1. Introduction
Wheat stands as the most extensively cultivated grain globally, with 20% of the world’s human dietary intake of calories and protein. By 2050, the world’s population is projected to exceed 9 billion, leading to a 60% increase in wheat demand as compared to 2020 [
1]. N is a key component of various basic functions of plants, such as chlorophyll synthesis, protein formation, nucleic acid production, enzyme activity, alkaloid synthesis, hormone regulation, and vitamin synthesis, all of which play important roles. N is usually the main yield-limiting factor in crop production. Properly using nitrogen fertilizer can greatly enhance crop yield, while overapplying it not only raises production costs but also leads to significant environmental pollution issues. Enhancing nitrogen use NUE of wheat emerges as a sustainable approach to achieve environmentally friendly and safety of wheat production. This highlighted the significance of breeding wheat varieties with high NUE in China. Various research studies have demonstrated that the efficiency of crop NUE largely depends on RSA, and the presence and spread of external nitrogen sources can, in return, modify the RSA of crops [
2]. Therefore, the wheat RSA response to external N levels may represent a pivotal avenue to enhance NUE. Historically, breeders have primarily assessed NUE based on above-ground phenotypic traits, over-looking the critical role of underground roots. This limited perspective has hindered advancements in high-yield wheat breeding to some extent [
3,
4,
5,
6].
Many studies have shown that the RSA is crucial in the growth and development of wheat [
3,
7,
8]. RSA encompasses the structural and spatial characteristics of the root system, encompassing traits such as total root length (TRL), total root surface area (TRS), total root volume (TRV), number of root tips (NRT), and average root diameter (ARD) [
3,
5]. RSA is intricately linked to physiological aspects like N absorption and transport [
4,
6,
9], and exhibit a significant positive correlation with plant N utilization capacity [
6,
10]. Thus, RSA has been used as an important index for associating N efficient varieties in rice [
11], wheat [
12,
13], and maize [
14,
15].
RSA is a common complex quantitative trait influenced by multiple genes. Combined GWAS and linkage map-ping are powerful tools for detecting genes and linkage analysis under complex traits [
16]. Because of the restricted genetic diversity in the parental populations, numerous QTLs are unable to be identified [
17,
18,
19]. Compared with linkage analysis, GWAS are more efficient and provide higher resolution in identifying gene loci. Moreover, combining linkage analysis with GWAS can improve the credibility of the results. This approach has been used to study agronomic characteristics, abiotic stress factors, and genes related to disease resistance in maize [
20], cucumber [
21], and Brassica napus [
22]. In the two latest studies on drought resistance in winter wheat, GWAS and linkage analysis were simultaneously used to identify genomic regions related to drought resistance [
23,
24]. Herein, our study con-ducted a comprehensive analysis of the natural population for GWAS, and the RIL population was utilized for link-age examination and confirmation of the hereditary loci distinguished by GWAS under the conditions of normal N (NN) and low N (LN) supply, and the important gene regions related to N response in wheat were identified through GWAS and linkage analysis. Study on RSA related traits of different genotypes at seedling stage under two nitrogen hydroponic environments of NN and LN. Different characteristics of RSA were analyzed to pinpoint genes associated with NUE, offering insight for developing high NUE wheat cultivars.
2. Materials and Methods
2.1. Plant Materials
This study utilized two panels made of wheat. Panel I: A total of 243 wheat varieties native to the Yellow and Huai Valley regions of China were utilized for GWAS (
Table S1) [
25]. Panel 2: The RIL population of 123 lines derived from the cross between Avocet and Chilero, both of them are the wheat backbone parent from the Internation-al Maize and Wheat Improvement Center (CIMMYT). Using the single seed descent method [
26]. It was previously utilized to describe resistance to leaf rust and stripe rust [
27]. Our study found that the RSA of Avocet and Chilero were quite different in NN and LN environments. The seeds of the parents and the RIL population were utilized for linkage examination and confirmation of the hereditary loci distinguished by GWAS.
2.2. Experimental Design and Trail Management
The nutrient solution culture method was used in a seedling stage test. The nutrient solution was referred to Hoagland et al. [
28] nutrient solution and appropriately modified according to the nutritional characteristics of wheat (
Table 1). Two treatments of normal N (NN, nutrient solution formula is shown in
Table 1) and low N (LN) were set in the laboratory. The nutrient solution difference was N supply with N in LN treatment are 0.4 mmol/L Ca(NO
3)
2 and 0 mmol/L (NH
4)
2SO
4.The uniform wheat grains were soaked in a 10% H
2O
2 solution for 10 minutes, then rinsed three times with deionized water before being placed on wet filter paper for culturing. Once the seedlings reached the one-leaf stage and exhibited similar growth, they were fixed in seedling trays and grown in NN and LN nutrient solutions. The nutrient solutions were aerated continuously with an oxygen pump, and changed every 3 days.
2.3. Trait Measurements
Following a 21 days of cultivation, the roots were washed with deionized water before being trimmed at the segmented points using scissors. Which were placed in a transparent tray for testing. Five individual plants were randomly selected from each sample, and the lateral roots of each plant were separated one by one, placed in a transparent root tray containing deionized water in a non overlapping state, and the root system was scanned by the WinRHIZOLA6400XL perspective scanning system to obtain the root system pictures of each individual plant. Then the WinRHIZOPro software was used to analyze the phenotypic data of each sample, such as TRL, TRS, TRV, NRT, and ARD, and the phenotypic data value of the material was determined by calculating the average of three replicates Subsequently, the relative values of each trait under two different N conditions were calculated, namely, RNRT, RTRL, RTRS, RTRV and RARD. Origin 2021 was used to statistically analyze the data, and the correlation coefficient was calculated.
2.4. Principal Component Analysis (PCA) and Kinship Analysis
2.5. GWAS
The 660k gene chip data of the natural population were analyzed by Beijing CapitaBio Technology Co. Ltd (
http://capital.en.drugdu.com/). in the study of Fusarium nipponense crown rot resistance of wheat previously used. After data quality check, 395782 SNP markers in total were kept [
25]. The mixed linear model (MLM) in Tassel V5.0 software was used for GWAS. The model principle is as follows ([
25,
29,
30].
Phenomenal attributes are denoted by Y, genotype by X, and principal components matrix (Q) by the first three principal components; The kinship matrix is K; corresponding effects are α. β and μ; residual effects are represented by the matrix ε. The matrices X and Q were regarded as fixed effects, while the matrices K and ε were regarded as random effects.
When the SNP marker -log10 (P-value) > 5, it is considered that there is a significant association between the marker and the trait. Based on the attenuation distance of whole genome linkage disequilibrium (LD), the 10MB interval before and after significant SNPs were identified as QTL sites [
25]. Using the CMplot program in the R package, manhattan and quantile quantile (Q-Q) plots were made.
The MLM model in Tassel V5.0 software was applied to analyze the correlation of root architecture of the natural population with PCA and kinship as covariates, combined with the screened genotype data. In the results of GWAS analysis, when Log10 (P-value) > 5, there was a considerable correlation between the marker and the characteristic. Using the R package CMplot program, Manhattan plots and Q-Q plots were visualized.
