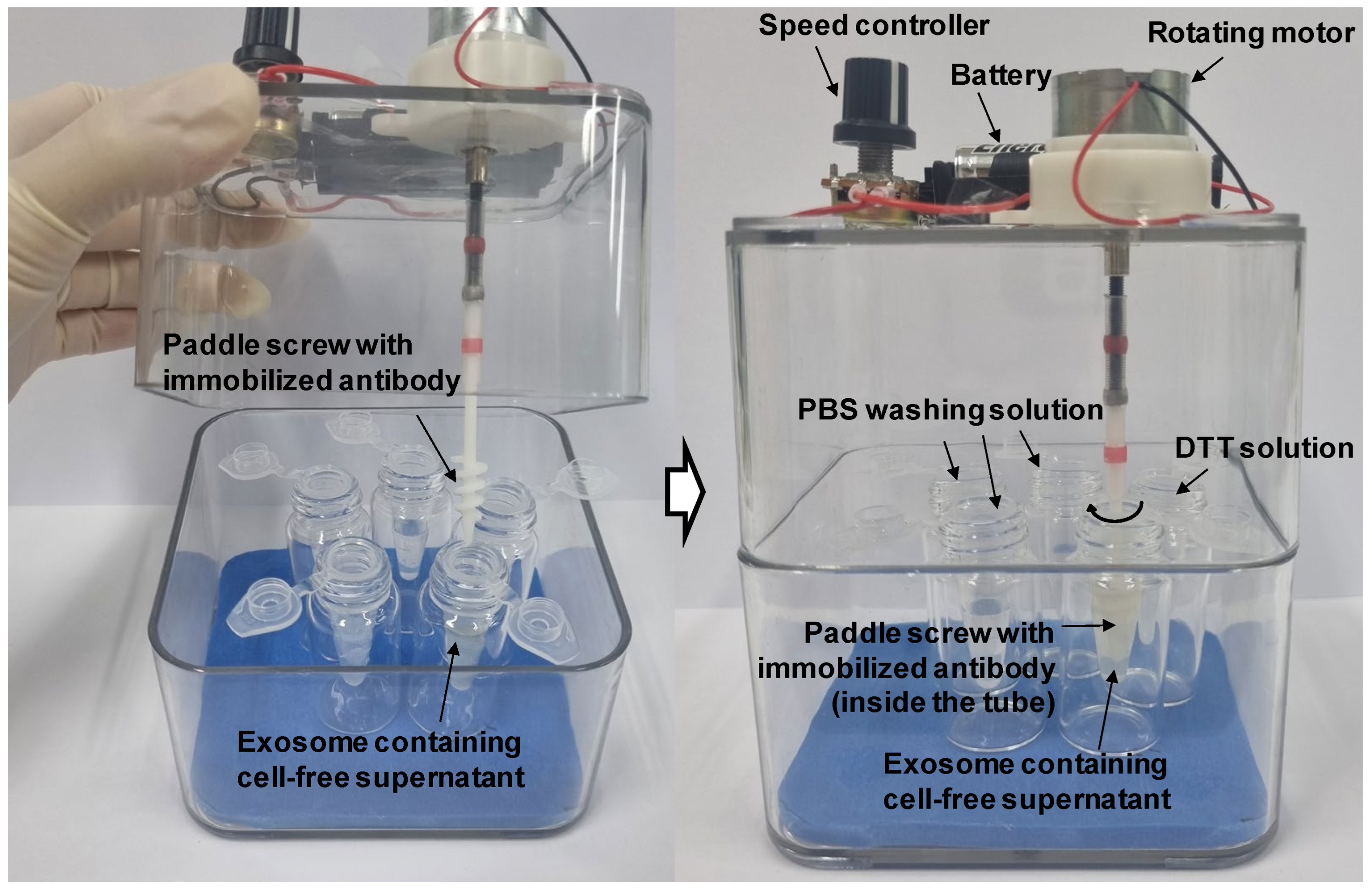
Paddle screws made of acrylonitrile butadiene styrene (ABS) were manufactured using a 3-D printing method. Among various shapes of 3D paddle structures, the paddle screw shown in the red box in
Figure 2 was selected because it was convenient to manufacture by 3-D printing. In addition, it matched well with a 1.5 ml tube. They were washed by sonication in ethanol and double-distilled water (each 3 times × 5 min at room temperature), dried under nitrogen, and stored in desiccator until use. These cleaned paddle screws were activated in a UV-ozone cleaner (144AX-220; Jelight Company, Inc., Irvine, CA, USA) for 10 min. Activated paddle screws were placed in a freshly prepared 3% (vol./vol.) 3-GPTES in ethanol for 1 h, then rindrf with ethanol for 2 min and dried under nitrogen. The silanized paddle screws were kept in an oven at 110°C for 1 h, then rinsed with ethanol for 2 min and dried under nitrogen. For immobilization of antibodies on the surface of paddle screws, 3-GPTES modified paddle screws were soaked in a mixed solution of anti-CD9 and anti-CD81 antibodies (each 100 μg/mL in pH 7.4 phosphate buffered saline (PBS)). These paddle crews were incubated at 4 °C overnight. After incubation, the paddle crews were rinsed and rotate at 500 rpm with PBS and deionized water to remove physically absorbed antibodies. Sequentially, empty sites where antibodies were not immobilized within the sensing area were blocked with 3% BSA in pH 7.4 PBS solution to prevent nonspecific binding of proteins in subsequent steps.
