1. Introduction
Metabolic engineering in plants represents a powerful approach to address global challenges in agriculture, including food security, climate change adaptation, and sustainable production of valuable compounds [
1]. The world faces an unprecedented challenge of feeding a growing population, projected to reach 9.7 billion by 2050, while simultaneously mitigating the impacts of climate change on agriculture [
2]. Metabolic engineering offers a promising solution by enabling targeted modifications of plant metabolism to enhance crop performance and resilience. By manipulating metabolic pathways, researchers can enhance crop yield, improve nutritional quality, increase stress tolerance, and produce novel metabolites with industrial or pharmaceutical applications [
3].
The advent of high-throughput sequencing technologies has provided unprecedented insights into plant genomes and metabolic networks. Synthetic biology approaches allow for the design and implementation of novel metabolic pathways, while precise gene editing tools like CRISPR-Cas9 enable targeted modifications of existing pathways with minimal unintended effects [
4]. These advancements have the potential to revolutionize agriculture, creating crops that are not only more productive but also more nutritious and better adapted to changing environmental conditions. Researchers can now fine-tune metabolic fluxes, redirect carbon flow to desired products, and even introduce entirely new biosynthetic capabilities into plants [
5]. This level of control over plant metabolism was unimaginable just a few decades ago and holds immense promise for addressing agricultural challenges. For instance, engineering efforts have successfully increased the vitamin content of staple crops, improved drought tolerance in cereals, and enabled plants to produce valuable pharmaceutical compounds [
6].
The field of plant metabolic engineering has rapidly advanced in recent years, driven by developments in genomics, synthetic biology, and gene editing technologies [
7]. These advancements have enabled more precise and efficient manipulation of plant metabolism, opening new avenues for crop improvement and biotechnology applications [
8]. This review aims to provide a comprehensive overview of current strategies and applications in plant metabolic engineering, highlighting recent successes and emerging trends. We will discuss the principles underlying metabolic pathway manipulation, the tools and techniques used for genetic modification, and the integration of systems biology approaches in metabolic engineering design [
9]. Additionally, we will explore the potential of metabolic engineering to address future agricultural challenges and contribute to sustainable food production [
10]. By providing a comprehensive overview of the current state and future directions of plant metabolic engineering, this study aims to illuminate the transformative potential of this field in shaping the future of agriculture and contributing to global food security and sustainability.
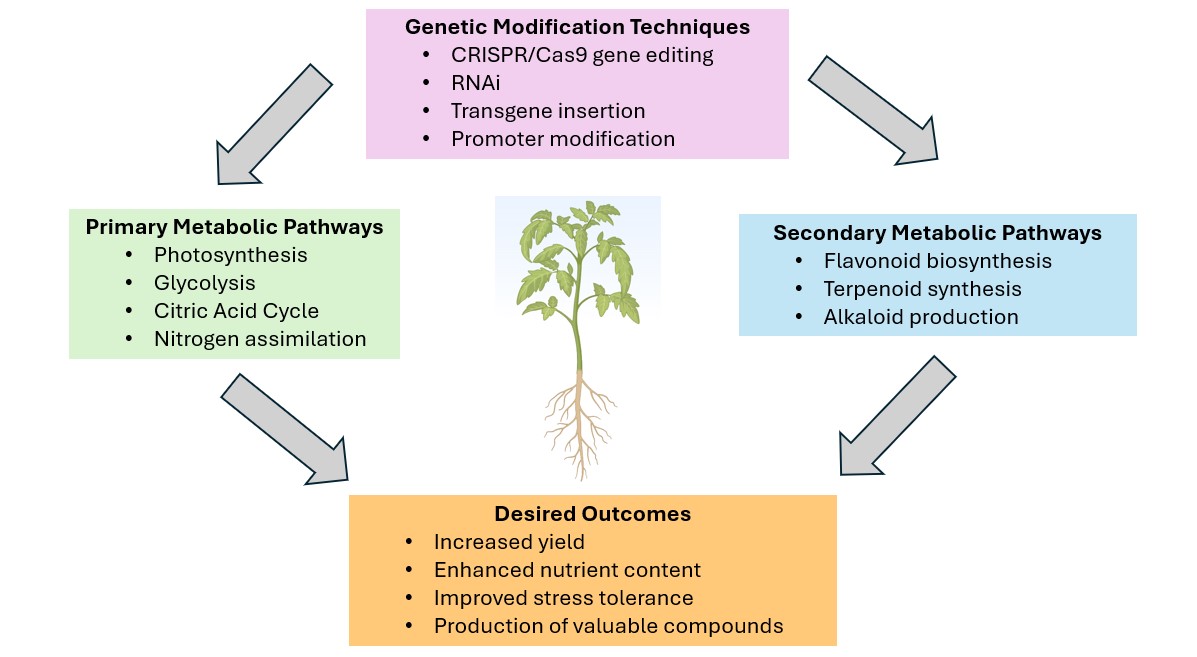
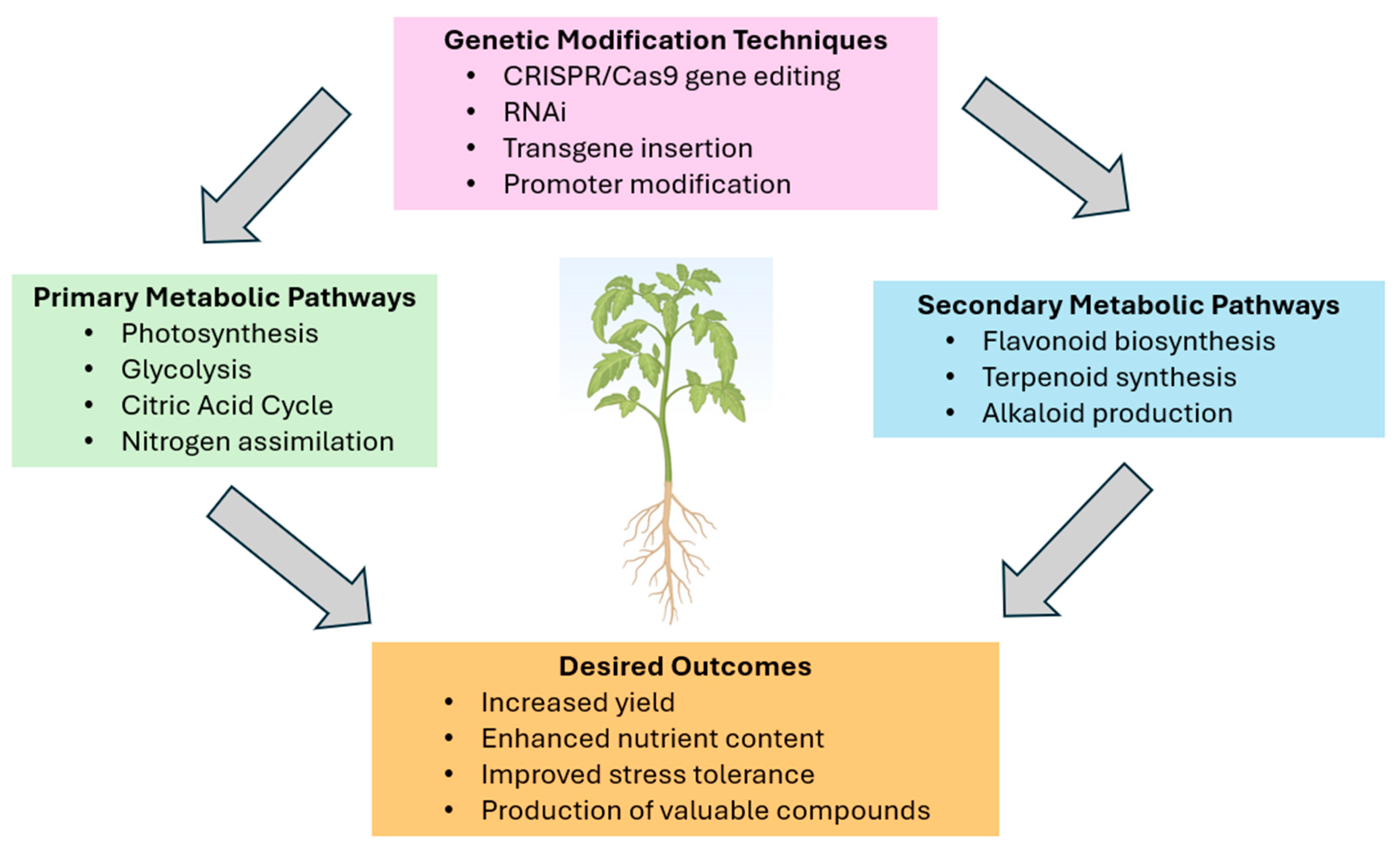
Figure 1.
Schematic overview of plant metabolic engineering.
Figure 1.
Schematic overview of plant metabolic engineering.
The diagram illustrates key components of metabolic engineering in plants. Central plant image represents the target organism. Primary metabolic pathways: photosynthesis, glycolysis, citric acid cycle, and nitrogen assimilation. Secondary metabolic pathways: flavonoid biosynthesis, terpenoid synthesis, and alkaloid production. Genetic modification techniques: CRISPR/Cas9 gene editing, RNAi, transgene insertion, and promoter modification. Desired outcomes: increased yield, enhanced nutrient content, improved stress tolerance, and production of valuable compounds. Arrows indicate how genetic modifications influence pathways to achieve specific outcomes. This diagram demonstrates the integrated approach of metabolic engineering in modifying plant metabolism for improved agronomic traits and novel product synthesis.
