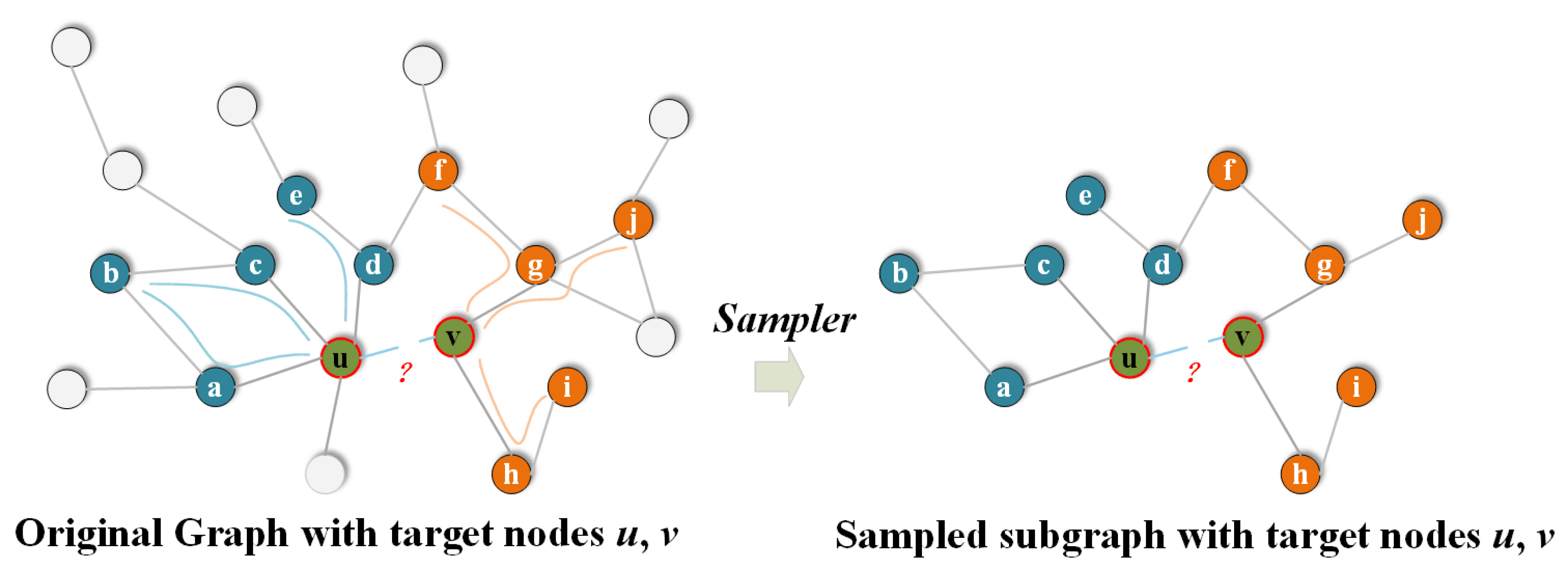
4.1. Datasets and Baselines
Datasets: We used nine real-world network datasets, including weighted and unweighted, undirected, attributed, and non-attributed graph data. The experiments divided these datasets into two categories: non-attributed and attributed datasets. For both attributed and non-attributed datasets, except for Cora, CiteSeer, and PubMed, which were divided into 70% training set, 10% validation set, and 20% test set according to specific experimental settings, the edges of the remaining datasets were randomly divided into 85% training set, 5% validation set, and 10% test set. The experimental datasets include NS [
55], a collaboration network of network science researchers, Power [
43], an electrical power grid of the western United States, Yeast [
56], a protein-protein interaction network, PB [
57], a political blog network, Cora [
58], a citation network in the field of machine learning, CiteSeer [
59], a scientific publication citation network, PubMed [
59], a diabetes-related scientific publication citation network, and Texas and Wisconsin [
60], web page datasets collected by computer science departments of different universities.
Table 1 details the statistical information of these datasets, with the first four being non-attributed networks and the last five being attributed networks. Node represents the number of nodes, Edge represents the number of edges, Avg Deg represents the average degree of the network, Feat represents the feature dimension of the nodes, and Type represents the network type.
Baselines: In this section, we experimentally analyze the proposed link prediction model AFD3S and compare it with nine existing advanced link prediction models on nine different real-world datasets. These include two message-passing graph neural network (MPGNNs) models: GCN [
61] and GIN [
62]; three autoencoder (AE) models: GAE, VGAE [
63], and GIC (Graph InfoClust) [
64]; and four SGRLs: SEAL [
44], WESLP [
51], Scaled [
45], and WalkPool [
52].
4.2. Experimental Setup
Experimental Environment: Equipped with AMD Ryzen 7 5800H CPU, 32GB memory The hardware environment of NVIDIA GeForce RTX 3070 Laptop GPU (8GB graphics memory) runs on the Windows 11 64-bit operating system, using PyCharm 2023.2.1 as the development tool, Python 3.10.9 as the development language, and PyTorch 1.12.1 and PyTorch Geometry 2.0.9 as the development framework.
Experimental Settings: For SGRLs and the AFD3S method on non-attributed datasets, the hop of the enclosing subgraphs,
h, is typically set to 2 (except for the WalkPool on the Power dataset, where
h is set to 3). For sparsely sampled subgraphs, the walk length
h is set to 2, and the number of walks
k is set to 50. On attributed datasets, the hop of the enclosing subgraphs,
h, is generally set to 3 (while the WalkPool sets it to 2). The settings for sparsely sampled subgraphs are the same as for non-attributed datasets. Additionally, in the AFD3S, the zero-one [
38] labeling scheme is uniformly adopted to label all datasets, while models like SEAL and Scaled use DRNL [
44] for labeling. The central common neighbor pooling readout function employs a simple mean aggregation approach. These settings and choices aim to ensure consistency and performance optimization of the models while accommodating the characteristics of different datasets and models. Moreover, for all datasets, the percentages of training, validation, and test sets across all models are uniformly set to 85%, 5%, and 10%, respectively, with a 1:1 sampling ratio for positive and negative samples.
