Submitted:
22 July 2024
Posted:
23 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Re-Identification of the Trichoderma Harzianum Strains Deposited in the GenBank
2.2. Evaluation of the Dominant HCC Species from South and Central America
3. Results
3.1. Re-identification of the Trichoderma harzianum strains deposited in the GenBank
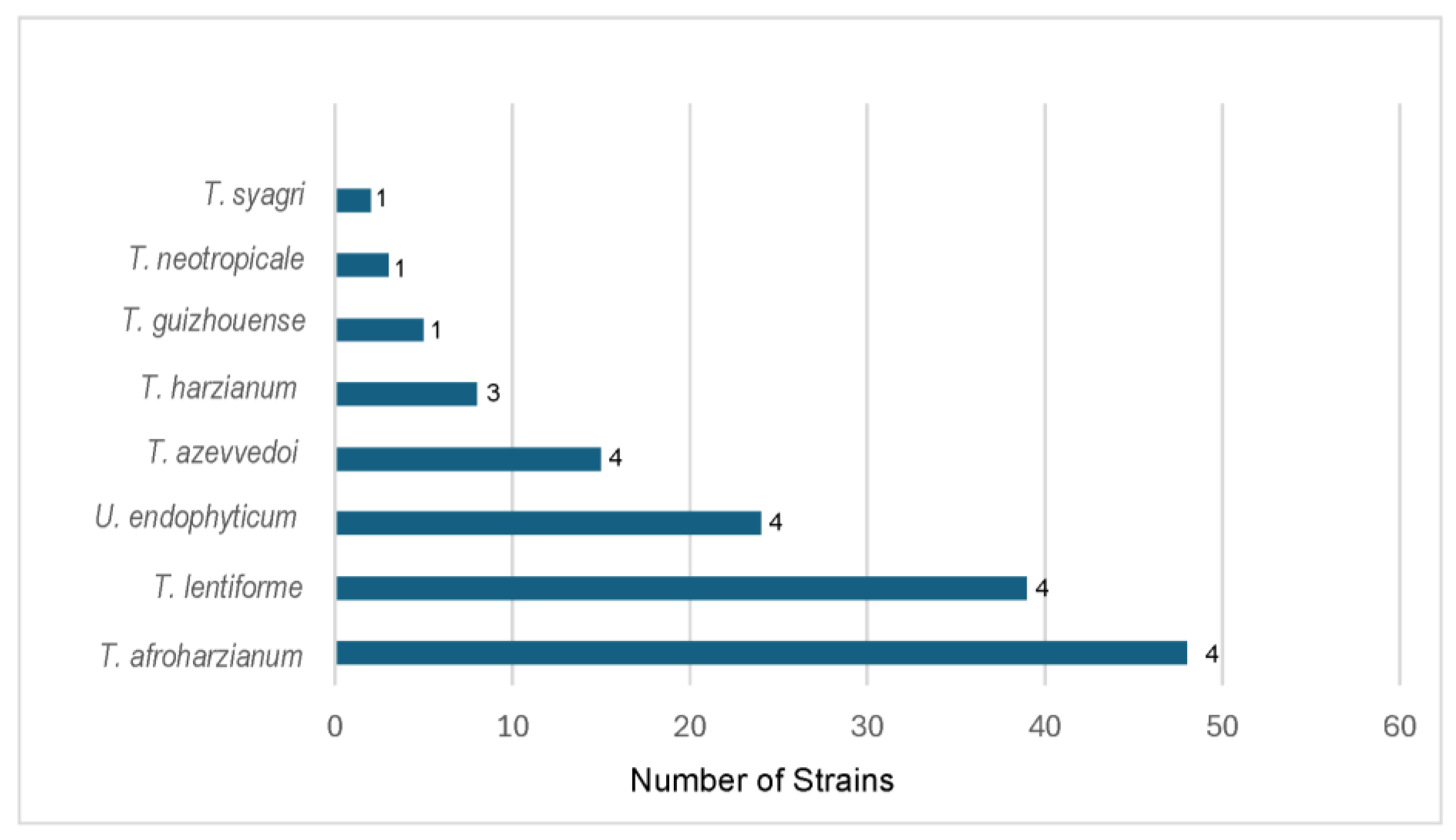
3.2. Evaluation of the dominant HCC species from South and Central America
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Samuels, G.J.; Hebbar, P.K. Trichoderma: Identification and Agricultural Applications; APS Press, 2015.