2. Principles of Metabolic Engineering in Plants
2.1. Overview of Plant Metabolic Pathways
Plant metabolism encompasses a complex network of biochemical reactions that are essential for growth, development, and adaptation to environmental conditions [
11]. Primary metabolic pathways, such as photosynthesis, respiration, and nitrogen assimilation, provide the basic building blocks and energy for cellular processes. Secondary metabolic pathways produce specialized compounds that play roles in plant defense, signaling, and adaptation [
12]. Understanding the regulation and flux through these pathways is crucial for successful metabolic engineering. Recent advances in metabolomics and flux analysis techniques have provided unprecedented insights into the dynamics of plant metabolism [
13,
14].
2.2. Strategies for Manipulating Metabolic Pathways
Metabolic engineering strategies in plants involve sophisticated approaches to modify the expression and activity of key enzymes or regulatory proteins in target pathways [
15]. These strategies aim to optimize metabolic flux, enhance desired metabolite production, or introduce novel metabolic capabilities. Gene overexpression is a common approach, increasing the activity of rate-limiting enzymes to enhance metabolic flux. This strategy has been successfully employed to boost the production of various metabolites, including vitamins, antioxidants, and essential amino acids [
16,
17]. Conversely, gene silencing or knockout techniques are used to reduce competing pathways or undesirable metabolites, with RNA interference (RNAi) and CRISPR/Cas9-mediated gene editing being widely adopted for this purpose [
18]. The introduction of heterologous genes has enabled the incorporation of new metabolic capabilities from other organisms, allowing for the production of novel compounds in plants, such as biodegradable plastics and pharmaceutical proteins [
19,
20].
Modifying regulatory elements, such as transcription factors, promoters, or regulatory RNAs, allows for fine-tuning of metabolic pathways by altering the expression patterns or responsiveness of metabolic genes [
21]. Enzyme engineering, which involves modifying the catalytic properties or substrate specificity of key enzymes, has proven effective in improving pathway efficiency or introducing new functionalities [
22]. Additionally, metabolic channeling through the creation of synthetic enzyme complexes or scaffolds has been explored to improve the efficiency of multi-step reactions [
23]. Recent advances in synthetic biology and genome editing have significantly expanded the toolkit for metabolic engineering, allowing for more precise and complex pathway modifications [
24]. These diverse strategies, often used in combination, provide researchers with a powerful set of tools to manipulate plant metabolism for desired outcomes.
2.3. Tools and Techniques for Genetic Modification
The arsenal of tools and techniques for plant genetic modification has grown significantly, enabling more efficient and precise manipulations. Agrobacterium-mediated transformation remains a cornerstone technique, widely used for stable integration of transgenes. Recent improvements have expanded its host range and transformation efficiency [
25]. For species recalcitrant to Agrobacterium transformation, particle bombardment continues to be a valuable alternative, with advances in particle coating and delivery systems improving transformation success rates [
26]. The CRISPR/Cas9 gene editing system has revolutionized plant biotechnology, enabling precise modifications to endogenous genes. Various Cas9 orthologs and engineered variants have been developed to expand editing capabilities [
27]. Building on this, base editing and prime editing technologies allow for precise nucleotide changes without double-strand breaks, reducing the risk of unintended mutations [
28].