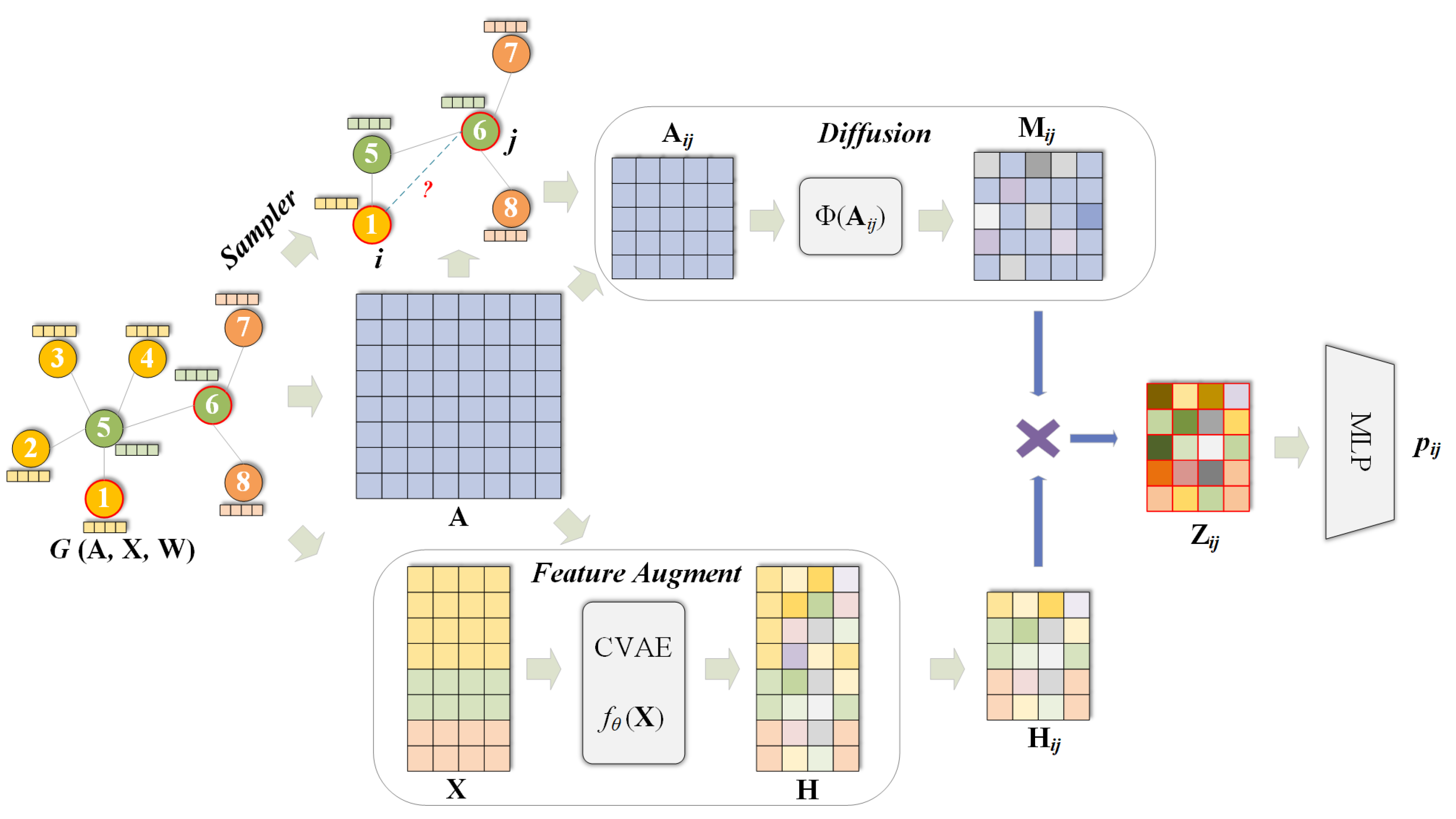
In the AFD3S model, the neural network utilizes SIGN [
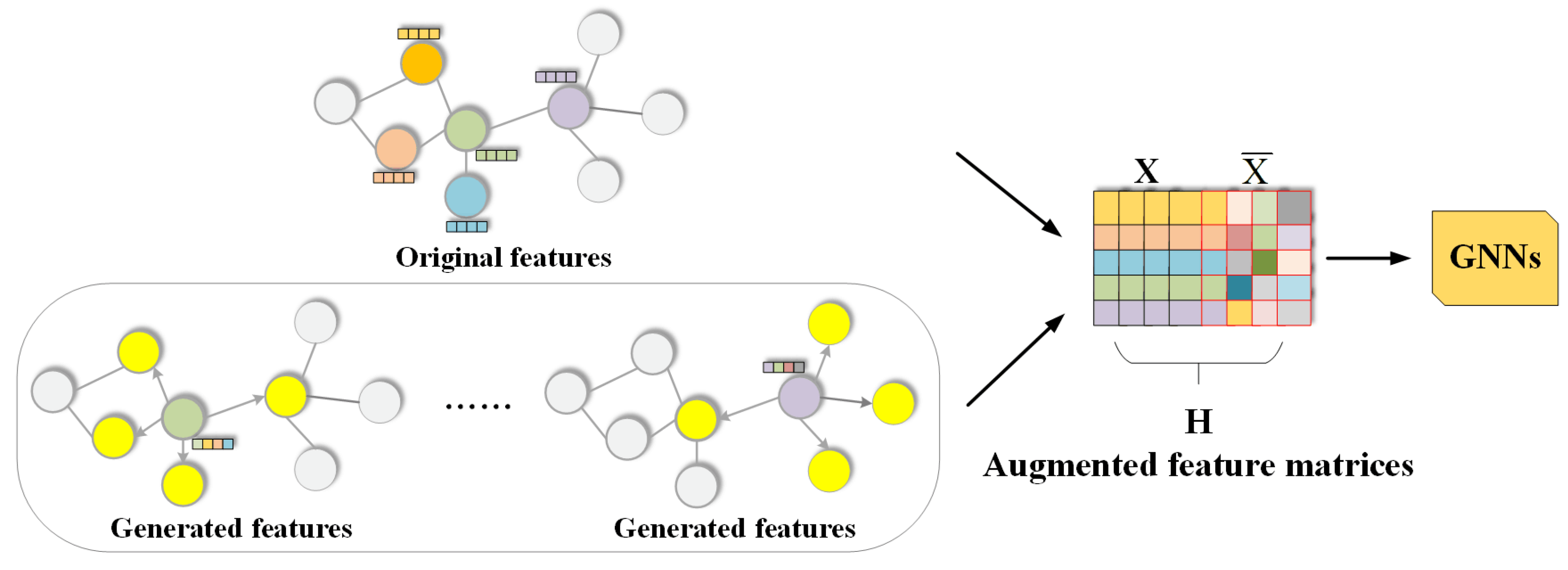
65], and for the non-attributed datasets, Node2Vec is employed to generate 256-dimensional feature vectors for each node. In the process of feature augmentation,
uniformly adopts concatenation as the augmentation method. For all datasets, the hidden dimension after pooling in Equation (
8) is set to 256, and an MLP with a 256-dimensional hidden layer is adopted in the experiments. To maintain consistency, the dropout rate is set to 0.5 for all models, the learning rate is set to 0.0001, and the Adam optimizer