- Weindling, R. Trichoderma lignorum as a Parasite of Other Soil Fungi. Phytopathology 1932, 22, 837–845. [Google Scholar]
- Lindsey, D.L.; Baker, R. Effect of Certain Fungi on Dwarf Tomatoes Grown under Gnotobiotic Conditions. Phytopathology 1967, 57, 1262–1263. [Google Scholar]
- Chang, Y.-C.; Chang, Y.-C.; Baker, R.; Kleifeld, O.; Chet, I. Increased Growth of Plants in the Presence of the Biological Control Agent Trichoderma harzianum. Plant Dis. 1986, 70, 145–148. [Google Scholar] [CrossRef]
- Harman, G.E. Trichoderma—Not Just for Biocontrol Anymore. Phytoparasitica 2011, 39, 103–108. [Google Scholar] [CrossRef]
- Zin, N.A.; Badaluddin, N.A. Biological Functions of Trichoderma Spp. for Agriculture Applications. Ann. Agric. Sci. 2020, 65, 168–178. [Google Scholar] [CrossRef]
- Rifai, M.A. A Revision of the Genus Trichoderma. Mycol Pap. 1969, 116, 1–56. [Google Scholar]
- Chaverri, P.; Castlebury, L.A.; Samuels, G.J.; Geiser, D.M. Multilocus Phylogenetic Structure within the Trichoderma harzianum/Hypocrea lixii Complex. Mol. Phylogenet. Evol. 2003, 27, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Bissett, J.; Druzhinina, I.; Kullnig-Gradinger, C.; Szakacs, G. Genetic and Metabolic Diversity of Trichoderma: A Case Study on South-East Asian Isolates. Fungal Genet. Biol. 2003, 38, 310–319. [Google Scholar] [CrossRef] [PubMed]
- Chaverri, P.; Branco-Rocha, F.; Jaklitsch, W.; Gazis, R.; Degenkolb, T.; Samuels, G.J. Systematics of the Trichoderma harzianum Species Complex and the Re-Identification of Commercial Biocontrol Strains. Mycologia 2015, 107, 558–590. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Wang, R.; Sun, Q.; Wu, B.; Sun, J.-Z. Four New Species of Trichoderma in the Harzianum Clade from Northern China. MycoKeys 2020, 73, 109–132. [Google Scholar] [CrossRef]
- del Carmen, H. Rodríguez, M.; Evans, H.C.; de Abreu, L.M.; de Macedo, D.M.; Ndacnou, M.K.; Bekele, K.B.; Barreto, R.W. New Species and Records of Trichoderma Isolated as Mycoparasites and Endophytes from Cultivated and Wild Coffee in Africa. Sci. Rep. 2021, 11, 5671. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Qiao, M.; Lv, Y.; Du, X.; Zhang, K.-Q.; Yu, Z. New Species of Trichoderma Isolated as Endophytes and Saprobes from Southwest China. J. Fungi 2021, 7, 467. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.-T.; Zhuang, W.-Y. Seven New Species of Trichoderma Hypocreales in the Harzianum and Strictipile Clades. Phytotax 2017, 305, 121–139. [Google Scholar] [CrossRef]
- Zhang, Y.-B.; Zhuang, W.-Y. New Species of Trichoderma in the Harzianum, Longibrachiatum and Viride Clades. Phytotaxa 2018, 379, 131–142. [Google Scholar] [CrossRef]
- Cao, Z.-J.; Qin, W.-T.; Zhao, J.; Liu, Y.; Wang, S.-X.; Zheng, S.-Y. Three New Trichoderma Species in Harzianum Clade Associated with the Contaminated Substrates of Edible Fungi. J. Fungi 2022, 8, 1154. [Google Scholar] [CrossRef] [PubMed]
- Maddison, W.P. Mesquite: A Modular System for Evolutionary Analysis. Version 3.18. 2007. http://www.mesquiteproject.