RNA interference (RNAi) remains a powerful tool for targeted gene silencing, with improved vector designs and delivery methods enhancing its efficiency and specificity [
29]. Virus-induced gene silencing (VIGS) offers a rapid method for transient gene silencing, particularly useful for functional genomics studies [
30]. Emerging technologies such as optogenetic and chemogenetic tools are enabling spatiotemporal control of gene expression and protein activity in plants, offering new possibilities for metabolic regulation [
31]. Nanomaterial-mediated transformation, utilizing nanoparticles for DNA delivery, shows promise in improving transformation efficiency and tissue specificity [
32]. The integration of these tools with high-throughput phenotyping and genome-wide association studies is accelerating the pace of metabolic engineering in plants [
33]. This expanding toolkit provides researchers with unprecedented precision and flexibility in manipulating plant genomes and metabolism, driving rapid advancements in crop improvement and biotechnology.
3. Applications in Crop Improvement
3.1. Enhancing Photosynthetic Efficiency
Improving photosynthetic efficiency is a major target for increasing crop yield potential, as even small improvements in this fundamental process can lead to significant gains in productivity. Strategies to enhance photosynthesis through metabolic engineering are diverse and multifaceted. One primary approach focuses on optimizing Rubisco kinetics and regeneration. Rubisco, the key enzyme in carbon fixation, is notoriously inefficient due to its oxygenase activity and slow catalytic rate. Researchers have explored various strategies to improve Rubisco performance, including engineering its catalytic subunits, optimizing its assembly, and enhancing its activation state [
34]. Recent work has also focused on improving the regeneration of RuBP, the substrate for Rubisco, by manipulating enzymes in the Calvin-Benson cycle.
Another promising avenue is the enhancement of carbon concentrating mechanisms. While some plants, such as C4 and CAM species, have evolved natural carbon concentrating mechanisms, efforts are underway to introduce these features into C3 crops. This includes attempts to engineer C4 photosynthesis into rice and other C3 plants, as well as developing synthetic carbon concentrating mechanisms inspired by cyanobacterial systems [
35]. Improving light harvesting and energy transfer is another key area of research. This involves optimizing the light-harvesting complexes, enhancing electron transport chains, and improving the distribution of light within the canopy. Strategies such as altering chlorophyll content, modifying leaf architecture, and engineering non-photochemical quenching mechanisms have shown promise in improving light use efficiency [
36].
Engineering photorespiratory bypasses has emerged as a powerful approach to reduce energy losses associated with photorespiration. By introducing alternative metabolic routes to process the products of Rubisco oxygenation, researchers have demonstrated significant improvements in biomass production under field conditions [
37]. The work of Xu et al. (2021) has provided new insights into the metabolic origins of non-photorespiratory CO2 release during photosynthesis, highlighting additional targets for optimization [
13]. These diverse approaches to enhancing photosynthetic efficiency, often used in combination, hold great promise for developing the next generation of high-yielding crops capable of meeting the growing global demand for food and biomass.
3.2. Improving Nutrient Use Efficiency
Enhancing nutrient uptake, assimilation, and utilization is a critical goal in crop improvement, aiming to increase yields while reducing the environmental impact of fertilizer use. Metabolic engineering approaches in this area focus on various aspects of plant nutrition, with particular emphasis on macronutrients such as nitrogen and phosphorus. Modifying nitrogen uptake and assimilation pathways has been a major focus, given the central role of nitrogen in plant growth and the significant environmental concerns associated with nitrogen fertilizer use. Strategies include enhancing the expression of nitrate and ammonium transporters to improve uptake, optimizing the enzymes involved in nitrogen assimilation such as nitrate reductase and glutamine synthetase, and improving the efficiency of nitrogen remobilization during senescence [
38]. These approaches have shown promise in improving nitrogen use efficiency in various crop species.
Enhancing phosphorus acquisition and utilization is another key area of research, given the limited global reserves of phosphate and its critical role in plant metabolism. Metabolic engineering efforts have focused on improving phosphate uptake through enhanced expression of phosphate transporters, increasing the production and exudation of organic acids and phosphatases to solubilize soil phosphate, and optimizing internal phosphate use efficiency through improved allocation and recycling [
39]. Some successful strategies have included the expression of bacterial phosphatases in plant roots and the engineering of plants to produce more organic acids in phosphate-deficient conditions.
Improving micronutrient accumulation and biofortification has gained significant attention as a means to address human nutritional deficiencies through crop improvement. Metabolic engineering approaches have been successful in enhancing the content of various micronutrients in edible plant tissues. Notable examples include golden rice, engineered to accumulate β-carotene in the grain, and high-iron rice varieties. Strategies involve enhancing uptake mechanisms, improving transport and accumulation in edible tissues, and increasing the synthesis of chelating agents to improve micronutrient bioavailability [
40]. These biofortification efforts not only aim to improve crop nutritional quality but also contribute to global health initiatives addressing micronutrient deficiencies.
3.3. Enhancing Stress Tolerance
Metabolic engineering can significantly improve crop resilience to various abiotic and biotic stresses, a crucial goal in the face of climate change and evolving pest pressures [
41,
42,
43]. Engineering osmolyte accumulation for drought and salinity tolerance has been a successful strategy in many crops. Osmolytes such as glycine betaine, proline, and trehalose help maintain cellular osmotic balance and protect cellular structures under stress conditions. By enhancing the biosynthesis or accumulation of these compounds through genetic engineering, researchers have developed crops with improved tolerance to drought and salt stress [
44]. For instance, overexpression of genes involved in glycine betaine synthesis has conferred enhanced drought tolerance in several crop species.
Enhancing antioxidant systems for oxidative stress tolerance is another key approach, as many environmental stresses lead to the accumulation of reactive oxygen species (ROS) in plant cells. Metabolic engineering strategies have focused on boosting the production of enzymatic antioxidants such as superoxide dismutase, catalase, and ascorbate peroxidase, as well as non-enzymatic antioxidants like ascorbic acid and glutathione [
45]. These approaches have shown success in improving plant tolerance to various stresses, including heat, cold, and high light intensity.