2.6. Isolation and Characterization of Cancer Cell-Derived Exosomes
Isolation of exosomes from an MCF-7 human breast carcinoma cell line was performed via two different immunoaffinity capture methods. The isolation using Dyna Beads
®-CD9 and CD81 (Invitrogen, USA) followed the manufacturer’s instructions. Exosome isolation using paddle screws was accomplished according to the following protocol. After MCF-7 cells were cultured for 24 h, harvested cell culture media were centrifuged at 2,000 x g for 10 min and then at 15,000 x g for 15 min to remove floating cells and cellular debris, followed by syringe filtration through a 0.22 µm membrane (Millipore, Billerica, MA, USA). The cell-free supernatant was then immediately transferred to a 1.5 ml tube. After adding the antibody-modified paddle crew to the tube, the paddle screw was rotated at 200 rpm for 30 min. The paddle crew was then transferred to a new 1.5 ml tube containing PBS buffer and washed by rotating at 500 rpm for 5 minutes. Next, the paddle crew was transferred to a 1.5 ml tube containing a dithiothreitol (DTT) solution (50 mM) and rotated at 200 rpm for 15 minutes to break the disulfide bond, thereby releasing captured exosomes from the paddle screw. All of the above processes using paddle screws were accomplished through a custom-made rotating system as shown in
Figure 3. The exosome-released solution was centrifuged at 10,000 × g for 1 hour at 4°C. After centrifugation, the supernatant was aspirated and discarded. The pellet at the bottom of the tube contained exosomes. The pellet was suspended in 1.0 ml of 1X PBS buffer for downstream analysis. Nanoparticle tracking analysis (NTA) measurements were performed using a NanoSight NS300 (Malvern Pananalytical, Malvern, UK) with specific parameters according to the manufacturer’s user manual. Captures and analysis were achieved using the built-in NanoSight Software NTA3.3.301. Protein concentrations of exosomes were measured using a bicinchoninic acid assay (BCA) kit obtained from Thermo Fisher Scientific. Western blot was carried out to detect expression levels of exosomal surface markers, CD9 and CD81.
2.7. RT-qPCR for Detecting miR-106b Using Total RNA Extracted from MCF-7 Human Breast Carcinoma Cell Line
Total RNA was extracted from exosomes isolated from MCF-7 human breast carcinoma cell line using an RNeasy Mini Kit (Qiagen, Maryland, MD, USA) following the manufacturer’s instructions. Total RNA samples obtained were diluted to various concentrations (0.01 ng/ml to 100 μg/mL). An RT-qPCR assay for detecting miR-106b was conducted for each concentration of the total RNA sample using an HB miR Multi Assay kit™ (HeimBiotek, Pankyo, Korea). First, we performed polyadenylation and reverse transcription (RT) at 37 °C for 30 min using a 20 μL reaction volume including nucleic mix I (20 mM dNTP, ATP, and RT primer), RT buffer, and enzyme mix (0.5 U poly (A) polymerase and 0.5 U reverse transcriptase). The temperature was then raised to 95 °C and held