is used for 50 training epochs. During the training process, except for the MPGNN model, which uses full-batch training on the input graph, the batch size for other models is set to 32. These settings ensure the experiments’ fairness and comparability while fully utilizing the potential of the AFD3S model.
Evaluation Metrics: This paper adopts AUC and AP as the evaluation standards for model performance, aiming to accurately assess the performance of the AFD3S in solving the link prediction problem. Additionally, to fully demonstrate the computational efficiency and scalability of the AFD3S, this study further compares the performance of the AFD3S with existing popular SGRLs in terms of average preprocessing time, average training time, average inference time, and total running time.
4.3. Results and Analysis
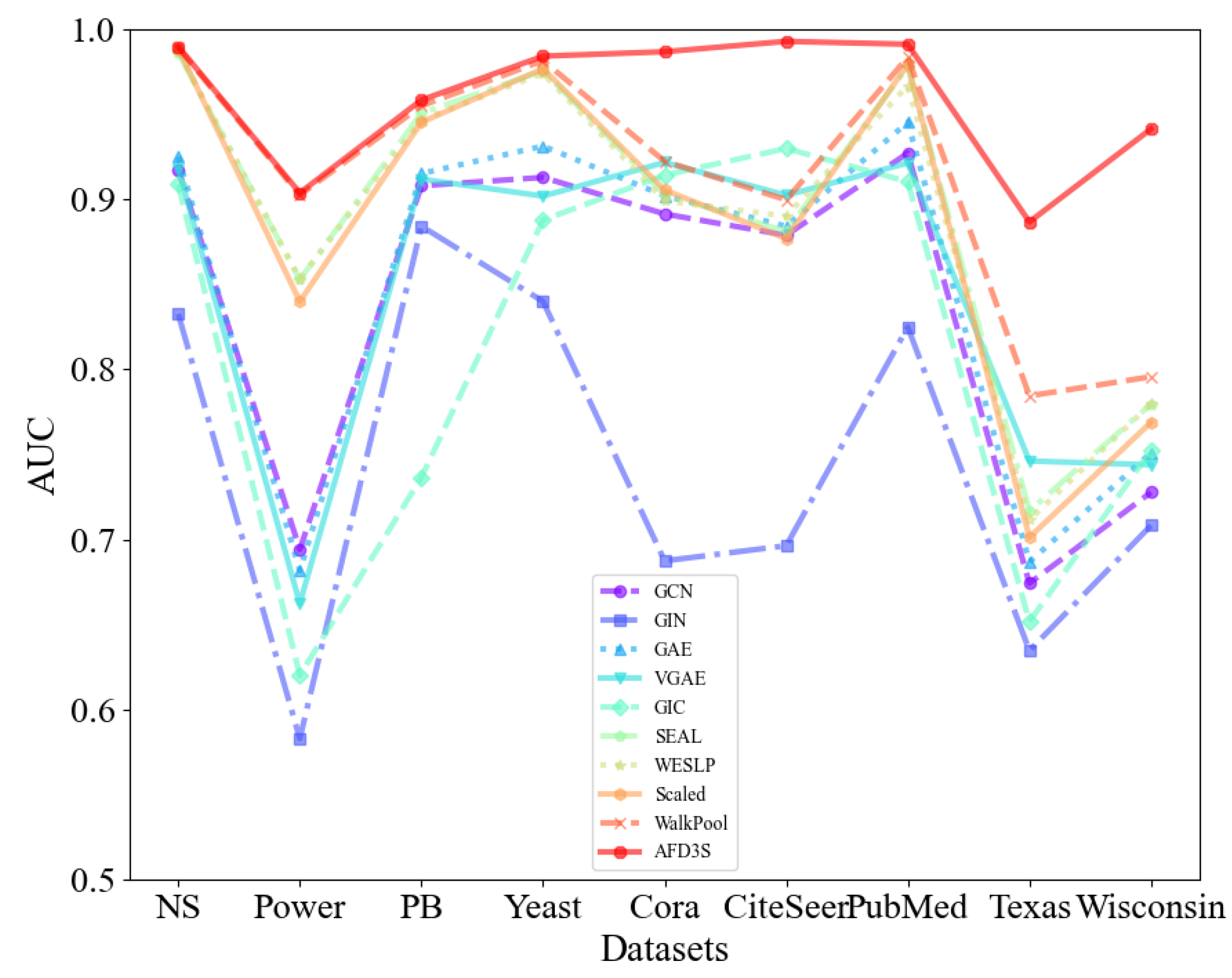
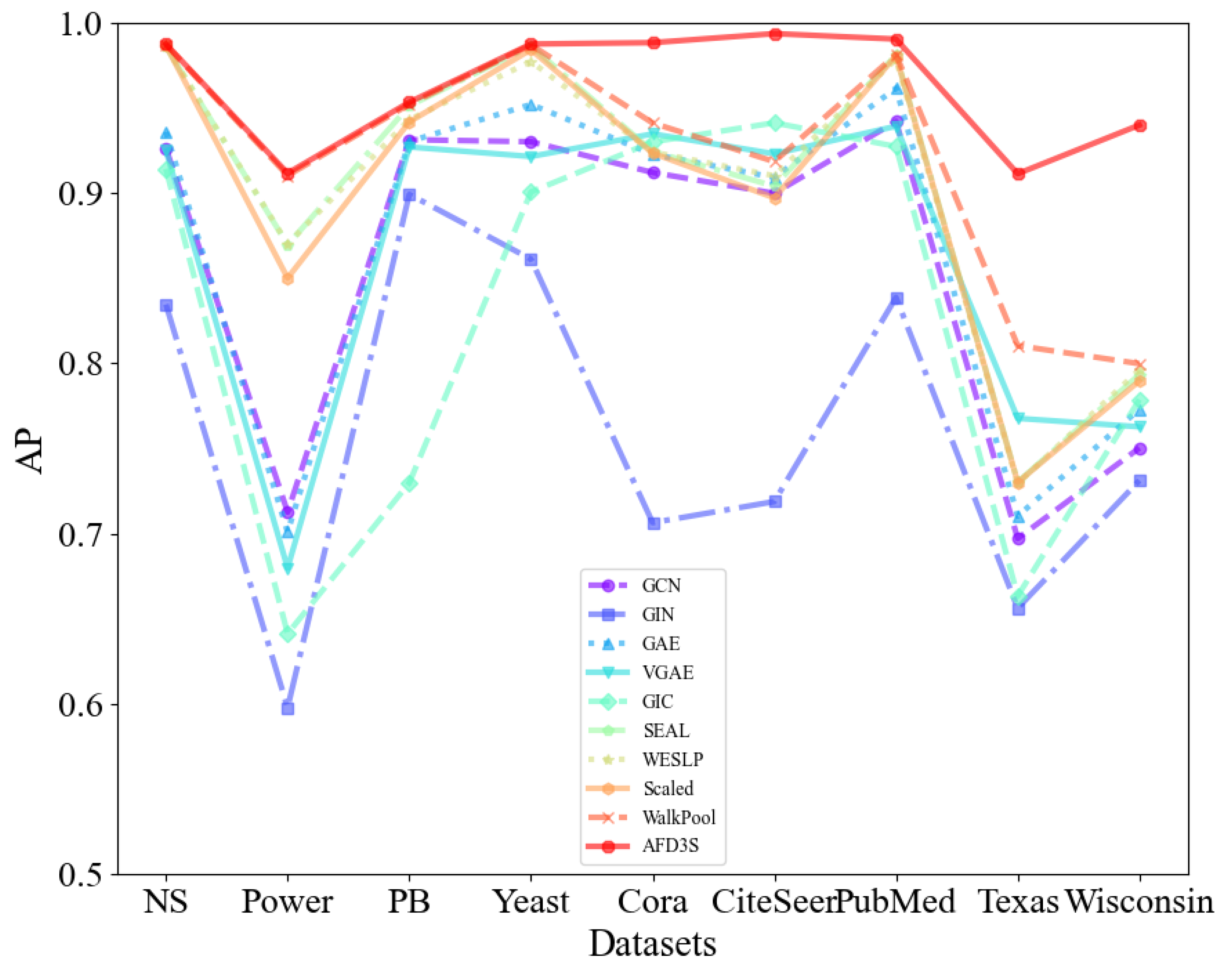
Link Prediction: For all models, on both attributed and non-attributed datasets, this study presents the average AUC and AP scores over 10 runs with different fixed random seeds on the test data.
Table 2 displays the AUC results for both non-attribute and attribute datasets, while
Table 3 displays the AP results for both non-attribute and attribute datasets. The optimal values are marked in bold.