Modifying hormone signaling pathways for broad stress tolerance has emerged as a powerful strategy, given the central role of plant hormones in coordinating stress responses. Engineering the biosynthesis, perception, or signaling of hormones such as abscisic acid (ABA), ethylene, and salicylic acid can enhance plant resilience to multiple stresses simultaneously [
46]. For example, modifying ABA signaling components has led to improved drought tolerance in several crop species. The work of Xu and Fu (2022) provided a comprehensive metabolomics view of how plant central metabolism is reprogrammed in response to abiotic stresses, offering valuable insights for engineering stress-tolerant crops [
47]. This systems-level understanding of stress responses is guiding more targeted and effective metabolic engineering strategies for developing resilient crop varieties.
3.4. Production of Valuable Metabolites
Plants can be engineered as biofactories for the production of high-value compounds, leveraging their sophisticated biosynthetic capabilities and scalability. The production of nutraceuticals and health-promoting compounds in plants has gained significant attention. Metabolic engineering approaches have successfully enhanced the production of various bioactive compounds, including flavonoids, carotenoids, and omega-3 fatty acids, in crop plants. For instance, tomatoes and soybeans have been engineered to produce increased levels of flavonoids with potential health benefits [
48]. These efforts not only aim to improve the nutritional quality of crops but also offer a sustainable source of valuable compounds for the food and supplement industries.
The use of plants for the production of industrial precursors and biomaterials represents another exciting application of metabolic engineering. Researchers have engineered plants to produce various polymers, including biodegradable plastics like polyhydroxyalkanoates (PHAs). The production of novel biomaterials in plants offers a renewable and potentially more environmentally friendly alternative to petroleum-based products [
49]. Additionally, plants are being engineered to produce precursors for various industrial applications, such as specialty fatty acids and biofuels, leveraging the plant's ability to harness solar energy for carbon fixation [
50].
Perhaps one of the most promising areas is the production of pharmaceutical compounds and vaccines in plants. This approach, often termed "molecular farming," offers several advantages, including lower production costs, ease of scalability, and reduced risk of contamination with human pathogens. Metabolic engineering has enabled the production of various therapeutic proteins, antibodies, and vaccine antigens in plants [
51]. Notable examples include the production of anti-cancer antibodies in tobacco and the development of plant-made vaccines against infectious diseases. The ability to produce complex pharmaceutical compounds in plants not only offers a novel production platform but also has the potential to improve global access to important medicines and vaccines.
These diverse applications of plant metabolic engineering for the production of valuable metabolites highlight the versatility and potential of plants as biofactories. As our understanding of plant metabolism deepens and engineering techniques become more sophisticated, the range of compounds that can be produced in plants is likely to expand, offering new opportunities for sustainable production of valuable materials.
4. Emerging Trends and Future Perspectives
4.1. Synthetic Biology Approaches
The application of synthetic biology principles is revolutionizing plant metabolic engineering, offering unprecedented control over metabolic pathways and cellular processes. The design of synthetic metabolic pathways has emerged as a powerful approach, allowing for the creation of novel routes for metabolite production or the enhancement of existing pathways through the rational assembly of enzymes from diverse organisms [
52]. This strategy has enabled the production of valuable compounds in plants that were previously difficult or impossible to obtain. Complementing this, the development of synthetic regulatory circuits has made it possible to engineer complex gene networks and feedback loops, achieving dynamic control of metabolic processes in response to environmental or developmental cues [
53]. These synthetic circuits can fine-tune metabolic fluxes, optimize resource allocation, and improve plant performance under various conditions [
54].
Another exciting frontier is the engineering of synthetic organelles for metabolite compartmentalization. By creating artificial cellular compartments, researchers can optimize pathway efficiency and prevent undesired metabolic interactions, leading to improved production of target metabolites [
53]. Genome-scale engineering approaches, employing large-scale DNA synthesis and assembly techniques, are pushing the boundaries of what's possible in plant metabolic engineering, allowing for the redesign of plant genomes for optimized metabolism and performance [
55]. The development of synthetic protein scaffolds has provided a means to co-localize enzymes and improve pathway flux, enhancing the efficiency of multi-step biosynthetic processes [
23,
56]. Additionally, efforts to engineer photosynthesis through the redesign of key components of the photosynthetic machinery are showing promise in enhancing carbon fixation and energy conversion efficiency [
57]. These synthetic biology approaches, collectively, are opening new avenues for crop improvement and the sustainable production of valuable plant-derived compounds.
4.2. Multi-Gene Trait Stacking
Combining multiple engineered traits in a single plant is becoming increasingly feasible, allowing for more complex and comprehensive crop improvements. This approach offers several advantages and is rapidly advancing the field of plant biotechnology. Synergistic trait combinations can lead to greater improvements than individual modifications alone, as different traits can complement and enhance each other's effects [
58]. For instance, stacking genes for drought tolerance with those for improved nutrient use efficiency can result in crops that perform better under a wider range of environmental conditions. Multigene pathway engineering has enabled the introduction or modification of entire biosynthetic pathways for complex metabolites or agronomic traits [
59]. This has been particularly valuable in biofortification efforts and in engineering plants to produce high-value compounds.
The pyramid of stress tolerance genes has emerged as a powerful strategy for creating crops with broad environmental adaptability. By combining multiple stress resistance mechanisms, researchers can develop plants capable of withstanding various abiotic and biotic stresses simultaneously [
60,
61]. In the realm of nutritional enhancement, stacking genes for various nutritional traits has led to the creation of more comprehensive biofortified crops, addressing multiple nutrient deficiencies in a single plant variety [
58]. Moreover, metabolic balancing through the fine-tuning of multiple pathway components simultaneously has proven crucial in optimizing flux and avoiding metabolic bottlenecks in engineered pathways [
62]. The development of advanced cloning techniques, such as Golden Gate assembly and Gibson Assembly, has greatly facilitated the creation of multi-gene constructs for trait stacking [
63]. As our understanding of plant metabolism and gene interactions deepens, multi-gene trait stacking is expected to play an increasingly important role in developing the next generation of improved crops.