2.5. Linkage Analysis
Zhao’s genetic linkage map data [
31] indicates that. After screening and de-redundancy, a high-density genetic linkage map was created from the whole genomes of the RIL population and its two parents using a wheat DArT array. The map covers 21 wheat chromosomes and is 8202.10 cM long. It features 3627 DArT markers spaced 2.26 cM apart on average. The QTL of the RSA trait was mapped using the compound interval mapping approach with the LOD threshold set to 2.5 using the BIP function of the QTL IciMapping V4.2 program. MapChart V2.32 software was used to map the QTL linkage group. QTLs that shared flanking markers or had a spacing of less than 1 cM were considered to be at the same locus [
32]. The QTL names were assigned using McCouch’s methodology [
33].
3. Results
3.1. Phenotypic Evaluation
The results showed that in the two populations, different RSA indexes of wheat showed certain variability, and the variation range of the variation coefficient was quite different. Under the NN condition, the variation coefficient of each character of the natural population and the RIL population ranged from 11.39% (ARD) -39.72% (TRL) and 14.29% (ARD) -48.98% (TRS), while under the LN condition, the variation coefficient of each character of the natural population and the RIL population ranged from 26.36% (ARD) -54.19% (NRT) and 16.39% (ARD) -55.09% (TRS) Ln conditions. Comparing the relative values, it was found that the relative values of all RSA traits were greater than 1 except RARD (0.98) in the natural population. The variation coefficients of each trait in the natural population and the RIL population ranged from 29.24% (RARD) -52.26% (NRT) and 14.78% (RARD) -46.62% (RNRT).