Based on the data in
Table 2 and
Table 3, it is evident that the proposed AFD3S demonstrates exceptional performance in terms of average AUC and AP results on both non-attributed and attributed datasets, achieving optimal levels. Specifically, on attributed datasets, compared to the advanced benchmark model WalkPool, the AUC results of the AFD3S show improvements of 6.44% on Cora, 9.32% on CiteSeer, 10.23% on Texas, and 14.57% on Wisconsin. Simultaneously, the AUC and AP results of the AFD3S on non-attributed datasets also exhibit a certain degree of improvement.
Figure 4.
The average AUC of all models on attributed and non-attributed datasets (over 10 runs).
Figure 4.
The average AUC of all models on attributed and non-attributed datasets (over 10 runs).
Figure 5.
The average AP of all models on attributed and non-attributed datasets (over 10 runs).
Figure 5.
The average AP of all models on attributed and non-attributed datasets (over 10 runs).
The significant advantage is the importance of node features in node classification and graph classification tasks. The node features of attributed datasets provide direct, rich, and semantically clear information whose expressive power is often superior to node features generated based on random walks. This direct utilization of original node features helps improve the interpretability and stability of the model while reducing additional computational costs. Furthermore, the AFD3S incorporates the neighboring node features of the central node during the local feature augmentation process, enabling it to capture complex relationships between nodes. This approach fully utilizes the multi-source information of graph data, providing superior performance for downstream tasks of GNNs. Therefore, the superior performance of the AFD3S on various datasets demonstrates its effectiveness and practicability in link prediction tasks.
Computational Efficiency: To further validate the computational efficiency and scalability of the AFD3S, this paper selects three currently popular and performance-advanced SGRLs—SEAL, GCN+DE (distance encoding) [
60], and WalkPool, and conducts comparative experiments on all datasets. The comparative experiments mainly focus on four key indicators: preprocessing time, average training time (50 epochs), average inference time, and total runtime (50 epochs), aiming to comprehensively demonstrate the computational efficiency of the AFD3S in practical applications.
Table 4 and
Table 5 present the experimental results on non-attributed and attributed datasets. In these tables, “Train” represents the average training time for 50 epochs, “Inference” represents the average inference time, “Preproc.” represents the preprocessing time, and “Runtime” represents the average runtime for 50 epochs. The fastest values are bolded, and the maximum (minimum) speedup ratio in “Speed up” refers to the ratio of the time required by the slowest (fastest) SGRL methods to the AFD3S model.