4.3. Integration of Omics and Computational Modeling
The integration of multi-omics data and advanced computational modeling is enhancing our ability to predict and optimize metabolic engineering outcomes. This systems biology approach is transforming the field by providing a more comprehensive understanding of plant metabolism and guiding engineering strategies. Genome-scale metabolic models have become powerful tools, offering comprehensive in silico representations of plant metabolism that can inform engineering decisions and predict the outcomes of genetic modifications [
64]. These models are continually refined with experimental data, improving their accuracy and predictive power. Metabolic flux analysis, using isotope labeling and sophisticated computational tools, allows researchers to quantify metabolic fluxes and identify rate-limiting steps in pathways of interest [
65]. This information is crucial for identifying targets for metabolic engineering and optimizing pathway performance.
Proteomics has emerged as a crucial tool in plant metabolic engineering, providing comprehensive insights into the protein landscape of engineered plants [
66,
67]. This high-throughput approach enables the identification and quantification of thousands of proteins simultaneously, offering a direct view of the functional state of cells[
15]. In metabolic engineering, proteomics can reveal unexpected changes in protein abundance or post-translational modifications resulting from genetic manipulations, helping to explain phenotypic outcomes and identify potential bottlenecks in engineered pathways[
20]. For instance, Fan et al. (2016) used stable isotope labeling and high-resolution mass spectrometry to analyze protein turnover in Arabidopsis, revealing how protein dynamics change in response to environmental stresses[
56]. Such approaches can be invaluable in assessing the impact of metabolic engineering on the broader cellular context, guiding refinements in engineering strategies, and uncovering new targets for manipulation. Moreover, integrating proteomic data with other omics datasets can provide a systems-level understanding of metabolic networks, essential for predicting and optimizing the outcomes of metabolic engineering efforts.
The application of machine learning and artificial intelligence in plant metabolic engineering is rapidly advancing, with advanced algorithms being employed to predict gene-phenotype relationships and optimize metabolic designs [
68]. These approaches can sift through vast amounts of data to identify patterns and relationships that might not be apparent through traditional analysis methods. Multi-omics data integration, combining transcriptomics, proteomics, metabolomics, and phenomics data, is providing a holistic understanding of metabolic regulation and plant responses to genetic modifications [
69,
70]. This integrated approach allows for a more nuanced understanding of the complex interplay between different levels of biological organization. Kinetic modeling of metabolic pathways, developing detailed mathematical models of enzyme kinetics and pathway dynamics, is informing engineering decisions by providing insights into the behavior of metabolic systems under various conditions [
71]. Additionally, genome-wide association studies (GWAS) are identifying natural genetic variants associated with metabolic traits, guiding engineering efforts by highlighting key genes and regulatory elements [
72]. These computational and systems biology approaches are enabling more rational and efficient metabolic engineering strategies, as demonstrated by recent work in elucidating carbon flux in plant metabolism [
18], and are poised to accelerate the development of improved crops and plant-based production systems.
4.4. Regulatory and Biosafety Considerations
As the field of plant metabolic engineering advances, addressing regulatory hurdles and public concerns regarding genetically modified organisms remains crucial for successful implementation in agriculture. The development of comprehensive risk assessment protocols is ongoing, with efforts focused on standardizing methods for evaluating the environmental and health impacts of metabolically engineered crops [
73]. These assessments aim to ensure the safety of new crop varieties while facilitating their approval and adoption. Gene containment strategies are being developed and implemented to prevent gene flow from engineered crops to wild relatives or non-GM crops, addressing concerns about the unintended spread of engineered traits [
74].
There is a growing discussion around shifting towards product-based regulation, focusing on the properties of the final product rather than the process of genetic modification [
74]. This approach could streamline the regulatory process for crops with traits that could have been achieved through traditional breeding methods. Public engagement and education efforts are crucial in improving communication and transparency about the benefits and risks of metabolic engineering in agriculture [
75]. These efforts aim to foster informed public discourse and build trust in the technology. International harmonization of regulatory frameworks is an ongoing challenge, with efforts directed towards creating consistent standards across countries to facilitate the global adoption of beneficial traits [
75].
Ethical considerations surrounding biodiversity, food sovereignty, and the socioeconomic impacts of engineered crops continue to be important topics of discussion [
75]. Addressing these concerns is essential for the responsible development and deployment of metabolically engineered crops. Traceability and labeling of products derived from metabolically engineered plants remain important issues, with ongoing development of methods for detection and clear communication to consumers [
76]. As the field continues to evolve, addressing these regulatory and societal challenges will be essential for realizing the full potential of plant metabolic engineering in addressing global agricultural and environmental challenges. The successful navigation of these considerations will be crucial in translating the scientific advancements in metabolic engineering into practical solutions for sustainable agriculture and food security.
5. Conclusions
The field of metabolic engineering in plants has made significant strides in recent years, offering promising solutions to global challenges in agriculture, nutrition, and sustainability. This review has highlighted the diverse strategies and applications of metabolic engineering, from enhancing photosynthetic efficiency and nutrient use to improving stress tolerance and producing valuable metabolites. The integration of advanced technologies, including CRISPR/Cas9 gene editing, synthetic biology approaches, and multi-omics data analysis, has dramatically expanded our ability to manipulate plant metabolism with unprecedented precision. These tools have enabled researchers to create crops with improved yield, nutritional quality, and resilience to environmental stresses, potentially revolutionizing agricultural practices in the face of climate change and population growth.
However, several challenges remain to be addressed as the field moves forward. The complexity of metabolic networks becomes increasingly apparent as our understanding of plant metabolism deepens. Future research should focus on systems-level approaches to better predict and manage the ripple effects of metabolic modifications. While many successes have been demonstrated in laboratory settings, translating these achievements to field-grown crops remains a significant challenge. More emphasis on field trials and real-world performance of engineered crops is needed. The development of clear, science-based regulatory frameworks and improved public communication strategies will be crucial for the widespread adoption of metabolically engineered crops. As new crop varieties are developed, careful consideration must be given to their long-term environmental impact and sustainability. Ensuring that the benefits of metabolic engineering reach smallholder farmers and developing regions will be essential for global food security.