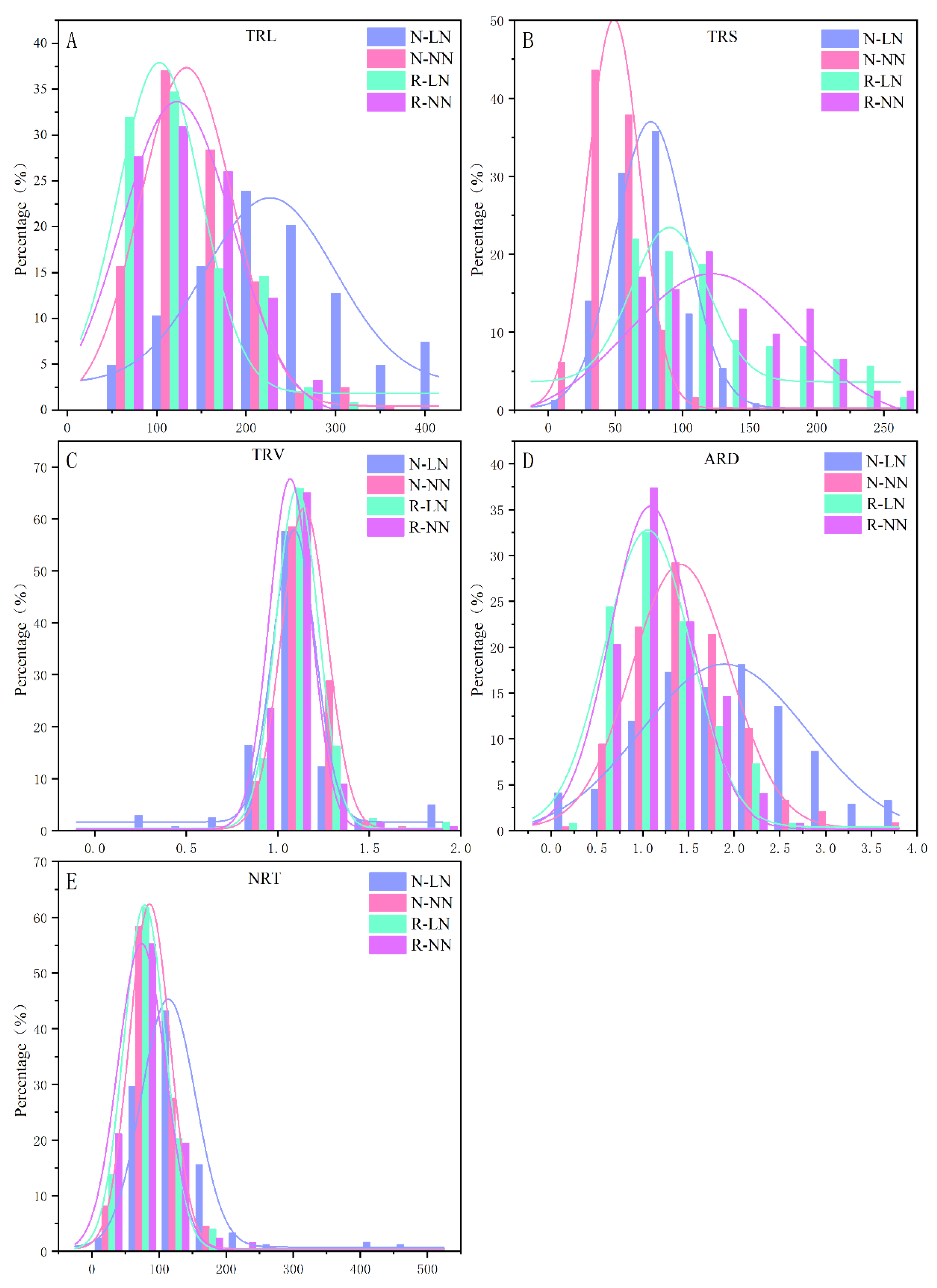
The RSA traits of the natural population and the RIL population showed continuous distribution under two N supply conditions, and the genetic basis was rich, which was appropriate for linkage analysis and GWAS analysis and linkage analysis (
Figure 1). The correlation analysis of RSA traits showed that LN supply could increase TRL, TRS and NRT, and the increase was more obvious in the natural population (
Table S1,
Table 2). Moreover,
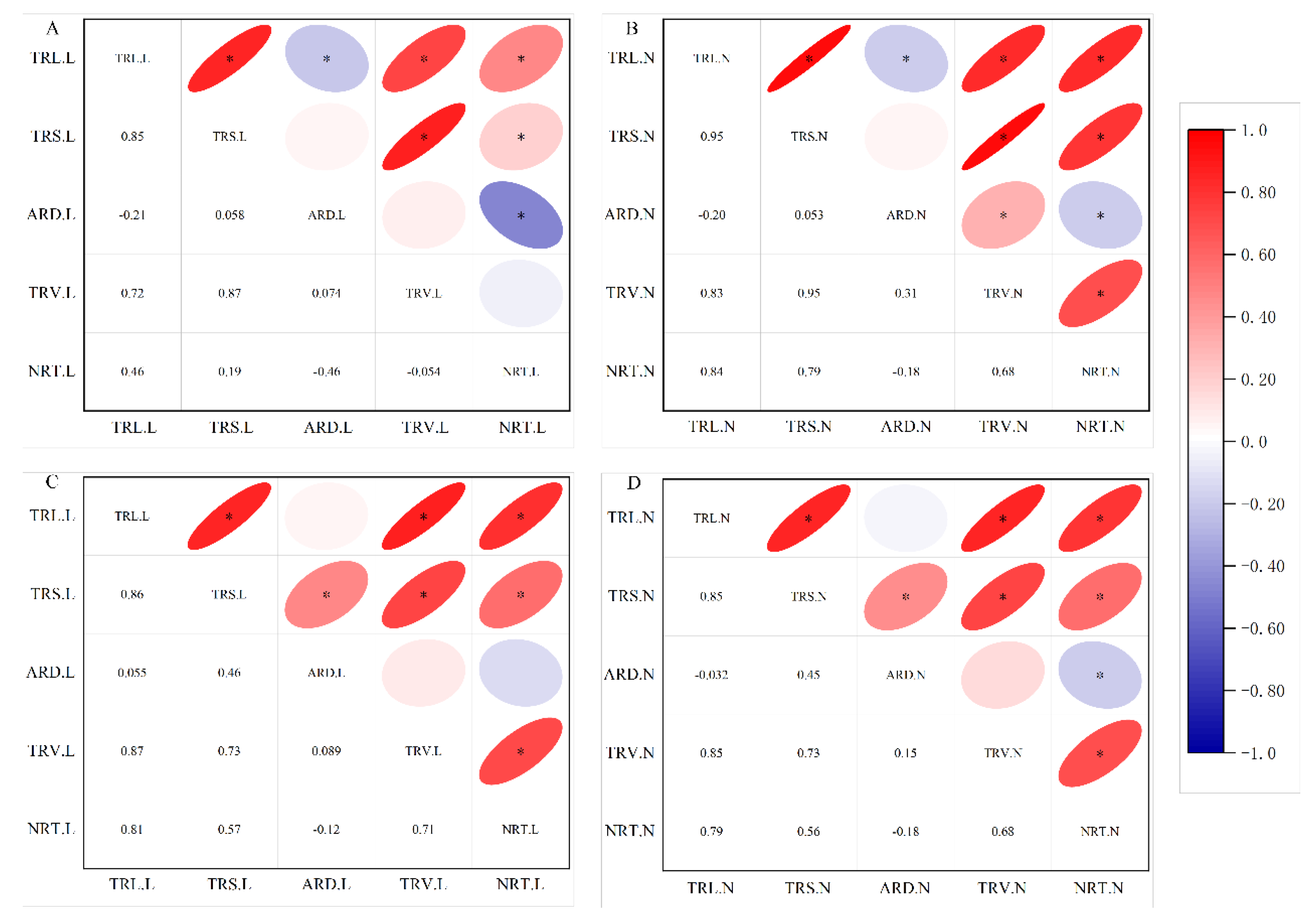
Figure 2 shows that the proportion of TRL, TRS, TRV, and NRT were significantly positively correlated in two populations and two N supply conditions (
P<0.05), with the correlation coefficient r ranging from 0.19 to 0.95. However, in the natural populations, the ARD was significantly negatively correlated with TRL and NRT in two N supply conditions (
P<0.05), and the correlation coefficient r ranging from -0.46 to -0.18, while in the RIL population, it was only negatively correlated in NN treatment (
P<0.05).
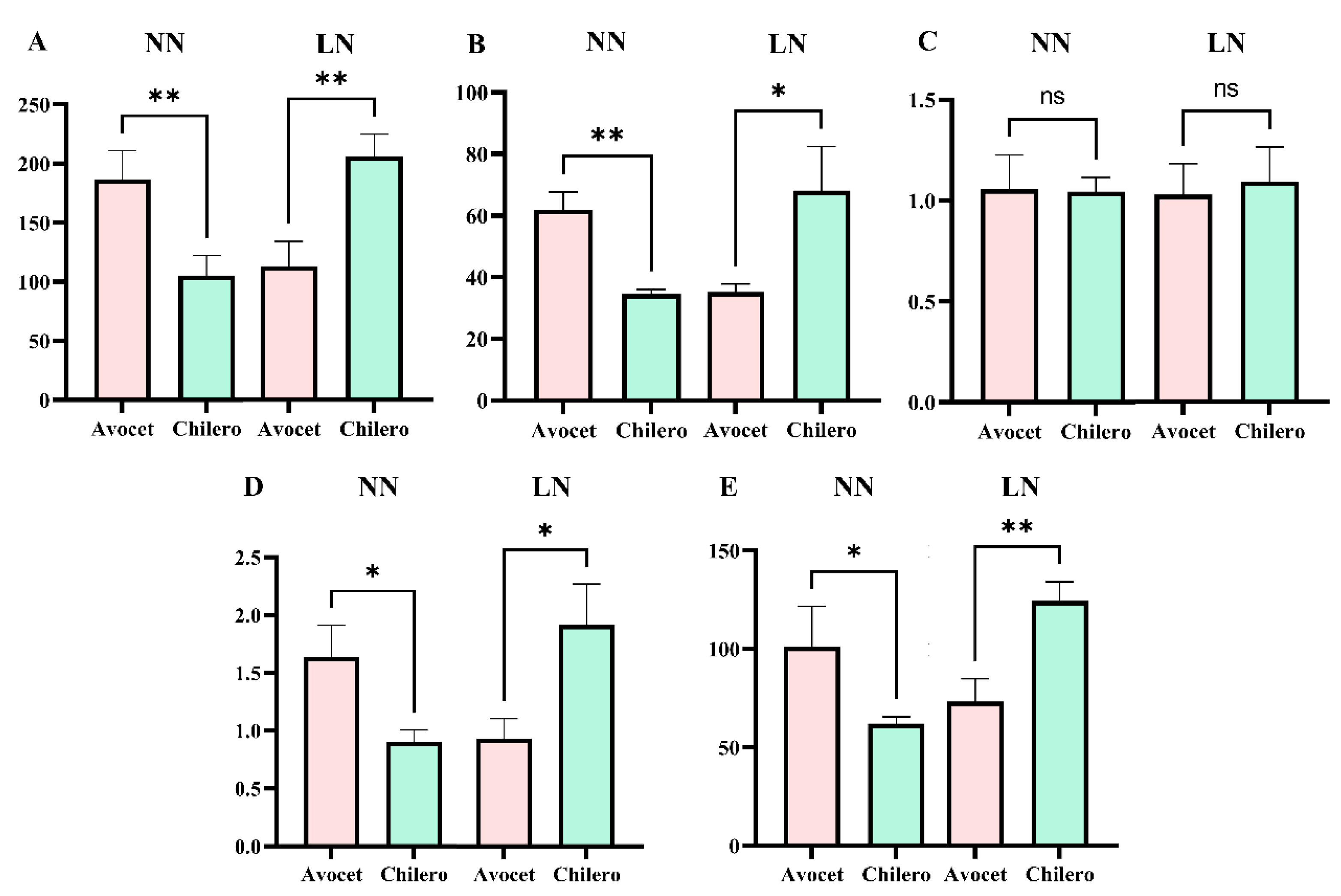
The T-test results of the RIL population parents showed that under the NN environment, the TRL, TRS, TRV, and NRT of Avocet were all greater than Chilero (P<0.05), however, there was an opposite tendency under the LN condition (P<0.05). This reflects the significant differences in the RSA responses of parents Avocet and Chilero under low nitrogen stress. (Figure. 3).