Through experimental results, we can observe that the AFD3S proposed in this paper has achieved significant acceleration in training, inference, and running time compared with other SGRLs on all datasets. Specifically, the training speed is improved by 3.34 to 17.95 times, the inference speed is accelerated by 3 to 61.05 times, and the overall running time is shortened by 2.27 to 14.53 times. Although the preprocessing time of the AFD3S model is relatively high, due to the significant improvement in training and inference speeds, this difference is effectively offset, making the maximum acceleration reach 14.53 times on the Yeast dataset. It is worth noting that as the scale of the dataset increases, the computational time acceleration effect of the AFD3S becomes particularly evident. The highest acceleration multiples are achieved on the three large PubMed, PB, and Yeast datasets, demonstrating the excellent performance of the AFD3S in computational efficiency and scalability.
This gain is primarily attributed to the innovative strategies employed by the AFD3S. Adopting a random walk-based strategy to sample sparse subgraphs instead of enclosing subgraphs significantly reduces subgraphs’ storage and computational overhead. As the graph size increases, the scale of extracted subgraphs decreases from exponential to linear, reducing computational complexity and improving model efficiency. Additionally, the randomness in random walks brings additional regularization benefits to the model, further enhancing its performance. Meanwhile, the AFD3S utilizes easily pre-computed subgraph-level diffusion operators to replace expensive message-passing schemes through feature diffusion, significantly improving training and inference speeds. These optimization measures collectively enable the AFD3S to demonstrate excellent computational efficiency and scalability in link prediction tasks.
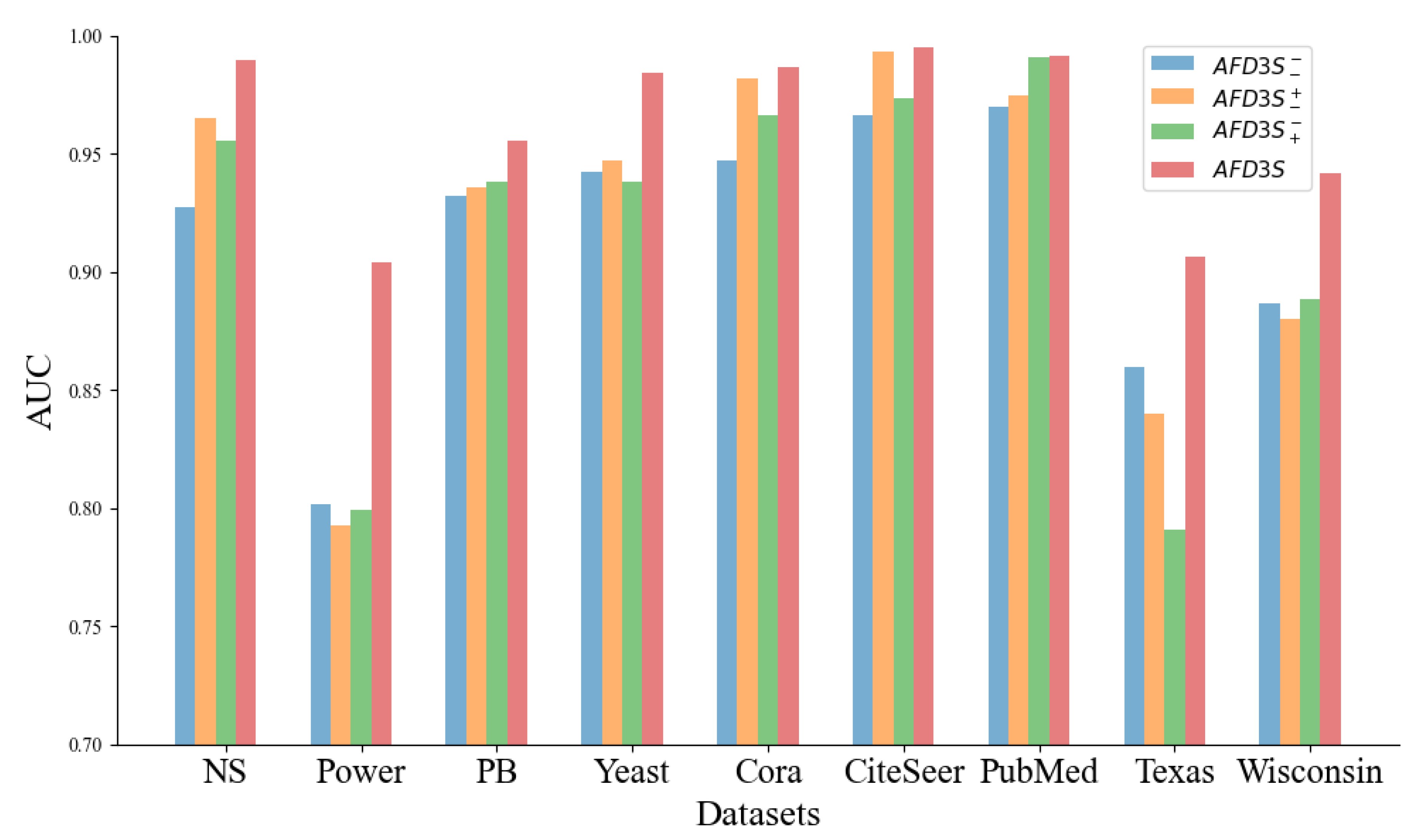
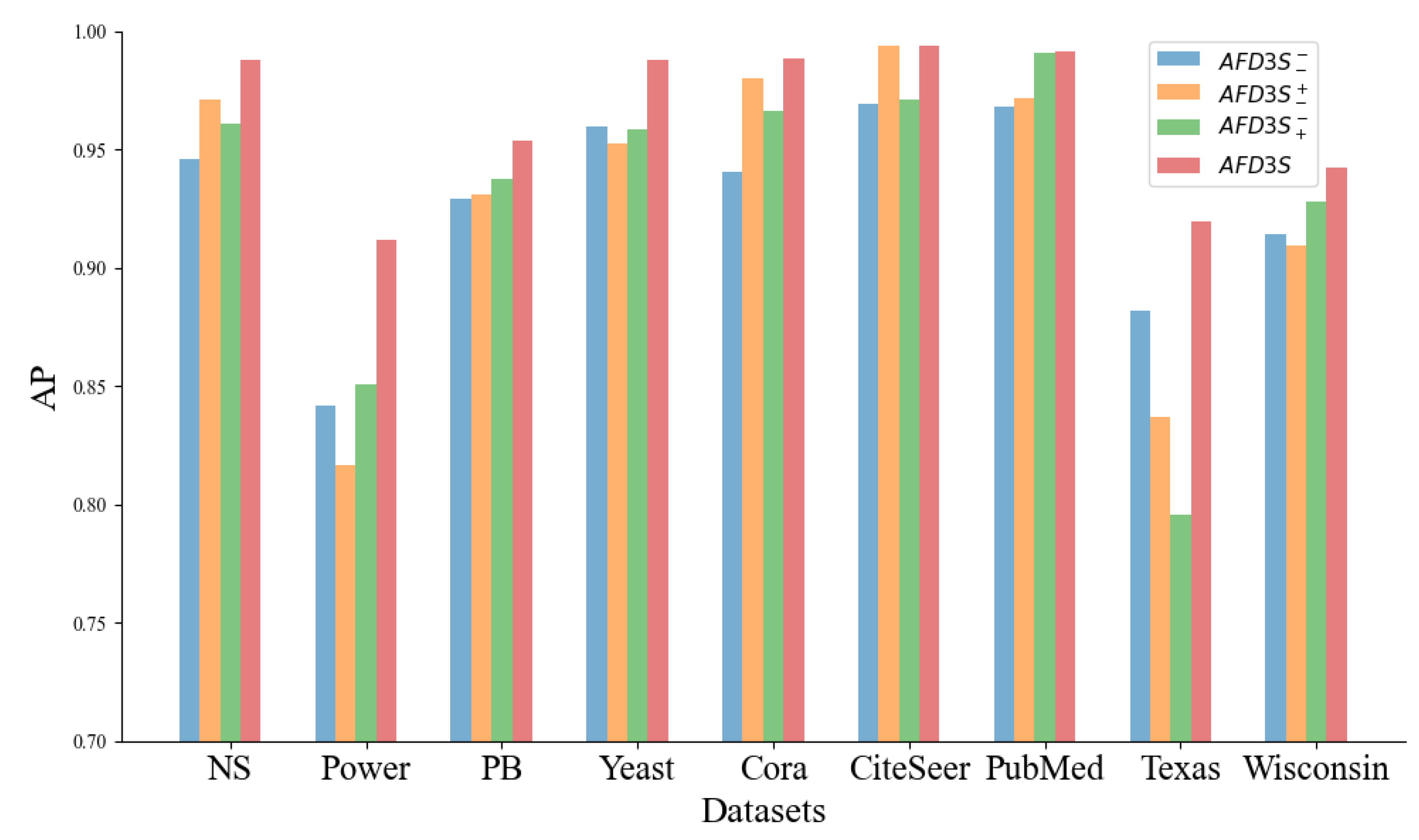
Ablation Study: We conducted an ablation study to further explore the key factors contributing to the performance gains of the AFD3S, specifically verifying the effectiveness of local feature augmentation and graph diffusion operations on the predictive performance of the AFD3S. Three variants of the AFD3S were designed and compared with the original AFD3S in terms of AUC and AP metrics for link prediction across all datasets. The three variants are: 1) Variant
, which does not perform local feature augmentation and graph diffusion operations; 2) Variant
, which performs local feature augmentation but does not perform graph diffusion operations; 3) Variant
, which performs graph diffusion operations but does not perform local feature augmentation. The experimental results are presented in
Figure 6 and
Figure 7.