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Hoyos-Carvajal, L.; Orduz, S.; Bissett, J. Genetic and Metabolic Biodiversity of Trichoderma from Colombia and Adjacent Neotropic Regions. Fungal Genet. Biol. 2009, 46, 615–631. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.; Beltrán, C.A.; Kusunoki, M.; Cotes, A.M.; Motohashi, K.; Kondo, T.; Deguchi, M. Diversity of Soil-Dwelling Trichoderma in Colombia and Their Potential as Biocontrol Agents against the Phytopathogenic Fungus Sclerotinia sclerotiorum (Lib.) de Bary. J. Gen. Plant Pathol. 2013, 79, 74–85. [Google Scholar] [CrossRef]
- Druzhinina, I.S.; Kopchinskiy, A.G.; Komoń, M.; Bissett, J.; Szakacs, G.; Kubicek, C.P. An Oligonucleotide Barcode for Species Identification in Trichoderma and Hypocrea. Fungal Genet. Biol. 2005, 42, 813–828. [Google Scholar] [CrossRef] [PubMed]
- Inglis, P.W.; Mello, S.C.; Martins, I.; Silva, J.B.; Macêdo, K.; Sifuentes, D.N.; Valadares-Inglis, M.C. Trichoderma from Brazilian Garlic and Onion Crop Soils and Description of Two New Species: Trichoderma azevedoi and Trichoderma peberdyi. PloS One 2020, 15, e0228485. [Google Scholar] [CrossRef] [PubMed]
- Barrera, V.A.; Iannone, L.; Romero, A.I.; Chaverri, P. Expanding the Trichoderma harzianum Species Complex: Three New Species from Argentine Natural and Cultivated Ecosystems. Mycologia 2021, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Hawksworth, D.L.; Crous, P.W.; Redhead, S.A.; Reynolds, D.R.; Samson, R.A.; Seifert, K.A.; Taylor, J.W.; Wingfield, M.J.; Abaci, Ö.; Aime, C.; et al. The Amsterdam Declaration on Fungal Nomenclature. IMA Fungus 2011, 2, 105–111. [Google Scholar] [CrossRef]
- Kubicek, C.P.; Steindorff, A.S.; Chenthamara, K.; Manganiello, G.; Henrissat, B.; Zhang, J.; Cai, F.; Kopchinskiy, A.G.; Kubicek, E.M.; Kuo, A.; et al. Evolution and Comparative Genomics of the Most Common Trichoderma species. BMC Genomics 2019, 20, 485. [Google Scholar] [CrossRef] [PubMed]
- van Lenteren, J. C.; Bueno, V. H. P.; Luna, M. G. Biological control in Latin America and the Caribbean: information sources, organizations, types and approaches in biological control. In J. C. V. Lenteren, V. H. P. Bueno, M. G. Luna, & Y. C. Colmeneraz (Eds.), Biological control in Latin America and the Caribbean: its rich history and bright future. (pp. 1-20). CABI 2020. [CrossRef]