3.2. GWAS Analysis
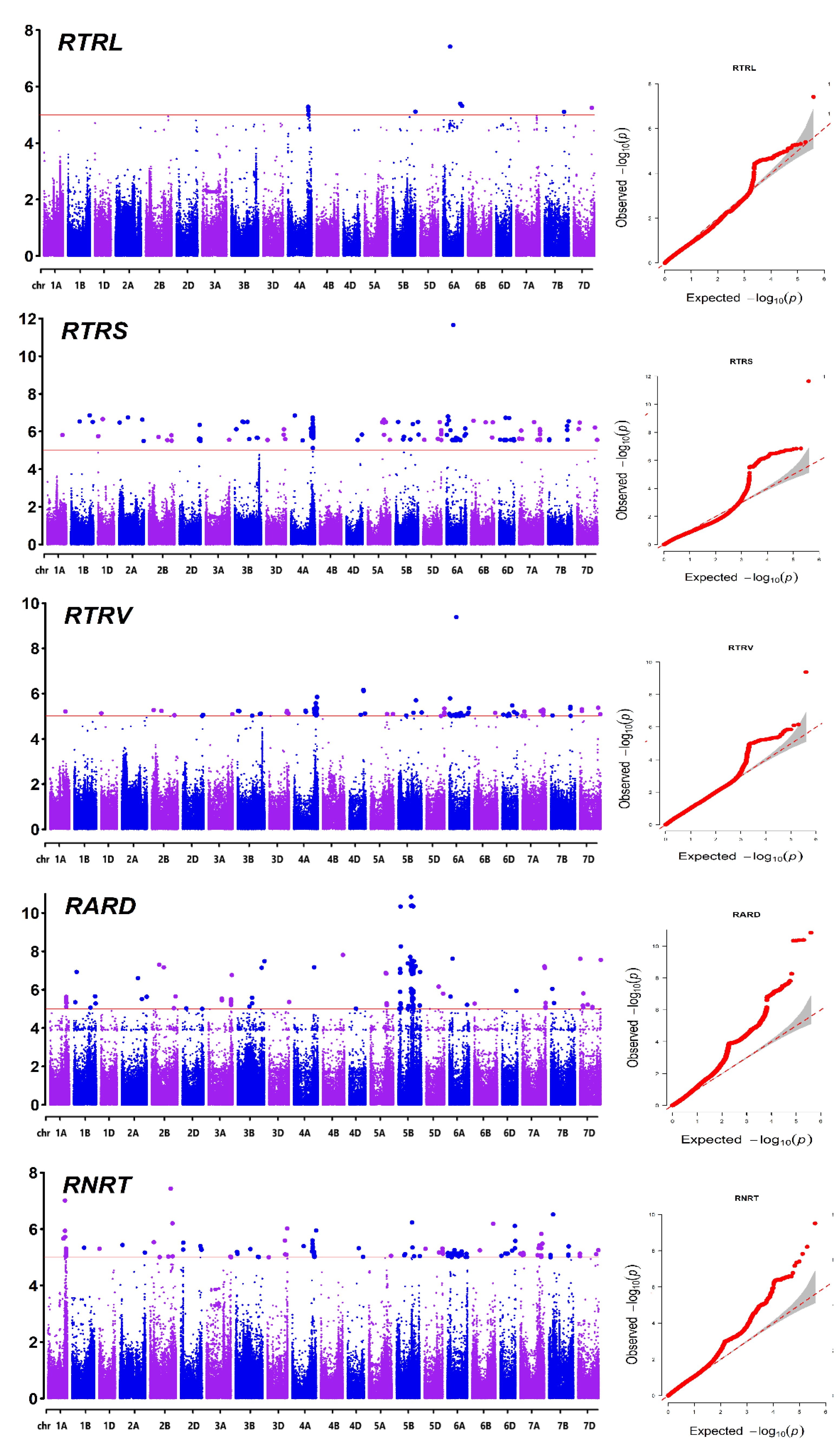
The quantile-quantile (Q-Q) plots indicated that the false positives were well controlled in GWAS models (
Figure 4). There were found to be 598 SNP markers that were substantially linked to features relevant to nitrogen utilization efficiency, distributed across 160 genetic loci, based on the ratios of RSA-related traits under two distinct nitrogen levels. (Figure.4 and
Table S2). Ten loci were linked to RTRL, accounting for 8.0%–15.6% of the phenotypic variation; ninety loci were linked to RTRS, accounting for 9.1%–26.9% of the variation; sixty-seven loci were linked to RTRV, accounting for 9.1%–19.8% of the variation; sixty-three loci were linked to RARD, accounting for 8.2%–24.7% of the variation; and seventy-nine loci were linked to RNRT, accounting for 8.9%–15.2% of the phenotypic variation. There was a high correlation between the ratio of RSA traits under two N supply conditions. Correspondingly, in the association results, we found that multiple relative traits have the same genetic loci. Among these one locus is associated with RTRS and RTRV, four loci are associated with RTRS and RARD, four loci are associated with RTRS and RNRT, and three loci are associated with RARD and RNRT, one locus is associated with RTRS, RTRV, and RARD, 31 loci are associated with RTRS, RTRV, and RNRT, one locus is associated with RTRS, RARD, and RNRT, six loci are associated with RTRL, RTRS, RTRV, and RNRT, 12 loci are associated with RTRS, RTRV, RARD, and RNRT, and three loci are associated with RTRL, RTRS, RTRV, RARD, and RNRT.
3.3. Linkage Analysis
Three QTLs in all were found to be linked to RTRL, and with LOD values ranging from 3.15 to 5.16, the unit point might account for 7.58%–15.04% of the phenotypic variation. The LOD values were 3.71-4.16 for the two QTLs that were determined to be linked to RTRS. These QTLs could account for 10.22%–15.92% of the variation in phenotype for each locus. Two QTLs in all were found to be linked to RARD, and with LOD values ranging from 2.52 to 3.95, the unit point might account for 8.16%–13.72% of the phenotypic variation. One QTL in all were found to be linked to RNRT, and with LOD values was 2.89, the unit point might account for 10.3% of the phenotypic variation (
Figure 5 and
Table 3).
3.4. Colocalized Gene Regions
Using GWAS and linkage mapping, three important RSA-related Quantitative Trait Locus (QTL) were found. (
Table 4). The significant marker AX-95160997 obtained through GWAS analysis of RTRL loci is located within the significant QTL QRtrl.haust-3D obtained through linkage analysis, with a physical interval of 39.61–43.74 MB. The significant marker AX-109592379 associated with RNRT is located within the significance QTL QRnrt.haust-5A, with a physical interval of 649.97-661.55 MB. The significant markers AX-110924288 associated with RTRL and RTRS are located in QRtrl.aust-7D and QRtrs.haust-7D, with a physical interval of 592.44-605.36 Mb.