for 5 min to inactivate the enzyme. Next, we measured real-time PCR products using fluorescence intensities of SYBR Green in a 20 µL reaction volume including nucleic mix II (extension sequence and primer mix), 2X SYBR Green Master Mix, and a cDNA template obtained from the first step. PCR cycling conditions were: 95°C for 15 min for initial activation, followed by 40 cycles of 95°C for 10 s and 60°C for 40 s. We conducted reverse transcription and real-time PCR on a T100™ Thermal Cycler and Bio-Rad CFX96™.
3. Results and Discussion
3.1. Sensing Principle of miR-106b Using the SAW Biosensor
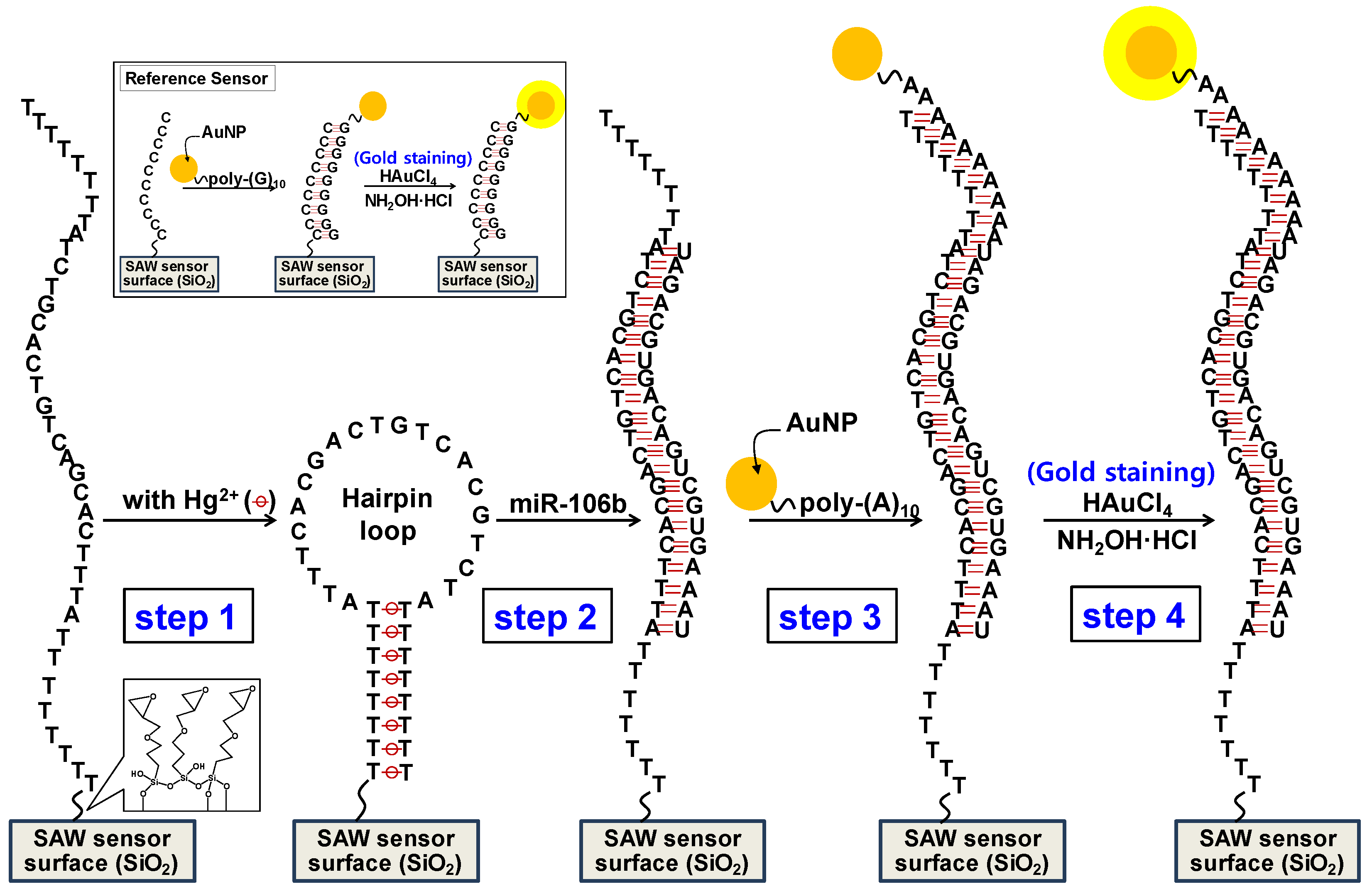
Scheme 1 presents the detection of miR-106b based on sandwich hybridization between a Hg
2+ ion-assisted hairpin loop capture probe and a target miR-106b in addition to a detection probe conjugated with AuNPs and a subsequent gold staining signal amplification on the SAW biosensor. In detail, firstly, a capture probe (5′-H
2N-(CH
2)
6-TTT TTT TTA TTT CAC GAC TGT CAC GTC TAT TTT TTT T-3′) was covalently immobilized to the 3-GPTES-coated SAW sensor surface. Following the addition of Hg
2+ ions, consecutive thymine bases at the 3′- and 5′-ends formed a T-Hg
2+-T pair, resulting in a stable hairpin-loop structure on the sensor surface (step 1). The hairpin loop formation was almost completely achieved with Hg
2+ concentration at 10 μM (
Figure S3). In this study, using a hairpin loop by a T-Hg
2+-T pair as a capture probe has the following advantages: (1) it is suitable for designing oligonucleotide probes immobilized on the sensor surface for nucleic acid detection (that is, the introduction of multiple thymines at both 3′- and 5′-ends can be free from overlap with the entire miRNA sequence and can thus be applied to all desired miRNA detection depending on sequence changes other than multiple thymines); (2) the simultaneous detection of multiple nucleic acids using an array sensor by sandwich hybridization requires only the design of a universal detecting oligonucleotide probe (poly A
10-(CH
2)
6-S-AuNP in this assay); and (3) the formation and destruction of hairpin loops can be easily manipulated. Hairpin loops are easily formed by the addition of Hg
2+ ions. Hg
2+ ions can be removed by treatment with mercury scavengers such as thiol compounds. Thus, hairpin loops can be easily destroyed.
Secondly, the addition of target miR-106b can destroy the hairpin structure, resulting in partial hybridization between the capture probe and miR-106b (step 2). Sandwich hybridization then occurred between the target miR-106b and complementary two probes after introducing the oligonucleotides detecting probe with the attached AuNPs (5′-AAA AAA AAA A-(CH
2)
6-S-AuNP) (step 3). Finally, size enlargement of the AuNPs by the gold staining process through reagents consisting of gold (III) chloride as an Au
3+ source and hydroxylamine hydrochloride as a reducing agent caused a large amplification of the signal intensity (step 4).
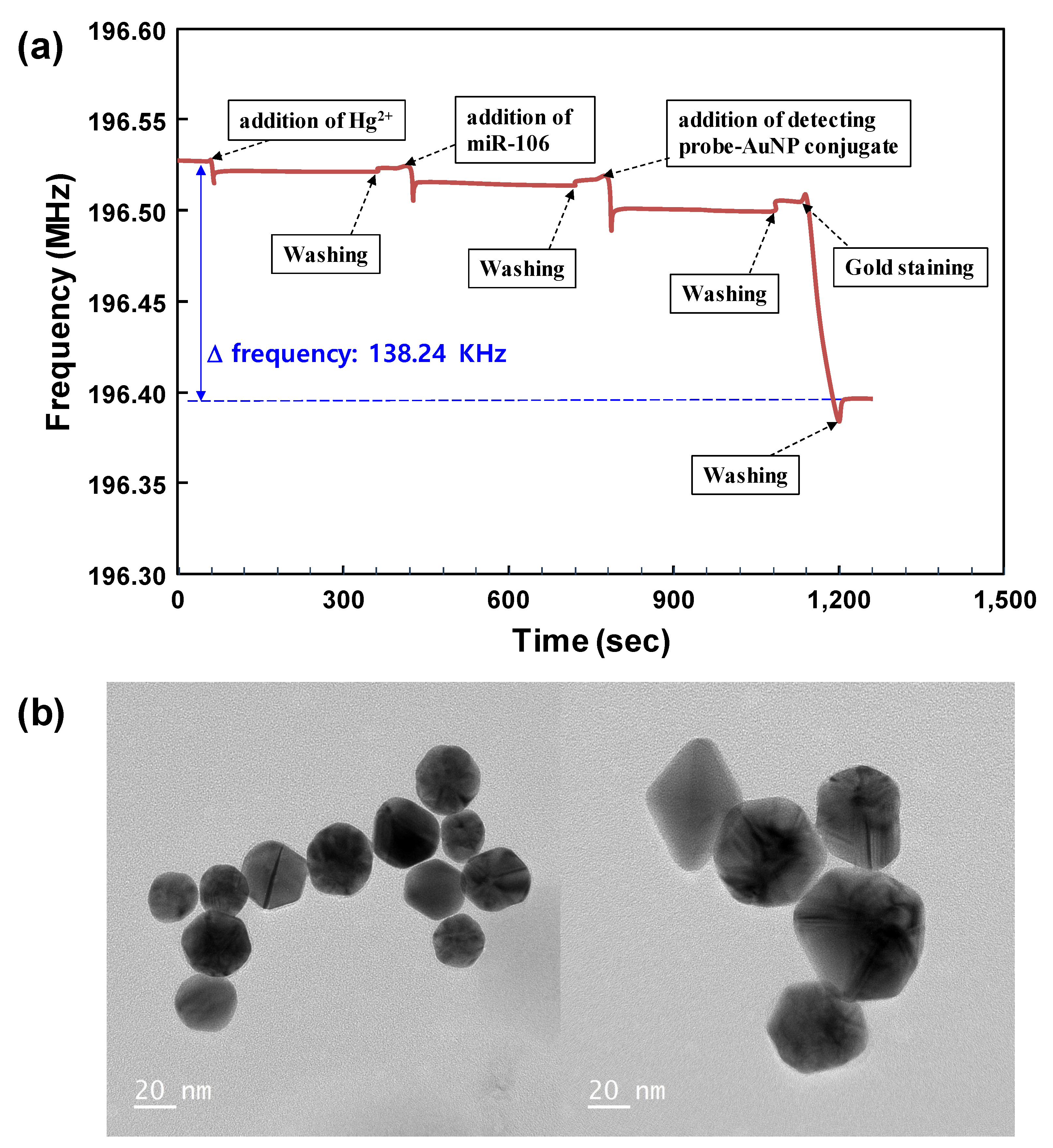