Looking ahead, several exciting avenues for future research emerge. The design of entirely novel metabolic pathways and regulatory circuits through synthetic biology approaches holds immense potential for creating crops with radically improved traits or new capabilities. As climate change intensifies, developing crops that can thrive under extreme conditions will become increasingly important. Further refinement of plants as production platforms for pharmaceuticals, biofuels, and industrial compounds could revolutionize multiple industries. Continued efforts in biofortification could have significant impacts on global nutrition and health. Combining metabolic engineering with other emerging technologies, such as nanotechnology or artificial intelligence-driven breeding programs, could yield synergistic benefits.
In conclusion, metabolic engineering in plants stands at the forefront of efforts to address some of the most pressing challenges of our time. By harnessing the power of plant metabolism, we have the potential to create a more sustainable and food-secure future. However, realizing this potential will require continued innovation, interdisciplinary collaboration, and responsible development and deployment of these powerful technologies. As we move forward, it will be crucial to balance the immense potential of metabolic engineering with careful consideration of its broader impacts on ecosystems, economies, and societies. With thoughtful development and application, metabolic engineering in plants promises to play a pivotal role in shaping a more resilient and sustainable agricultural future.
References
- Lee, S. Y., Nielsen, J. & Stephanopoulos, G. Metabolic Engineering: Concepts and Applications. (2021).
- van Dijk, M.; Morley, T.; Rau, M.L.; Saghai, Y. A meta-analysis of projected global food demand and population at risk of hunger for the period 2010–2050. Nat. Food 2021, 2, 494–501, . [CrossRef]
- Antoniewicz, M.R. A guide to metabolic flux analysis in metabolic engineering: Methods, tools and applications. Metab. Eng. 2020, 63, 2–12, . [CrossRef]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013, 8, 2281–2308, doi:10.1038/nprot.2013.143.
- Sweetlove, L.J.; Nielsen, J.; Fernie, A.R. Engineering central metabolism – a grand challenge for plant biologists. Plant J. 2016, 90, 749–763, . [CrossRef]
- Hefferon, K.L. Nutritionally Enhanced Food Crops; Progress and Perspectives. Int. J. Mol. Sci. 2015, 16, 3895–3914, . [CrossRef]
- Petrie, J.R.; Shrestha, P.; Belide, S.; Kennedy, Y.; Lester, G.; Liu, Q.; Divi, U.K.; Mulder, R.J.; Mansour, M.P.; Nichols, P.D.; et al. Metabolic Engineering Camelina sativa with Fish Oil-Like Levels of DHA. PLOS ONE 2014, 9, e85061, . [CrossRef]
- Boghigian, B.A.; Seth, G.; Kiss, R.; Pfeifer, B.A. Metabolic flux analysis and pharmaceutical production. Metab. Eng. 2010, 12, 81–95, . [CrossRef]
- Henry, C.S.; Broadbelt, L.J.; Hatzimanikatis, V. Thermodynamics-Based Metabolic Flux Analysis. Biophys. J. 2007, 92, 1792–1805, . [CrossRef]
- Dai, Z.; Locasale, J.W. Understanding metabolism with flux analysis: From theory to application. Metab. Eng. 2016, 43, 94–102, . [CrossRef]
- Xu, Y. Metabolomics study on Arabidopsis thaliana abiotic stress responses for priming, recovery, and stress combinations. (2018).
- Holms, H. Flux analysis and control of the central metabolic pathways in Escherichia coli. FEMS Microbiol. Rev. 19, 85–116 (1996).
- Xu, Y.; Fu, X.; Sharkey, T.D.; Shachar-Hill, Y.; Walker, B.J. The metabolic origins of non-photorespiratory CO2 release during photosynthesis: a metabolic flux analysis. Plant Physiol. 2021, 186, 297–314, . [CrossRef]
- Lee, D.-Y.; Yun, H.; Park, S.; Lee, S.Y. MetaFluxNet: the management of metabolic reaction information and quantitative metabolic flux analysis. Bioinformatics 2003, 19, 2144–2146, . [CrossRef]
- Fan, K.-T.; Xu, Y.; Hegeman, A.D. Elevated Temperature Effects on Protein Turnover Dynamics in Arabidopsis thaliana Seedlings Revealed by 15N-Stable Isotope Labeling and ProteinTurnover Algorithm. Int. J. Mol. Sci. 2024, 25, 5882, . [CrossRef]
- Long, C.P.; Antoniewicz, M.R. High-resolution 13C metabolic flux analysis. Nat. Protoc. 2019, 14, 2856–2877, . [CrossRef]
- Quek, L.-E.; Wittmann, C.; Nielsen, L.K.; O Krömer, J. OpenFLUX: efficient modelling software for 13C-based metabolic flux analysis. Microb. Cell Factories 2009, 8, 25–25, . [CrossRef]
- Xu, Y.; Wieloch, T.; Kaste, J.A.M.; Shachar-Hill, Y.; Sharkey, T.D. Reimport of carbon from cytosolic and vacuolar sugar pools into the Calvin–Benson cycle explains photosynthesis labeling anomalies. Proc. Natl. Acad. Sci. 2022, 119, . [CrossRef]
- Quek, L.-E.; Dietmair, S.; Krömer, J.O.; Nielsen, L.K. Metabolic flux analysis in mammalian cell culture. Metab. Eng. 2010, 12, 161–171, . [CrossRef]
- Fan KaiTing, F. K. et al. Application of data-independent acquisition approach to study the proteome change from early to later phases of tomato pathogenesis responses. (2019).
- Wittmann, C. & Heinzle, E. Mass spectrometry for metabolic flux analysis. Biotechnol. Bioeng. 62, 739–750 (1999).
- Bar-Even, A.; Noor, E.; Savir, Y.; Liebermeister, W.; Davidi, D.; Tawfik, D.S.; Milo, R. The Moderately Efficient Enzyme: Evolutionary and Physicochemical Trends Shaping Enzyme Parameters. Biochemistry 2011, 50, 4402–4410, . [CrossRef]
- E Dueber, J.; Wu, G.C.; Malmirchegini, G.R.; Moon, T.S.; Petzold, C.J.; Ullal, A.V.; Prather, K.L.J.; Keasling, J.D. Synthetic protein scaffolds provide modular control over metabolic flux. Nat. Biotechnol. 2009, 27, 753–759, . [CrossRef]
- López-Calcagno, P. E. et al. Stimulating photosynthetic processes increases productivity and water-use efficiency in the field. Nat. Plants 6, 1054–1063 (2020).