4. Discussion
4.1. Effect of RSA on NUE
Wheat yield is significantly impacted by RSA [
2]. Breeders partially completed the selection of wheat root architecture by indirect means during the artificial breeding process to aggregate high-yield genes and choose high-yield materials [
34]. In this trial, we found that the RSA were different among materials of the two wheat populations, and there was a high correlation between the ratio of RSA traits under two N supply conditions. Therefore, it is very necessary to indirectly explore NUE related loci by carrying out the identification of RSA under different N conditions to promote the selection and breeding of wheat with high NUE.
The developed root system is the basis of efficient N absorption and utilization. The more developed the root, the thinner the average diameter of the root [
35]. The more fine roots, the more root tips, the longer the TRL, and the larger the TRS are, the more conducive it is for crops to effectively capture N in the soil [
36,
37]. Crops with different N efficiency not only have different performance in dry matter weight and N accumulation, but also have different root morphological and quantitative properties [
2,
38]. Numerous research has demonstrated that wheat that is N efficient has higher TRS [
2,
39]. N efficient rice had substantially higher TRL, TRS, and TRV than N inefficient rice [
40,
41]. Cassman et al [
42] discovered that the N efficient rice’s TRV offered more benefits. These findings suggested that the roots of the N efficient genotype were more developed.
This study demonstrated that reduced N supply enhanced the TRL, TRS, TRV, and NRT of seedlings compared to normal N supply, which is consistent with the prior research results [
2,
38]. Moreover, the change amplitude of natural populations is more obvious than the RIL populations, which may be related to the richer genetic diversity of natural populations. This proves that the elongation of most materials’ roots can be promoted under low nitrogen stress. Besides ARD, all coefficient of variations (CV) are greater than 20%, indicating that this is closely related to the genotype of the material. Moreover, through correlation analysis, this study found that ARD was negatively correlated with TRL, TRS, TRV and, NRT, suggesting that the average diameter of roots is smaller in better developed roots. Crops have an easier time efficiently absorbing nitrogen from the soil when there are more fine roots, more root tips, longer total root length, and greater root absorption area. Which is in line with the findings of earlier studies [
35,
36]. Moreover, through correlation analysis, this study found that ARD was negatively correlated with TRL, TRS, TRV, and NRT, which indicated that the more developed roots are, the thinner the average diameter of the roots is. The more fine roots, the more root tips, the longer the total root length and the larger the root absorption area are, the more conducive it is for crops to effectively capture nitrogen in the soil. Which is consistent with previous research results.
To mine QTL loci relevant to nitrogen efficiency, the response of wheat root RSA to low nitrogen stress was investigated in this work using the relative value approach of various nitrogen circumstances. This approach has been widely utilized to mine wheat NUE genes [
2,
43]. It can more correctly mine RSA genes affected by low nitrogen and better depict the reaction of plant seedling roots to low nitrogen environment. The CV of the relative proportion of RSA of the natural population and the RIL population is more than 40% except for RARD, which proves that under low nitrogen stress, there are great differences in RSA of wheat genotypes, and different genotypes have different responses to low nitrogen.
4.2. Linkage Analysis and GWAS together Offer a Novel Method for Identifying the Genes Causing RSA
Linkage analysis, sometimes referred to as QTL mapping, is a common technique for examining how quantitative traits are inherited. QTL loci for traits related to NUE were mapped under field conditions, however, relatively limited genetic variation in the parental population resulted in a large physical distance [
44,
45]. To reduce QTL intervals by raising marker density inside candidate segments, fine mapping is typically necessary [
46,
47]. Compared with traditional linkage analysis, GWAS takes natural populations as materials, with a wide range of sources (wild species, local varieties, modern varieties, and high generation strains), high gene polymorphisms, and no need to build a parental population.
Its benefits include low cost, excellent resolution, and high efficiency. The process of genetically analyzing quantitative traits has been greatly aided in recent years by the invention and application of wheat gene chips [
48], Numerous QTLs and QTL clusters have also been found in various wheat N treatment trials [
2,
38,
49,
50,
51,
52,
53,
54]. In this study, the wheat 660k SNP chip and DArT technology were used to analyze the inheritance of RSA during the wheat seedling stage employing GWAS and QTL mapping under different N levels We found that the total number of loci significantly associated with RSA is distributed on 21 chromosomes. Some of these loci overlapped or coincided with multiple RSA related loci discovered by Jin et al. [
2], Xiong et al. [
38], Fan et al. [
54], Meng-jiao Yang et al. [
52], and Ren et al. [
53] (
Table S3). In which the mapped area contains numerous known NUE related genes such as NRT2s genes on chromosome 6A, which can be associated with RTRL, RTRS, RNRT, and other traits. In addition, NPF6.2, FD-GOGAT, and the GWAS and QTL mapped region contain the GS2 genes [
55,
56,
57].
Combined GWAS and linkage mapping, a significant locus
AX-95160997/QRtrl.haust-3D (
Table 4) was detected, which may be an important NUE locus. According to its colocalized physical location, it is between 39.61-43.74MB named as
qRtrl-3D. In this interval, the Chinese Spring 2.0 database was used to screen high-confidence genes. There are a total of 61 annotated genes. Through screening, we found a possible candidate gene
TraesCS3D02G090300, which is located at 0.05Mb flanking the significance marker
DarT1145273 (
QRtrl.haust-3D). By querying the gene annotation, we found that it encodes a MADS box transcription factor. The MADS box gene family plays an irreplaceable role in the process of plant growth and development and signaling. Studies have shown that the MADS box transcription factor is involved in regulating lateral root growth in Arabidopsis [
58,
59]. In Arabidopsis, the MADS box transcription factor is mainly expressed in roots and regulates lateral root development by regulating auxin synthesis in roots [
60]. Recent studies have shown that, the MADS box transcription factor family is involved in regulating NUE in wheat and plays an important role in nitrate supply in roots, which provides new clues for further exploring the function of this gene [
61,
62].
This study also found two colocalized intervals
AX-109592379/QRnrt.haust-5A and
AX-110924288/ QRtrl.haust-7D/QRtrs.haust-7D (
Table 4). According to the physical location of the colocalization of these two sites, it is between 649.97-661.55 MB and 592.44-605.36 MB, which is called
qRnrt-5A and
qRtrl-7D. Compared with previous studies, no overlapping interval was found, which may be two new loci. Candidate genes were screened in the
qRtrl-7D interval, and some possible candidate genes were listed, such as the TraesCS7D02G538000 gene, which encodes an E3 ubiquitin protein ligase. The multitype protein family E3 ubiquitin protein ligase is crucial for root and bud growth and development, as well as plant N absorption. [
63,
64]. Candidate genes were screened in the
q Rnrt-5A interval, and some possible candidate genes were listed, such as the TraesCS5A02G519300 gene, which encodes a protein with the NAC binding domain (
Table 5). Studies have shown that overexpression of the NAC transcription factor significantly increases nitrate influx rate, N uptake, and other traits [
65], but it cannot be identified as a candidate gene due to the large candidate interval.