Observing the experimental results, it can be seen that on most datasets, among the three variants of AFD3S, the link prediction performance of Variant using local augmentation alone and Variant using graph diffusion alone both show some degree of improvement compared to Variant without any augmentation. This proves the effectiveness of the two augmentation strategies proposed in this paper for AFD3S in link prediction tasks. Furthermore, when local feature augmentation and graph diffusion operations are used simultaneously (i.e., the original AFD3S), the performance is improved on all datasets, with significant improvements on most datasets. The main reason is that after local feature augmentation, the subsequent graph diffusion operation amplifies this augmentation effect, significantly improving subsequent prediction performance.
Specifically, since the AFD3S extracts sparsely sampled subgraphs of target node pairs to reduce subgraphs’ storage and computational efficiency, it cannot, in most cases, include all h-hop neighbor nodes of the target node pairs as enclosing subgraphs can. However, the local feature augmentation operation is based on the original input graph, fusing features from other neighbor nodes of the central node through feature augmentation, thus compensating for the shortcomings of sparse subgraphs. This augmentation is further amplified by the graph diffusion operation, and it can capture the structural relationships and similarities between nodes. At the same time, it simulates the information diffusion process between nodes through feature diffusion to simplify the information transmission and aggregation operations in subgraph representation learning, accelerating the training and inference speed of downstream tasks.