Figure 4a shows real-time response results of hairpin loop formation, hybridization, and signal amplification processes to detect a 1.0 nM concentration of synthetic miR-106b on the SAW biosensor surface. It was clear that gold staining was the main cause of the decrease in SAW resonance frequency. Therefore, most of the total changes in SAW resonance frequency were generated by this signal amplification process. In this regard, we performed transmission electron microscopy (TEM) analysis to confirm the size enhancement of AuNPs by the gold staining reaction.
Figure 4b show typical TEM images of bare AuNPs and size-enhanced AuNPs, respectively. Results of the gold staining reaction on AuNPs showed a thick deposition of metallic gold on the underlying AuNPs during the staining process. On the other hand, in the reference SAW biosensor, a capture probe [5′-NH
2-(CH
2)
6-CCC CCC CCC C-3′] was immobilized and a detecting probe conjugated with AuNPs [5′-GGG GGG GGG G-(CH
2)
6-S-AuNP] was hybridized to the capture probe. A subsequent gold staining reaction occurred. Hybridization of poly C
10 and poly G
10 occurred perfectly in the reference sensor. Since the reference sensor was adjacent to working sensors, side reactions such as non-specific adsorption that occurred in working sensors also occurred in the reference sensor at the same level.
3.2. Sensitivity and Selectivity of the SAW Biosensor for miR-106b Detection
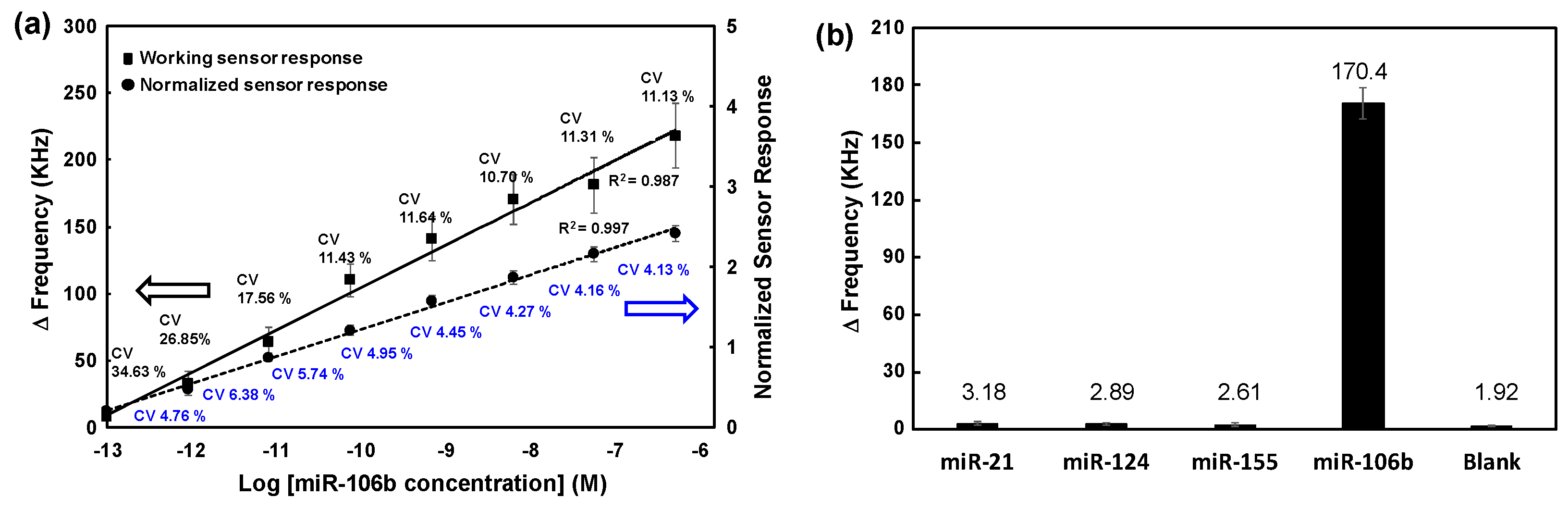
A series of ten-fold serially diluted solutions at concentrations ranging from 0.1 pM to 1.0 μM of the synthetic miR-106b were prepared and effects of concentration on SAW biosensor responses were analyzed based on previously mentioned process and detection mechanism. As expected, as the concentration of added miR-106b increased, the change in resonance frequency of the SAW biosensor also increased logarithmically due to an increase in the effective mass. As the concentration of miR-106b increased, more sandwich hybridization occurred, which increased the number of AuNPs on the sensor surface. Thus, changes in SAW resonance frequency increased due to a mass loading effect of AuNPs and their dramatic size enlargement. The above experiments were performed in quadruplicate for each concentration of synthetic miR-106b. Average results are shown as solid lines in
Figure 5a. Blank subtraction was performed for all delta frequency values for each concentration of miR-106b. Here, the blank refers to the result of all subsequent processes being performed without adding the target miR-106b to the hairpin loop formed by adding Hg
2+ to the capture probe. The delta (Δ) frequency value of blank was 2.42 ± 0.15 KHz. The limit of detection (LOD) of miR-106b was calculated to be 0.084 pM according to three times the standard deviation of the blank sample, a commonly accepted criterion. When the x-axis was in a log scale, the correlation coefficient was 0.987 over the entire range, indicating a good linearity.