NUE was significant differences among species or subspecies and different plant organs. With the release of wheat genome sequence, gene function verification and exploration have become an important topic in the post genome era of wheat. However, the widespread phenomenon of one cause and multiple effects in organisms also makes the determination of gene function difficult, which still requires solid gene function verification. At present, bioinformatic analysis based on reliable association markers is an effective method to explore candidate genes related to complex agronomic traits. To provide more candidate gene loci for N efficient molecular breeding, targeted improvement of specific crop subspecies or creation of excellent new varieties, and providing references for improving NUE, we should deeply explore the key regulatory genes and excellent allelic variation that control NUE different N application levels.
5. Conclusions
In this study, based on exploring the ratio of RSA related traits of the 243 natural population and 123 RIL population to normal and low N supply under two N supply conditions, a total of 598 SNP markers, which were significantly associated with RSA related traits, were detected at 160 genetic loci identified by GWAS, and a total of 10 QTL loci related to RSA were discovered identified by linkage mapping. Finally, three significant RSA related QTL genetic loci, namely qRtrl-3D, qRnrt-3D, and qRtrl-7D, were mapped according to the responses of root architecture related traits. Which may be the new NUE related locus in wheat. These results will be helpful for marker assisted selection and further research on the cloning of NUE genes in Wheat.
Supplementary Materials
The following supporting information can be downloaded at the website of the paper posted on Preprints.org, Table S1: 243 natural population names; Table S2 Marker-trait associations for nitrogen use efficiency related traits (P ≤ 0.0001) ; Table S3 Compare the results of previous studies.
Author Contributions
YJ: Conceptualization, Data curation, Formal analysis, Writing-original draft, Writing–review & editing. NX: Data curation, Formal analysis, Writing–original draft. JZ: Investigation, Validation, Writing–original draft. KR: Investigation, Validation, Writing–original draft. JW: Investigation, Validation, Writing–original draft. CW: Project administration, Resources, Writing–review & editing. YL: Funding acquisition, Project administration, Resources, Supervision, Writing–review & editing. MH: Project administration, Resources, Supervision, Writing–review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by National Key Research and Development Program of China (under Grant No. 2022YFD2300800), the Science and Technology Research Project of Henan, China (under Grant No. 232102111009).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available in the main body of the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Langridge, P.; Alaux, M.; Almeida, N.F.; Ammar, K.; Baum, M.; Bekkaoui, F.; Bentley, A.R.; Beres, B.L.; Berger, B.; Braun, H. Meeting the challenges facing wheat production: The strategic research agenda of the Global Wheat Initiative. Agronomy 2022, 12, 2767. [CrossRef]
- Jin, Y.; Liu, J.; Liu, C.; Jia, D.; Liu, P.; Wang, Y. Genome-wide association study of nitrogen use efficiency related traits in common wheat (Triticum aestivum L.). Acta Agronomica Sinica 2021, 47, 394-404. [CrossRef]
- Maccaferri, M.; El-Feki, W.; Nazemi, G.; Salvi, S.; Canè, M.A.; Colalongo, M.C.; Stefanelli, S.; Tuberosa, R. Prioritizing quantitative trait loci for root system architecture in tetraploid wheat. J Exp Bot 2016, 67, 1161-1178. [CrossRef]
- de Dorlodot, S.; Forster, B.; Pagès, L.; Price, A.; Tuberosa, R.; Draye, X. Root system architecture: opportunities and constraints for genetic improvement of crops. Trends Plant Sci 2007, 12, 474-481. [CrossRef]
- Kabir, M.R.; Liu, G.; Guan, P.; Wang, F.; Khan, A.A.; Ni, Z.; Yao, Y.; Hu, Z.; Xin, M.; Peng, H. Mapping QTLs associated with root traits using two different populations in wheat (Triticum aestivum L.). Euphytica 2015, 206, 175-190. [CrossRef]
- Bishopp, A.; Lynch, J.P. The hidden half of crop yields. Nat Plants 2015, 1, 1-2. [CrossRef]
- Chen, J.; Zhang, Y.; Tan, Y.; Zhang, M.; Zhu, L.; Xu, G.; Fan, X. Agronomic nitrogen-use efficiency of rice can be increased by driving Os NRT 2.1 expression with the Os NAR 2.1 promoter. Plant Biotechnol J 2016, 14, 1705-1715. [CrossRef]
- Ruffel, S.; Gojon, A.; Lejay, L. Signal interactions in the regulation of root nitrate uptake. J Exp Bot 2014, 65, 5509-5517. [CrossRef]
- Paez-Garcia, A.; Motes, C.M.; Scheible, W.; Chen, R.; Blancaflor, E.B.; Monteros, M.J. Root traits and phenotyping strategies for plant improvement. Plants 2015, 4, 334-355. [CrossRef]
- Osmont, K.S.; Sibout, R.; Hardtke, C.S. Hidden branches: developments in root system architecture. Annu Rev Plant Biol 2007, 58, 93-113. [CrossRef]
- Ju, C.; Buresh, R.J.; Wang, Z.; Zhang, H.; Liu, L.; Yang, J.; Zhang, J. Root and shoot traits for rice varieties with higher grain yield and higher nitrogen use efficiency at lower nitrogen rates application. Field Crop Res 2015, 175, 47-55. [CrossRef]
- Duncan, E.G.; O’Sullivan, C.A.; Roper, M.M.; Palta, J.; Whisson, K.; Peoples, M.B. Yield and nitrogen use efficiency of wheat increased with root length and biomass due to nitrogen, phosphorus, and potassium interactions. J Plant Nutr Soil Sc 2018, 181, 364-373. [CrossRef]
- Ping, W.; WANG, Z.; CAI, R.; Yong, L.I.; CHEN, X.; YIN, Y. Physiological and molecular response of wheat roots to nitrate supply in seedling stage. Agricultural sciences in China 2011, 10, 695-704. [CrossRef]
- Zhan, A.; Lynch, J.P. Reduced frequency of lateral root branching improves N capture from low-N soils in maize. J Exp Bot 2015, 66, 2055-2065. [CrossRef]