In summary, AFD3S improves the link prediction performance and reduces subgraphs’ storage and computational overhead, enhancing the model’s computational efficiency and scalability. It provides an efficient and practical solution for graph analysis tasks.
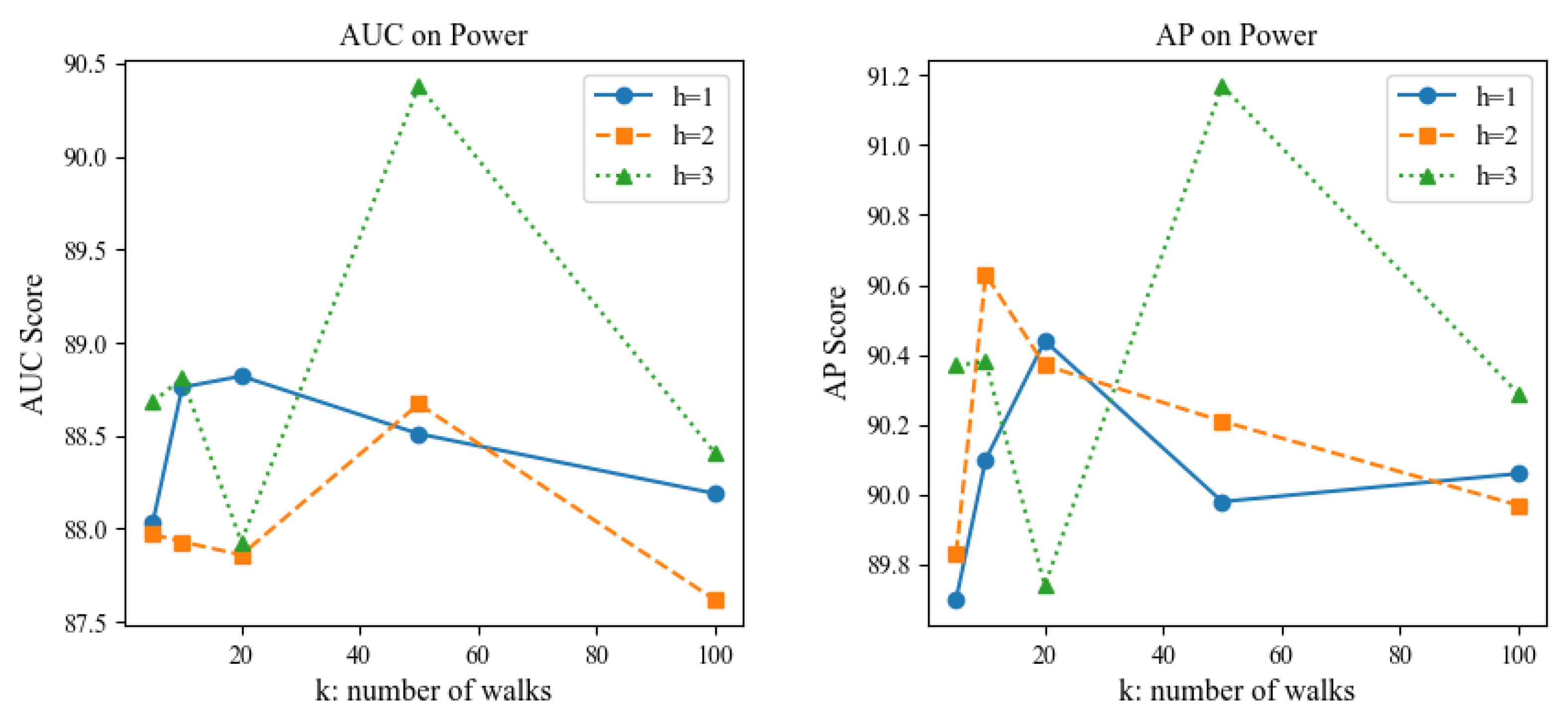
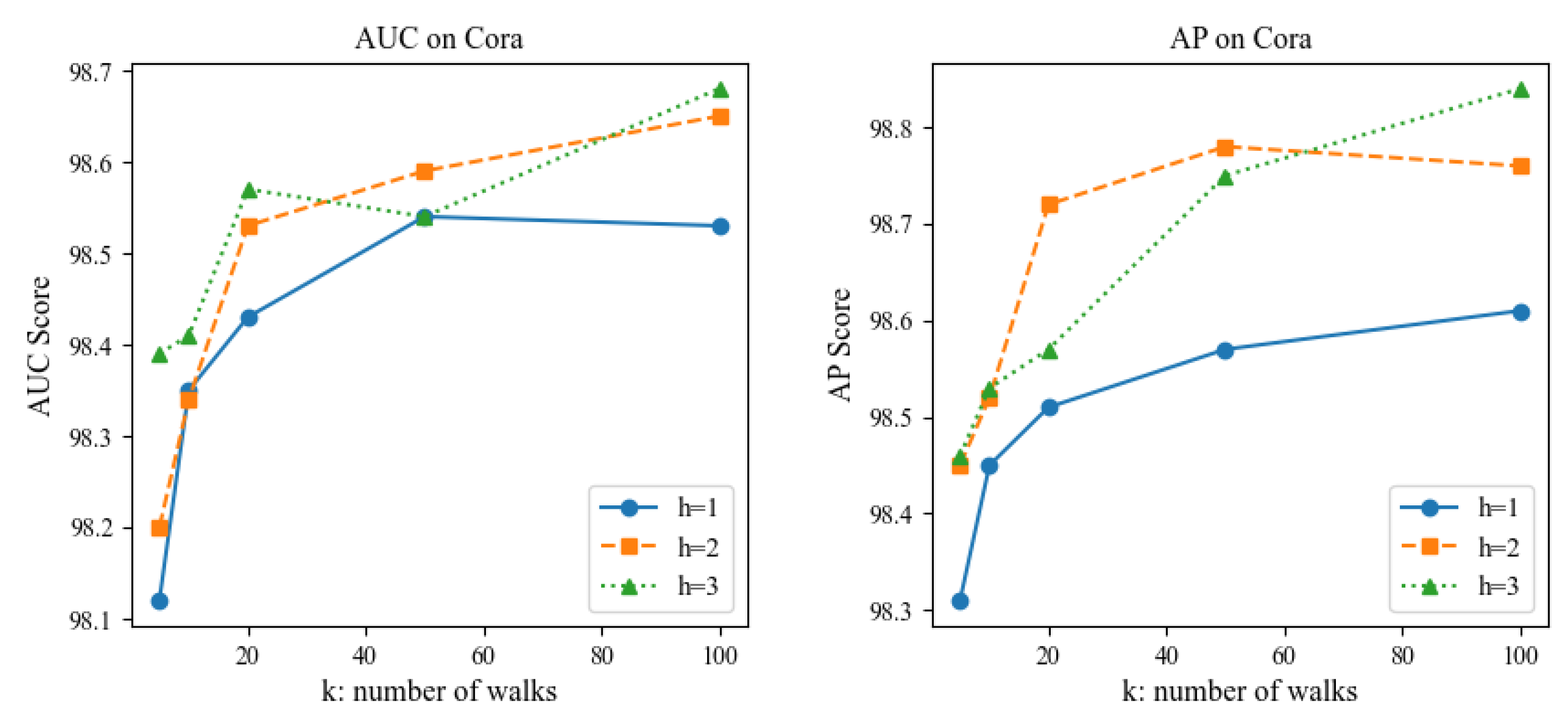
Parameter Sensitivity: We also conducted a sensitivity analysis on the two hyperparameters of the extracted sparsely sampled subgraph, namely the walk length
h and the number of walks
k, to investigate the impact of the size of the sampled subgraph on the link prediction performance of the AFD3S. Due to certain similarities across different datasets, experiments were performed on the Power non-attribute and Cora attribute datasets as examples. All other parameters remained unchanged in the experiments except for the test variable to maintain fairness. The experimental results are shown in
Figure 8 and
Figure 9.
The experimental results show that the AFD3S performs excellent prediction even when using smaller h and k values. Notably, when the extracted subgraph is too large, the prediction performance decreases compared to a smaller subgraph. This is likely because a larger subgraph may contain nodes and edges irrelevant to the target link prediction task. Such noise and irrelevant information may interfere with the model’s learning process, leading the model to capture incorrect patterns and thus reducing prediction accuracy. This finding further proves that the AFD3S can extract key information from sparse and small sampled subgraphs, significantly improving computational efficiency while ensuring prediction performance. Additionally, it demonstrates the superiority and scalability of AFD3S in handling large-scale graph data. The AFD3S can more effectively cope with large-scale graph data by reducing computational and storage overhead, providing strong support for practical applications.