- Gelvin, S. B. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol. Mol. Biol. Rev. 67, 16–37 (2003).
- Christou, P. Particle gun mediated transformation. Curr. Opin. Biotechnol. 1993, 4, 135–141, . [CrossRef]
- Rojo, F.P.; Nyman, R.K.M.; Johnson, A.A.T.; Navarro, M.P.; Ryan, M.H.; Erskine, W.; Kaur, P. CRISPR-Cas systems: ushering in the new genome editing era. Bioengineered 2018, 9, 214–221, . [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157, . [CrossRef]
- Kaplan, F.; Guy, C.L. RNA interference of Arabidopsis beta-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant J. 2005, 44, 730–743, . [CrossRef]
- Liu, Y., Schiff, M. & Dinesh-Kumar, S. P. Virus-induced gene silencing in tomato. Plant J. 31, 777–786 (2002).
- Khakhar, A., Leydon, A. R., Lemmex, A. C., Klavins, E. & Nemhauser, J. L. Synthetic hormone-responsive transcription factors can monitor and re-program plant development. Elife 7, e34702 (2018).
- Weber, W.; Fussenegger, M. Emerging biomedical applications of synthetic biology. Nat. Rev. Genet. 2011, 13, 21–35, . [CrossRef]
- Urano, K., Kurihara, Y., Seki, M. & Shinozaki, K. ‘Omics’ analyses of regulatory networks in plant abiotic stress responses. Curr. Opin. Plant Biol. 13, 132–138 (2010).
- Parry, M.A.J.; Andralojc, P.J.; Scales, J.C.; Salvucci, M.E.; Carmo-Silva, A.E.; Alonso, H.; Whitney, S.M. Rubisco activity and regulation as targets for crop improvement. J. Exp. Bot. 2012, 64, 717–730, . [CrossRef]
- Adebiyi, A.O.; Jazmin, L.J.; Young, J.D. 13C flux analysis of cyanobacterial metabolism. Photosynth. Res. 2014, 126, 19–32, . [CrossRef]
- Long, S.P.; Marshall-Colon, A.; Zhu, X.-G. Meeting the Global Food Demand of the Future by Engineering Crop Photosynthesis and Yield Potential. Cell 2015, 161, 56–66, . [CrossRef]
- South, P.F.; Cavanagh, A.P.; Liu, H.W.; Ort, D.R. Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 2019, 363, 45–+, . [CrossRef]
- Forde, B.G.; Lea, P.J. Glutamate in plants: metabolism, regulation, and signalling. J. Exp. Bot. 2007, 58, 2339–2358, . [CrossRef]
- Veneklaas, E.J.; Lambers, H.; Bragg, J.; Finnegan, P.M.; Lovelock, C.E.; Plaxton, W.C.; Price, C.A.; Scheible, W.; Shane, M.W.; White, P.J.; et al. Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 2012, 195, 306–320, . [CrossRef]
- Garcia-Oliveira, A.L.; Tan, L.; Fu, Y.; Sun, C. Genetic Identification of Quantitative Trait Loci for Contents of Mineral Nutrients in Rice Grain. J. Integr. Plant Biol. 2008, 51, 84–92, . [CrossRef]
- Igarashi, D.; Bethke, G.; Xu, Y.; Tsuda, K.; Glazebrook, J.; Katagiri, F. Pattern-Triggered Immunity Suppresses Programmed Cell Death Triggered by Fumonisin B1. PLOS ONE 2013, 8, e60769, . [CrossRef]
- Li, T.; Pang, N.; He, L.; Xu, Y.; Fu, X.; Tang, Y.; Shachar-Hill, Y.; Chen, S. Re-Programing Glucose Catabolism in the Microalga Chlorella sorokiniana under Light Condition. Biomolecules 2022, 12, 939, . [CrossRef]
- Chen, Y., Fan, K., Hung, S. & Chen, Y. The role of peptides cleaved from protein precursors in eliciting plant stress reactions. New Phytol. 225, 2267–2282 (2020).
- Umezawa, T.; Fujita, M.; Fujita, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Engineering drought tolerance in plants: discovering and tailoring genes unlock the future. Curr. Opin. Biotechnol. 2006, 17, 113–122, doi:10.1016/j.copbio.2006.02.002.
- Asada, K. Production and Scavenging of Reactive Oxygen Species in Chloroplasts and Their Functions. Plant Physiol. 2006, 141, 391–396, . [CrossRef]
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop. J. 2016, 4, 162–176, . [CrossRef]
- Xu, Y.; Fu, X. Reprogramming of Plant Central Metabolism in Response to Abiotic Stresses: A Metabolomics View. Int. J. Mol. Sci. 2022, 23, 5716, . [CrossRef]
- Malik, M.R.; Tang, J.; Sharma, N.; Burkitt, C.; Ji, Y.; Mykytyshyn, M.; Bohmert-Tatarev, K.; Peoples, O.; Snell, K.D. Camelina sativa, an oilseed at the nexus between model system and commercial crop. Plant Cell Rep. 2018, 37, 1367–1381, . [CrossRef]
- Poirier, Y.; Nawrath, C.; Somerville, C. Production of Polyhydroxyalkanoates, a Family of Biodegradable Plastics and Elastomers, in Bacteria and Plants. Nat. Biotechnol. 1995, 13, 142–150, . [CrossRef]
- Xu, Y. Metabolic Flux Redistribution in Engineered Microorganisms for Biofuel Production. Innov. Life Sci. J. 7, 1–12 (2021).