- Saengwilai, P.; Strock, C.; Rangarajan, H.; Chimungu, J.; Salungyu, J.; Lynch, J.P. Root hair phenotypes influence nitrogen acquisition in maize. Ann Bot-London 2021, 128, 849-858. [CrossRef]
- Neumann, K.; Kobiljski, B.; Denčić, S.; Varshney, R.K.; Börner, A. Genome-wide association mapping: a case study in bread wheat (Triticum aestivum L.). Mol Breeding 2011, 27, 37-58. [CrossRef]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 2013, 9, 1-9. [CrossRef]
- Li, F.; Xie, J.; Zhu, X.; Wang, X.; Zhao, Y.; Ma, X.; Zhang, Z.; Rashid, M.A.; Zhang, Z.; Zhi, L. Genetic basis underlying correlations among growth duration and yield traits revealed by GWAS in rice (Oryza sativa L.). Front Plant Sci 2018, 9, 650. [CrossRef]
- Li, X.; Guo, T.; Wang, J.; Bekele, W.A.; Sukumaran, S.; Vanous, A.E.; McNellie, J.P.; Tibbs-Cortes, L.E.; Lopes, M.S.; Lamkey, K.R. An integrated framework reinstating the environmental dimension for GWAS and genomic selection in crops. Mol Plant 2021, 14, 874-887. [CrossRef]
- Li, X.; Zhou, Z.; Ding, J.; Wu, Y.; Zhou, B.; Wang, R.; Ma, J.; Wang, S.; Zhang, X.; Xia, Z. Combined linkage and association mapping reveals QTL and candidate genes for plant and ear height in maize. Front Plant Sci 2016, 7, 833. [CrossRef]
- Bo, K.; Wei, S.; Wang, W.; Miao, H.; Dong, S.; Zhang, S.; Gu, X. QTL mapping and genome-wide association study reveal two novel loci associated with green flesh color in cucumber. Bmc Plant Biol 2019, 19, 1-13. [CrossRef]
- He, Y.; Wu, D.; Wei, D.; Fu, Y.; Cui, Y.; Dong, H.; Tan, C.; Qian, W. GWAS, QTL mapping and gene expression analyses in Brassica napus reveal genetic control of branching morphogenesis. Sci Rep-Uk 2017, 7, 15971. [CrossRef]
- Guo, J.; Guo, J.; Li, L.; Bai, X.; Huo, X.; Shi, W.; Gao, L.; Dai, K.; Jing, R.; Hao, C. Combined linkage analysis and association mapping identifies genomic regions associated with yield-related and drought-tolerance traits in wheat (Triticum aestivum L.). Theor Appl Genet 2023, 136. [CrossRef]
- Sallam, A.; Eltaher, S.; Alqudah, A.M.; Belamkar, V.; Baenziger, P.S. Combined GWAS and QTL mapping revealed candidate genes and SNP network controlling recovery and tolerance traits associated with drought tolerance in seedling winter wheat. Genomics 2022, 114, 110358. [CrossRef]
- Yang, X.; Pan, Y.; Singh, P.K.; He, X.; Ren, Y.; Zhao, L.; Zhang, N.; Cheng, S.; Chen, F. Investigation and genome-wide association study for Fusarium crown rot resistance in Chinese common wheat. Bmc Plant Biol 2019, 19, 1-14. [CrossRef]
- Basnet, B.R.; Singh, R.P.; Ibrahim, A.; Herrera-Foessel, S.A.; Huerta-Espino, J.; Lan, C.; Rudd, J.C. Characterization of Yr54 and other genes associated with adult plant resistance to yellow rust and leaf rust in common wheat Quaiu 3. Mol Breeding 2014, 33, 385-399. [CrossRef]
- Ponce-Molina, L.J.; Huerta-Espino, J.; Singh, R.P.; Basnet, B.R.; Alvarado, G.; Randhawa, M.S.; Lan, C.X.; Aguilar-Rincón, V.H.; Lobato-Ortiz, R.; García-Zavala, J.J. Characterization of leaf rust and stripe rust resistance in spring wheat ‘Chilero’. Plant Dis 2018, 102, 421-427. [CrossRef]
- 28. Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Circular. California agricultural experiment station 1950, 347.
- Lv, G.; Dong, Z.; Wang, Y.; Geng, J.; Li, J.; Lv, X.; Sun, C.; Ren, Y.; Zhang, J.; Chen, F. Identification of genetic loci of black point in Chinese common wheat by genome-wide association study and linkage mapping. Plant Dis 2020, 104, 2005-2013. [CrossRef]
- Niaz, M.; Zhang, L.; Lv, G.; Hu, H.; Yang, X.; Cheng, Y.; Zheng, Y.; Zhang, B.; Yan, X.; Htun, A. Identification of TaGL1-B1 gene controlling grain length through regulation of jasmonic acid in common wheat. Plant Biotechnol J 2023, 21, 979-989. [CrossRef]
- Zhao, Y.; Yan, X.; Zeng, Z.; Zhao, D.; Chen, P.; Wang, Y.; Chen, F.; Wang, C. Integrated genome-wide association study and QTL mapping reveals qSa-3A associated with English grain aphid, Sitobion avenae (Fabricius) resistance in wheat. Pest Manag Sci 2023. [CrossRef]
- Mapchart, V.E. softwarefortheGraphical PresentationofLinkageMapsandQTLs. Journalof Heredity 2002, 93, 77r-78r.
- McCouch, S.R.; CGSNL Committee On Gene Symbolization, N.A.L.R. Gene nomenclature system for rice. Rice 2008, 1, 72-84. [CrossRef]
- Wang, Y.; Hou, J.; Liu, H.; Li, T.; Wang, K.; Hao, C.; Liu, H.; Zhang, X. TaBT1, affecting starch synthesis and thousand kernel weight, underwent strong selection during wheat improvement. J Exp Bot 2019, 70, 1497-1511. [CrossRef]
- Kong, D.L.; Wang, J.J.; Kardol, P.; Wu, H.F.; Zeng, H.; Deng, X.B.; Deng, Y. Economic strategies of plant absorptive roots vary with root diameter. Biogeosciences 2016, 13, 415-424. [CrossRef]
- Xue, Y.; Zhang, W.; Liu, D.; Yue, S.; Cui, Z.; Chen, X.; Zou, C. Effects of nitrogen management on root morphology and zinc translocation from root to shoot of winter wheat in the field. Field Crop Res 2014, 161, 38-45. [CrossRef]
- Zhang, H.; Jennings, A.; Barlow, P.W.; Forde, B.G. Dual pathways for regulation of root branching by nitrate. Proceedings of the National Academy of Sciences 1999, 96, 6529-6534. [CrossRef]
- Xiong, H.; Guo, H.; Zhou, C.; Guo, X.; Xie, Y.; Zhao, L.; Gu, J.; Zhao, S.; Ding, Y.; Liu, L. A combined association mapping and t-test analysis of SNP loci and candidate genes involving in resistance to low nitrogen traits by a wheat mutant population. Plos One 2019, 14, e211492. [CrossRef]
- An, D.; Su, J.; Liu, Q.; Zhu, Y.; Tong, Y.; Li, J.; Jing, R.; Li, B.; Li, Z. Mapping QTLs for nitrogen uptake in relation to the early growth of wheat (Triticum aestivum L.). Plant Soil 2006, 284, 73-84. [CrossRef]
- Erenoglu, E.B.; Kutman, U.B.; Ceylan, Y.; Yildiz, B.; Cakmak, I. Improved nitrogen nutrition enhances root uptake, root-to-shoot translocation and remobilization of zinc (65Zn) in wheat. New Phytol 2011, 189, 438-448. [CrossRef]
- Wei, H.; Zhang, H.; Zhang, S.; Jie, H. Root Morphological and Physiological Characteristics in Rice Genotypes with Different N Use Efficiencies. ACTA AGRONOMICA SINICA 2008, 429-436. [CrossRef]