In addition to sensitivity, one of the important factors in biosensor performance is target selectivity. In order to verify the selectivity of target miRNA, we applied a target miR-106b and three control non-complementary miRNAs (miR-21, miR-124, miR-155). Measurement results are shown in
Figure 5b. Target miR-106b showed a significantly larger signal than results for three non-complementary targets (e.g., 53.0-fold greater signal than that of miR-155). When a mixture of the four miRNAs was applied, the change in resonance frequency was similar to that caused by miR-106b, indicating that no significant interference occurred between them.
Meanwhile, although blank subtraction was performed to reduce deviation from different measurements, the signal deviation at each concentration of miR-106 was somewhat large as shown in solid line in
Figure 5a. Thus, the dependence of normalized sensor responses due to sandwich hybridization and photocatalytic silver staining on the applied concentration of miR-106b was investigated. Normalized results of four measurements for each concentration of miR-106b are shown as dashed line in
Figure 5a. We compared coefficient of variation (CV) of normalized sensor responses with those of working sensor responses. CVs of normalized sensor responses were in the range of 4.1% to 6.4%, lower than those of working sensor responses in the range of 10.7% to 34.6%. In this assay, high standard deviations or CV of working sensor responses mainly resulted from various environmental factors such as temperature, pressure, and viscosity as well as non-specific adsorption and non-linear growth of metallic gold on AuNPs due to gold staining. Introducing an internal reference sensor adjacent to the working sensor reduced the CV of the assay, which improved the reproducibility of the SAW biosensor. This demonstrated that normalization could screen out background noise by manipulating data and minimize non-uniformity in the gold staining process by suppressing disturbances to signals of both working and reference sensors. As an effect of increasing sensitivity as much as reducing reproducibility of CV, the LOD of miR-106b detection using normalized signals was improved to 0.0034 pM. These results show that the LOD and linearity of the SAW biosensor for detecting miR-106b in this study are comparable to those of recently reported miRNA biosensors (
Table 2) [
18,
19,
20,
21,
22,
23,
24,
25,
26].
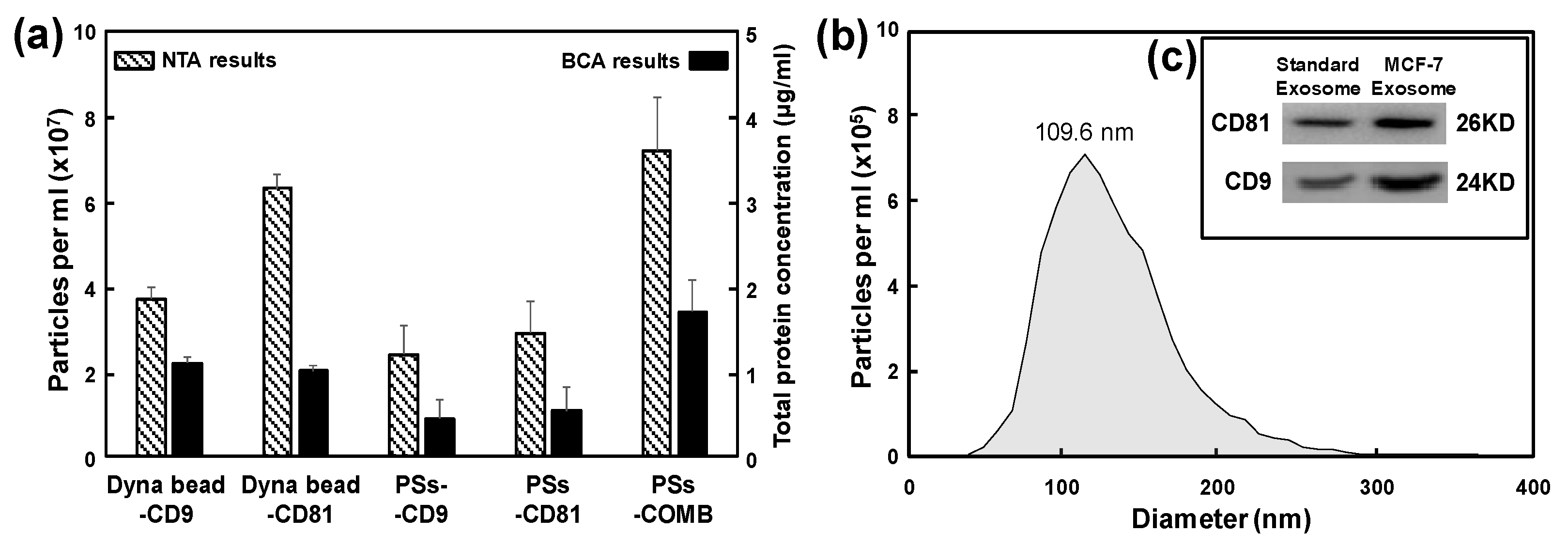
3.3. Exosome Isolation from MCF-7 Cell Line and Its Characterization
Here, two immunoaffinity-based approaches, commercially available magnetic beads conjugated with anti-CD9 and anti-CD81 antibodies (Dyna beads
®-CD9 and Dyna beads
®-CD81) and paddle screws conjugated with anti-CD9 and anti-CD81 antibody using a combination of two antibodies (PSs-CD9, PSs-CD81 and PSs-COMB), for isolating exosomes from cell-free supernatants obtained by pre-enrichment of cultured MCF-7 cell lines were compared. The MCF-7 cell line is routinely used as a model system for human breast cancer research. Additionally, upregulation of miR-106b is known to promote the proliferative capacity of MCF-7 cells in vivo [