- Kromdijk, J.; Long, S.P. One crop breeding cycle from starvation? How engineering crop photosynthesis for rising CO2and temperature could be one important route to alleviation. Proc. R. Soc. B: Biol. Sci. 2016, 283, 20152578, . [CrossRef]
- Bar-Even, A.; Noor, E.; Lewis, N.E.; Milo, R. Design and analysis of synthetic carbon fixation pathways. Proc. Natl. Acad. Sci. 2010, 107, 8889–8894, . [CrossRef]
- Medford, J.I.; Prasad, A. Plant synthetic biology takes root. Science 2014, 346, 162–163, . [CrossRef]
- Xu, Y.; A Koroma, A.; E Weise, S.; Fu, X.; Sharkey, T.D.; Shachar-Hill, Y. Daylength variation affects growth, photosynthesis, leaf metabolism, partitioning, and metabolic fluxes. Plant Physiol. 2023, 194, 475–490, . [CrossRef]
- Wang, H.H.; Isaacs, F.J.; Carr, P.A.; Sun, Z.Z.; Xu, G.; Forest, C.R.; Church, G.M. Programming cells by multiplex genome engineering and accelerated evolution. Nature 2009, 460, 894–898, . [CrossRef]
- Fan, K.-T.; Rendahl, A.K.; Chen, W.-P.; Freund, D.M.; Gray, W.M.; Cohen, J.D.; Hegeman, A.D. Proteome Scale-Protein Turnover Analysis Using High Resolution Mass Spectrometric Data from Stable-Isotope Labeled Plants. J. Proteome Res. 2016, 15, 851–867, . [CrossRef]
- Simkin, A.J.; McAusland, L.; Lawson, T.; Raines, C.A. Overexpression of the RieskeFeS Protein Increases Electron Transport Rates and Biomass Yield. Plant Physiol. 2017, 175, 134–145, . [CrossRef]
- Naqvi, S.; Zhu, C.; Farre, G.; Ramessar, K.; Bassie, L.; Breitenbach, J.; Conesa, D.P.; Ros, G.; Sandmann, G.; Capell, T.; et al. Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc. Natl. Acad. Sci. 2009, 106, 7762–7767, . [CrossRef]
- Xu, Z.; Li, J.; Guo, X.; Jin, S.; Zhang, X. Metabolic engineering of cottonseed oil biosynthesis pathway via RNA interference. Sci. Rep. 2016, 6, 33342, . [CrossRef]
- Mittler, R.; Blumwald, E. Genetic Engineering for Modern Agriculture: Challenges and Perspectives. Annu. Rev. Plant Biol. 2010, 61, 443–462, . [CrossRef]
- Fu, X.; Xu, Y. Dynamic Metabolic Changes in Arabidopsis Seedlings under Hypoxia Stress and Subsequent Reoxygenation Recovery. Stresses 2023, 3, 86–101, . [CrossRef]
- Sweetlove, L.J.; Fernie, A.R. The role of dynamic enzyme assemblies and substrate channelling in metabolic regulation. Nat. Commun. 2018, 9, 1–12, . [CrossRef]
- Iba, K. et al. A gene encoding a chloroplast ω-3 fatty acid desaturase complements alterations in fatty acid desaturation and chloroplast copy number of the fad7 mutant of Arabidopsis thaliana. J. Biol. Chem. 268, 24099–24105 (1993).
- Cheung, C.M.; Poolman, M.G.; Fell, D.A.; Ratcliffe, R.G.; Sweetlove, L.J. A Diel Flux Balance Model Captures Interactions between Light and Dark Metabolism during Day-Night Cycles in C3 and Crassulacean Acid Metabolism Leaves. Plant Physiol. 2014, 165, 917–929, . [CrossRef]
- Zamboni, N.; Fendt, S.-M.; Rühl, M.; Sauer, U. 13C-based metabolic flux analysis. Nat. Protoc. 2009, 4, 878–892, . [CrossRef]
- Fan, K.-T.; Hsu, Y.; Yeh, C.-F.; Chang, C.-H.; Chang, W.-H.; Chen, Y.-R. Quantitative Proteomics Reveals the Dynamic Regulation of the Tomato Proteome in Response to Phytophthora infestans. Int. J. Mol. Sci. 2021, 22, 4174, . [CrossRef]
- Fan, K.; Hsu, C.; Chen, Y. Mass spectrometry in the discovery of peptides involved in intercellular communication: From targeted to untargeted peptidomics approaches. Mass Spectrom. Rev. 2023, 42, 2404–2425, . [CrossRef]
- Schwartz, J.-M.; Gaugain, C.; Nacher, J.C.; de Daruvar, A.; Kanehisa, M. Observing metabolic functions at the genome scale. Genome Biol. 2007, 8, R123–R123, . [CrossRef]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol. Biochem. 2013, 72, 21–34, . [CrossRef]
- Xu, Y.; Freund, D.M.; Hegeman, A.D.; Cohen, J.D. Metabolic signatures of Arabidopsis thaliana abiotic stress responses elucidate patterns in stress priming, acclimation, and recovery. Stress Biol. 2022, 2, 1–16, . [CrossRef]
- Stanford, N.J.; Lubitz, T.; Smallbone, K.; Klipp, E.; Mendes, P.; Liebermeister, W. Systematic Construction of Kinetic Models from Genome-Scale Metabolic Networks. PLOS ONE 2013, 8, e79195, . [CrossRef]
- Lasanthi-Kudahettige, R.; Magneschi, L.; Loreti, E.; Gonzali, S.; Licausi, F.; Novi, G.; Beretta, O.; Vitulli, F.; Alpi, A.; Perata, P. Transcript Profiling of the Anoxic Rice Coleoptile. Plant Physiol. 2007, 144, 218–231, . [CrossRef]
- Harwood, W.; Ricroch, A.; Clairand, P. Use of CRISPR systems in plant genome editing: toward new opportunities in agriculture. Emerg. Top. Life Sci. 2017, 1, 169–182, . [CrossRef]
- Li, Y.; Hallerman, E.M.; Liu, Q.; Wu, K.; Peng, Y. The development and status of Bt rice in China. Plant Biotechnol. J. 2015, 14, 839–848, . [CrossRef]
- Zilberman, D., Kaplan, S., Kim, E. & Waterfield, G. Lessons from the California GM labelling proposition on the state of crop biotechnology. in Handbook on Agriculture, Biotechnology and Development 538–549 (Edward Elgar Publishing, 2014).
- Fraiture, M.-A.; Herman, P.; Taverniers, I.; De Loose, M.; Deforce, D.; Roosens, N.H. Current and New Approaches in GMO Detection: Challenges and Solutions. BioMed Res. Int. 2015, 2015, 1–22, . [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).