- Cassman, K.G.; Dobermann, A.; Walters, D.T. Agroecosystems, nitrogen-use efficiency, and nitrogen management. AMBIO: A Journal of the Human Environment 2002, 31, 132-140. [CrossRef]
- Shi, H.; Chen, M.; Gao, L.; Wang, Y.; Bai, Y.; Yan, H.; Xu, C.; Zhou, Y.; Xu, Z.; Chen, J. Genome-wide association study of agronomic traits related to nitrogen use efficiency in wheat. Theor Appl Genet 2022, 135, 4289-4302. [CrossRef]
- Zuo, W.; Chao, Q.; Zhang, N.; Ye, J.; Tan, G.; Li, B.; Xing, Y.; Zhang, B.; Liu, H.; Fengler, K.A. A maize wall-associated kinase confers quantitative resistance to head smut. Nat Genet 2015, 47, 151-157. [CrossRef]
- Zandipour, M.; Majidi Hervan, E.; Azadi, A.; Khosroshahli, M.; Etminan, A. A QTL hot spot region on chromosome 1B for nine important traits under terminal drought stress conditions in wheat. Cereal Res Commun 2020, 48, 17-24. [CrossRef]
- Tura, H.; Edwards, J.; Gahlaut, V.; Garcia, M.; Sznajder, B.; Baumann, U.; Shahinnia, F.; Reynolds, M.; Langridge, P.; Balyan, H.S. QTL analysis and fine mapping of a QTL for yield-related traits in wheat grown in dry and hot environments. Theor Appl Genet 2020, 133, 239-257. [CrossRef]
- Zhang, Q.; Wei, W.; Zuansun, X.; Zhang, S.; Wang, C.; Liu, N.; Qiu, L.; Wang, W.; Guo, W.; Ma, J. Fine mapping of the leaf rust resistance gene Lr65 in spelt Wheat ‘Altgold’. Front Plant Sci 2021, 12, 666921. [CrossRef]
- Hitz, K.; Clark, A.J.; Van Sanford, D.A. Identifying nitrogen-use efficient soft red winter wheat lines in high and low nitrogen environments. Field Crop Res 2017, 200, 1-9. [CrossRef]
- Cui, F.; Zhao, C.; Ding, A.; Li, J.; Wang, L.; Li, X.; Bao, Y.; Li, J.; Wang, H. Construction of an integrative linkage map and QTL mapping of grain yield-related traits using three related wheat RIL populations. Theor Appl Genet 2014, 127, 659-675. [CrossRef]
- Fontaine, J.; Ravel, C.; Pageau, K.; Heumez, E.; Dubois, F.; Hirel, B.; Le Gouis, J. A quantitative genetic study for elucidating the contribution of glutamine synthetase, glutamate dehydrogenase and other nitrogen-related physiological traits to the agronomic performance of common wheat. Theor Appl Genet 2009, 119, 645-662. [CrossRef]
- Guo, Y.; Kong, F.; Xu, Y.; Zhao, Y.; Liang, X.; Wang, Y.; An, D.; Li, S. QTL mapping for seedling traits in wheat grown under varying concentrations of N, P and K nutrients. Theor Appl Genet 2012, 124, 851-865. [CrossRef]
- Yang, M.; Wang, C.; Hassan, M.A.; Wu, Y.; Xia, X.; Shi, S.; Xiao, Y.; He, Z. QTL mapping of seedling biomass and root traits under different nitrogen conditions in bread wheat (Triticum aestivum L.). J Integr Agr 2021, 20, 1180-1192. [CrossRef]
- Ren, D.; Fang, X.; Jiang, P.; Zhang, G.; Hu, J.; Wang, X.; Meng, Q.; Cui, W.; Lan, S.; Ma, X. Genetic architecture of nitrogen-deficiency tolerance in wheat seedlings based on a nested association mapping (NAM) population. Front Plant Sci 2018, 9, 845. [CrossRef]
- Fan, X.; Zhang, W.; Zhang, N.; Chen, M.; Zheng, S.; Zhao, C.; Han, J.; Liu, J.; Zhang, X.; Song, L. Identification of QTL regions for seedling root traits and their effect on nitrogen use efficiency in wheat (Triticum aestivum L.). Theor Appl Genet 2018, 131, 2677-2698. [CrossRef]
- Saini, D.K.; Chopra, Y.; Pal, N.; Chahal, A.; Srivastava, P.; Gupta, P.K. Meta-QTLs, ortho-MQTLs and candidate genes for nitrogen use efficiency and root system architecture in bread wheat (Triticum aestivum L.). Physiol Mol Biol Pla 2021, 27, 2245-2267. [CrossRef]
- Yang, Y.; Amo, A.; Wei, D.; Chai, Y.; Zheng, J.; Qiao, P.; Cui, C.; Lu, S.; Chen, L.; Hu, Y. Large-scale integration of meta-QTL and genome-wide association study discovers the genomic regions and candidate genes for yield and yield-related traits in bread wheat. Theor Appl Genet 2021, 134, 3083-3109. [CrossRef]
- Liu, H.; Mullan, D.; Zhang, C.; Zhao, S.; Li, X.; Zhang, A.; Lu, Z.; Wang, Y.; Yan, G. Major genomic regions responsible for wheat yield and its components as revealed by meta-QTL and genotype–phenotype association analyses. Planta 2020, 252, 1-22. [CrossRef]
- Zhang, H.; Forde, B.G. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science 1998, 279, 407-409. [CrossRef]
- Remans, T.; Nacry, P.; Pervent, M.; Filleur, S.; Diatloff, E.; Mounier, E.; Tillard, P.; Forde, B.G.; Gojon, A. The Arabidopsis NRT1. 1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proceedings of the National Academy of Sciences 2006, 103, 19206-19211. [CrossRef]
- Yu, L.; Miao, Z.; Qi, G.; Wu, J.; Cai, X.; Mao, J.; Xiang, C. MADS-box transcription factor AGL21 regulates lateral root development and responds to multiple external and physiological signals. Mol Plant 2014, 7, 1653-1669. [CrossRef]
- Lei, L.; Li, G.; Zhang, H.; Powers, C.; Fang, T.; Chen, Y.; Wang, S.; Zhu, X.; Carver, B.F.; Yan, L. Nitrogen use efficiency is regulated by interacting proteins relevant to development in wheat. Plant Biotechnol J 2018, 16, 1214-1226. [CrossRef]
- Wang, M.; Zhang, P.; Liu, Q.; Li, G.; Di, D.; Xia, G.; Kronzucker, H.J.; Fang, S.; Chu, J.; Shi, W. TaANR1-TaBG1 and TaWabi5-TaNRT2s/NARs link ABA metabolism and nitrate acquisition in wheat roots. Plant Physiol 2020, 182, 1440-1453. [CrossRef]
- Craig, A.; Ewan, R.; Mesmar, J.; Gudipati, V.; Sadanandom, A. E3 ubiquitin ligases and plant innate immunity. J Exp Bot 2009, 60, 1123-1132. [CrossRef]
- Park, G.; Park, J.; Yoon, J.; Yu, S.; An, G. A RING finger E3 ligase gene, Oryza sativa Delayed Seed Germination 1 (OsDSG1), controls seed germination and stress responses in rice. Plant Mol Biol 2010, 74, 467-478. [CrossRef]
- He, X.; Qu, B.; Li, W.; Zhao, X.; Teng, W.; Ma, W.; Ren, Y.; Li, B.; Li, Z.; Tong, Y. The nitrate-inducible NAC transcription factor TaNAC2-5A controls nitrate response and increases wheat yield. Plant Physiol 2015, 169, 1991-2005. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).