34]. Pre-enrichment was done by ultracentrifugation. Exosomes isolated by the two different immunoaffinity-based methods were validated by nanoparticle tracking analysis (NTA). Total protein concentration was determined using bicinchoninic acid (BCA) assay (
Figure 6a). For the MCF-7 cell line, the PSs-COMB resulted in a relatively high yield (the exosome concentration was about 7.2 × 10
7 particles/mL) and purity of isolated exosomes. PSs-CD9 and PSs-CD81 using single CD9 or CD81 antibody showed lower yields than those using commercially available Dyna beads
® for exosome isolation. However, PSs-COMB, which combined two CD9 and CD81 antibodies at a 1:1 ratio, had a higher isolation yield than using a single antibody. Its yield was similar to the commercial method. It has been demonstrated that using combination of two types of exosome-specific antibodies rather than a single antibody is a more sensitive and specific strategy for exosome isolation and protein analysis. Results of this study showed that the paddle screw-based exosome isolation approach could offer an accessible, flexible and efficient method, with a recovery time of less than 1 hr without including a pre-enrichment step. In particular, it has an exosome isolation efficiency that is comparable to that of the commercially optimized Dyna-beads method (incubation time of 16-20 hr at 4 °C). It also shows a fast analysis time. The stirring process by active rotation of the paddle screw plays a crucial role. For example, the difference in mixing effect between two liquids resulting from simple diffusion and active stirring (200 rpm) based on computational fluid dynamics (CFD) simulations was about 13 times (
Figure S4). Meanwhile, the average diameter of isolated esosomes by PSs-COMB method was about 109.6 nm, as detected by the NTA characterization (
Figure 6b). Additionally, these isolated exosomes were validated through quantitative analysis of CD9 and CD81 as common exosome biomarkers using western blot. Western blot showed that CD9 and CD81 were expressed on MCF-7-derived exosomes (
Figure 6c), and these results indicated that these isolated vesicles with the paddle crews contained exosomal proteins CD9 and CD81. Thus, they were considered as genuine exosomes.
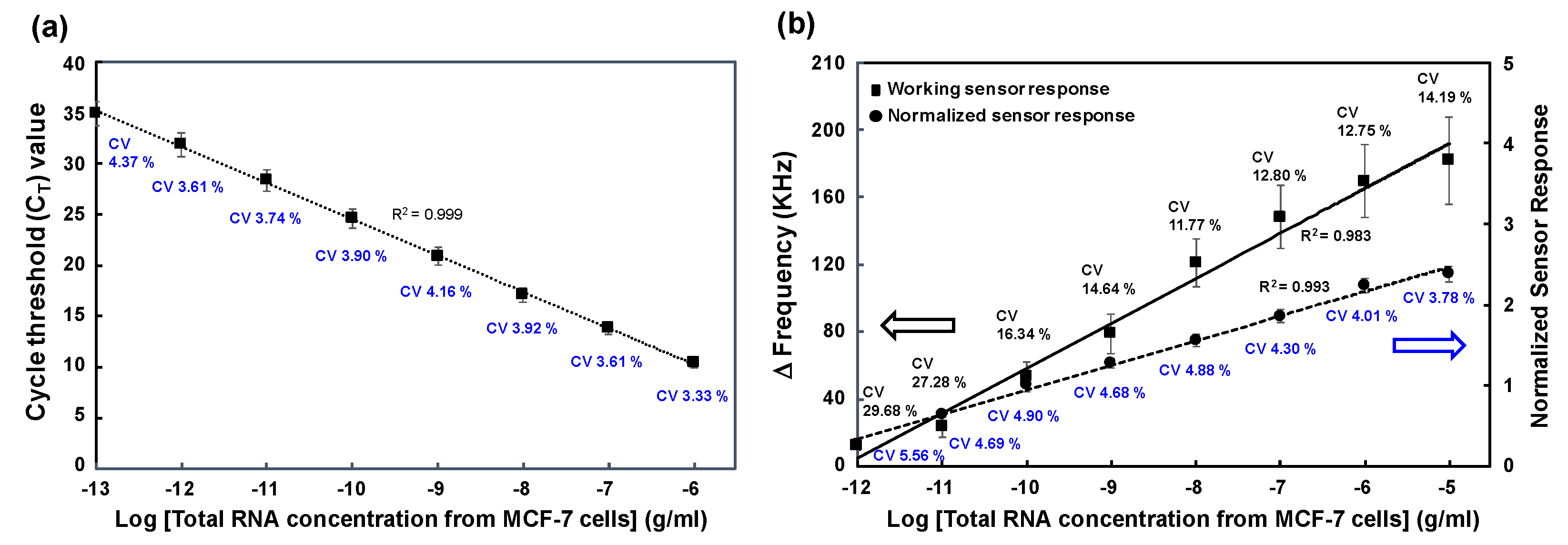
3.4. Comparison of miR-106b Detection in MCF-7 Cell-Derived RNA Samples Using RT-qPCR Assay and SAW Biosensor
To confirm the performance of the SAW biosensor for analyzing total RNA samples derived from cancer cells, we performed a miR-106b detection assay using total RNA extracted from exosomes isolated from the MCF-7 cell line with the PSs-COMB method. Total RNA extraction from isolated exosomes was carried out using the RNeasy Mini Kit (Qiagen). Extracted samples were serially diluted 10-fold to obtain various concentrations (0.1 pg/ml to 10ug/ml). Firstly, we conducted RT-qPCR analysis using the HB miR Multi Assay kit™ for the detection of miR-106b in total RNA extracted from isolated exosomes. Plotting logarithmic units of total RNA concentration versus cycle threshold (C
T) values showed an excellent linearity (correlation coefficient of 0.999) and sensitivity with LOD of 0.028 pg/ml of total RNA (
Figure 7a).
When the same samples used for RT-qPCR was applied to the SAW biosensor, normalized sensor responses are shown in
Figure 7b). SAW biosensor results also showed a good linearity (correlation coefficient of 0.993). The LOD was 0.064 pg/ml for total RNA extracted from isolated exosomes, which was 2.3 times higher than RT-qPCR results. Meanwhile, this result also showed a good sensitivity, about 1,000 times better than the experiment using commercial human breast reference total RNA (LOD of 62 pg/ml in
Figure S5). Generally, the concentration of miR-106b in total RNA extracted from cancer cells is known to be 100 to 1,000 times higher than the concentration of total RNA extracted from healthy people [
35]. Therefore, the amount of miR-106b contained in total RNA extracted from the MCF-7 cell line was higher than the amount of miR-106b present in the same concentration of reference human breast total RNA from healthy people. Therefore, the designed miRNA detection mechanism of the SAW biosensor worked sufficiently using cancer cell-derived RNA samples.
4. Conclusions
In this study, we successfully detected miR-106b with high sensitivity and specificity by applying a hybrid duplex formation of a target miRNA and two probe oligonucleotides with subsequent size enlargement of gold nanoparticles on a SAW biosensor. Through this process, the LOD for synthetic miR-106b was confirmed to be as low as 0.0034 pM when normalization by the reference sensor was applied. The SAW biosensor was highly selective for miR-106b without being significantly affected by other miRNAs. Additionally, this SAW biosensor could detect miR-106b in total RNA extracted from the MCF-7 cancer cell line using our unique paddle screw type devices, showing performance comparable to that of RT-qPCR analysis. This shows its potential to be used as a promising alternative to detect miR-106b in real samples for clinical applications. It is believed that this SAW-based miRNA biosensor is suitable for biomedical research, early detection of malignancies, and evaluation of the effectiveness of cancer treatment. Future research should focus on developing a SAW biosensor array device that can simultaneously detect multiple types of miRNAs and improve its detection sensitivity through methods such as increasing the efficiency of sensor surface modification.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: Optimization of the concentration of the immobilized capture probe; Figure S2: Sensing and fluidic blocks of the SAW sensor; Figure S3: Optimization of the concentration of added Hg2+ ions for the hairpin loop formation; Figure S4: Computational fluid dynamics (CFD) simulations of the mixing effect of two liquids; Figure S5: Detection of miR-106b in human breast total RNA
Author Contributions
Conceptualization, S.L.; methodology, S.H.; software, S.H.; validation, S.L., S.H.; formal analysis, S.H.; investigation, S.L.; resources, S.L.; data curation, S.H.; writing—original draft preparation, S.H.; writing—review and editing, S.L.; visualization, S.H.; supervision, S.L.; project administration, S.L.; funding acquisition, S.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF-2022R1F1A1067428).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Acknowledgments
This work was supported by Soonchunhyang University. This work was also the results of the Leaders in Industry-university Cooperation 3.0 Project, supported by the Ministry of Education and National Research Foundation of Korea (No. 1345356224).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P.K. Extracellular vesicles: Composition, biological relevance, and methods of study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef] [PubMed]
- Dilsiz, N. Role of exosomes and exosomal microRNAs in cancer. Future Sci. OA 2020, 6, FSO465. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Crow, J.; Roth, M.; Zeng, Y.; Godwin, A.K. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip 2014, 14, 3773–3780. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Shao, H.; Park, Y.I.; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 32, 490–495. [Google Scholar] [CrossRef]
- Ueda, K.; Ishikawa, N.; Tatsuguchi, A.; Saichi, N.; Fujii, R.; Nakagawa, H. Antibody-coupled monolithic silica microtips for highthroughput molecular profiling of circulating exosomes. Sci. Rep. 2014, 4, 6232. [Google Scholar] [CrossRef]
- Baranyai, T.; Herczeg, K.; Onódi, Z.; Voszka, I.; Módos, K.; Marton, N.; Nagy, G.; Mäger, I.; Wood, M.; Andaloussi, S.; Pálinkás, Z.; Kumar, V.; Nagy, P.; Giricz, Z. Isolation of exosomes from blood plasma: qualitative and quantitative comparison of ultracentrifugation and size exclusion chromatography methods. PLoS ONE 2015, 10, e0145686. [Google Scholar] [CrossRef]
- Livshits, M.; Khomyakova, E.; Evtushenko, E.; Lazarev, V.; Kulemin, N.; Semina, S.; Generozov, E.; Govorun, V. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 2015, 5, 17319. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Choi, M.; Lee, H.; Kim, Y.H.; Han, J.Y.; Lee, E.S.; Cho, Y. Direct isolation and characterization of circulating exosomes from biological samples using magnetic nanowires. J. Nanobiotechnology 2019, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Qasim, M.; Kim, J.-H. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells 2019, 8, 307. [Google Scholar] [CrossRef]
- Ciesla, M.; Skrzypek, K.; Kozakowska, M.; Loboda, A.; Jozkowicz, A.; Dulak, J. MicroRNAs as biomarkers of disease onset. Anal. Bioanal. Chem. 2011, 401, 2051–2061. [Google Scholar] [CrossRef]
- Rabinowits, G.; Gercel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Calin, G.A.; Liu, C.-G.; Ambs, S.; Cimmino, A.; Petrocca, F.; Visone, R.; Iorio, M.; Roldo, C.; Ferracin, M. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. 2006, 103, 2257–2261. [Google Scholar] [CrossRef]
- Speranza, M.C.; Frattini, V.; Pisati, F.; Kapetis, D.; Porrati, P. NEDD9, a novel target of miR-145, increases the invasiveness of glioblastoma. Oncotarget 2012, 3, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Sagar, S.K. miR-106b as an emerging therapeutic target in cancer. Genes & Diseases 2022, 9, 889–899. [Google Scholar]
- Yang, F.; Sun, Z.; Wang, D.; Du, T. MiR-106b-5p regulates esophageal squamous cell carcinoma progression by binding to HPGD. BMC Cancer 2022, 22, 308. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.J.; Kwak, J.; Lee, J.-H.; Lee, S.S. Real-time qRT-PCR assay for the detection of miRNAs using bi-directional extension sequences. Anal. Biochem. 2017, 536, 32–35. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.H.; Kwak, J.; Lee, J.-H.; Lee, S.S. Efficient and accurate analysis of microRNA using a specific extension sequence. Mol. Biol. Rep. 2018, 45, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, W.; Liu, Z.; Zhang, G.; Chen, Z.; Guo, L.; Lin, Z.; Qiu, B.; Chen, G. Enzyme-free fluorescent biosensor for miRNA-21 detection based on MnO2 nanosheets and catalytic hairpin assembly amplification. Anal. Methods 2016, 8, 8492–8497. [Google Scholar] [CrossRef]
- Hong, M.; Sun, H.; Xu, L.; Yue, Q.; Shen, G.; Li, M.; Tang, B.; Li, C.-Z. In situ monitoring of cytoplasmic precursor and mature microRNA using gold nanoparticle and graphene oxide composite probes. Anal. Chim. Acta 2018, 1021, 129–139. [Google Scholar] [CrossRef]
- Ou, S.; Xu, T.; Liu, X.; Yu, X.; Li, R.; Deng, J.; Yuan, J.; Chen, Y. Rapid and ultrasensitive detection of microRNA based on strand displacement amplification-mediated entropy-driven circuit reaction. Sens. Actuators B Chem. 2018, 255, 3057–3063. [Google Scholar] [CrossRef]
- Zouari, M.; Campuzano, S.; Pingarrón, J.M.; Raouafi, N. Amperometric biosensing of miRNA-21 in serum and cancer cells at nanostructured platforms using anti-DNA–RNA hybrid antibodies. ACS Omega 2018, 3, 8923–8931. [Google Scholar] [CrossRef]
- Koo, K.M.; Carrascosa, L.G.; Shiddiky, M.J.A.; Trau, M. Poly (A) extensions of miRNAs for amplification-free electrochemical detection on screen-printed gold electrodes. Anal. Chem. 2016, 88, 2000–2005. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, A.; Cheema, J.A.; Rajwar, D.; Ammanath, G.; Xiaohu, L.; Koon, L.S.; Yi, W.; Yildiz, U.H.; Liedberg, B. Polythiophene derivative on quartz resonators for miRNA capture and assay. Analyst 2015, 140, 7912–7917. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Lee, S.S. QCM sensing of miR-21 by formation of microRNA–DNA hybrid duplexes and intercalation on surface-functionalized pyrene. Analyst 2019, 144, 6936–6943. [Google Scholar] [CrossRef] [PubMed]
- Premaratne, G.; Mubaraka, Z.H.A.; Senavirathna, L.; Liu, L.; Krishnan, S. Measuring ultra-low levels of nucleotide biomarkers using quartz crystal microbalance and SPR microarray imaging methods: A comparative analysis. Sens. Actuators B Chem. 2017, 253, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Guven, B.; Dudak, F.C.; Boyaci, I.H.; Tamer, U.; Ozsoz, M. SERS-based direct and sandwich assay methods for mir-21 detection. Analyst 2014, 139, 1141–1147. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Das, P.K.; Bhethanabotla, V.R. Surface acoustic waves in biosensing applications. Sens. Actuators Rep. 2021, 3, 100041. [Google Scholar] [CrossRef]
- Han, S.B.; Lee, S.S. Simultaneous Detection of Exosomal microRNAs Isolated from Cancer Cells Using Surface Acoustic Wave Sensor Array with High Sensitivity and Reproducibility. Micromachines 2024, 15, 15–249. [Google Scholar] [CrossRef]
- Länge, K.; Rapp, B.E.; Rapp, M. Surface acoustic wave biosensors: a review. Anal. Bioanal. Chem. 2008, 391, 1509–1519. [Google Scholar] [CrossRef]
- Lee, H.J.; Namkoong, K.; Cho, E.C.; Ko, C.; Park, J.C.; Lee, S.S. Surface acoustic wave immunosensor for real-time detection of hepatitis B surface antibodies in whole blood samples. Biosens. Bioelectron. 2009, 24, 3120–3125. [Google Scholar] [CrossRef]
- Choi, Y.-S.; Lee, J.; Lee, Y.; Kwak, J.; Lee, S.S. Increase in detection sensitivity of surface acoustic wave biosensor using triple transit echo wave. Appl. Phys. Lett. 2018, 113, 083702. [Google Scholar] [CrossRef]
- Cheng, C.-H.; Yatsuda, H.; Goto, M.; Kondoh, J.; Liu, S.-H.; Wang, R.Y.L. Application of Shear Horizontal Surface Acoustic Wave (SH-SAW) Immunosensor in Point-of-Care Diagnosis. Biosensors 2023, 13, 605. [Google Scholar] [CrossRef]
- Branch, D.W.; Brozik, S.M. Low-level detection of a Bacillus anthracis simulant using Love-wave biosensors on 36° YX LiTaO3. Biosens. Bioelectron. 2004, 19, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Miao, Y.; Shan, Y.; Liu, B.; Li, Y.; Zhao, L.; Jia, L. MiR-106b and miR-93 regulate cell progression by suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell Death Dis. 2017, 8, e2796. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chaerkady, R.; Beer, M. A.; Mendell, J. T.; Pandey, A. Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics 2009, 9, 1374–1384. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).