- Xiao, Z.; Zhao, Q.; Li, W.; Gao, L.; Liu, G. Strain Improvement of Trichoderma harzianum for Enhanced Biocontrol Capacity: Strategies and Prospects. Front. Microbiol. 2023, 14, 1146210. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Wu, Z.; Zhong, Z.; Williams, J.; Jacobsen, S.E.; Sun, Z.; Tang, Y. Assessing the Biosynthetic Inventory of the Biocontrol Fungus Trichoderma afroharzianum T22. J. Agric. Food Chem. 2023, 71, 11502–11519. [Google Scholar] [CrossRef]
- Silva, L.G.; Camargo, R.C.; Mascarin, G.M.; Nunes, P.S. de O.; Dunlap, C.; Bettiol, W. Dual Functionality of Trichoderma: Biocontrol of Sclerotinia sclerotiorum and Biostimulant of Cotton Plants. Front. Plant Sci. 2022, 13, 983127. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhuang, W.-Y. Discovery from a Large-Scaled Survey of Trichoderma in Soil of China. Sci. Rep. 2017, 7, 9090. [Google Scholar] [CrossRef] [PubMed]
- Jambhulkar, P.P. , Singh, B., Raja, M. et al. Genetic diversity and antagonistic properties of Trichoderma strains from the crop rhizospheres in southern Rajasthan, India. Sci Rep 2024, 14, 8610. [Google Scholar] [CrossRef]
- Mirzaeipour, Z.; Bazgir, E.; Zafari, D.; Darvishnia, M. Isolation and identification of Harzianum clade species of Trichoderma from Khorramabad County. Mycologia Iranica 2023, 10, 67–78. [Google Scholar] [CrossRef]
- Tang, G.T.; Li, Y; Zhou, Y; Zhou, Y.; Zhu, Y.H.; Zheng, X.J.; Chang, X.L.; Zhang, S. R.; Gong, G.S. Diversity of Trichoderma species associated with soil in the Zoige alpine wetland of Southwest China. Sci. Rep. 2022, 12, 21709. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Lu, Q; Zhao, Q.; Wang, J.; Ouyang, H. Effects of alpine wetland landscapes on regional climate on the Zoige Plateau of China. Advances in Meteorology. 2013, 972430. 7pp. [CrossRef]
- du Plessis, I.L.; Druzhinina, I.S; Atanasova, L.; Yarden O.; Jacobs, K. The diversity of Trichoderma species from soil in South Africa, with five new additions, Mycologia, 2018. [CrossRef]
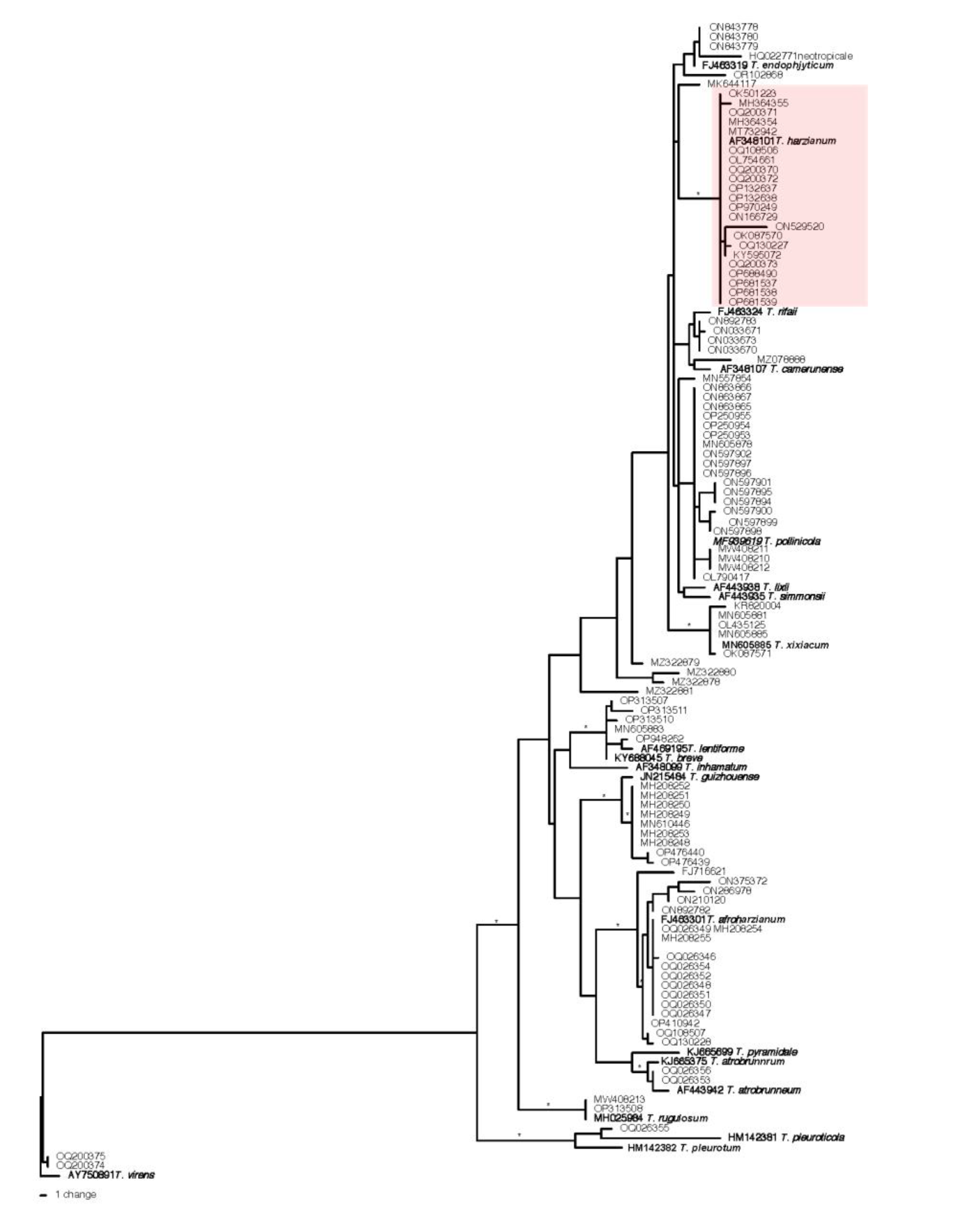
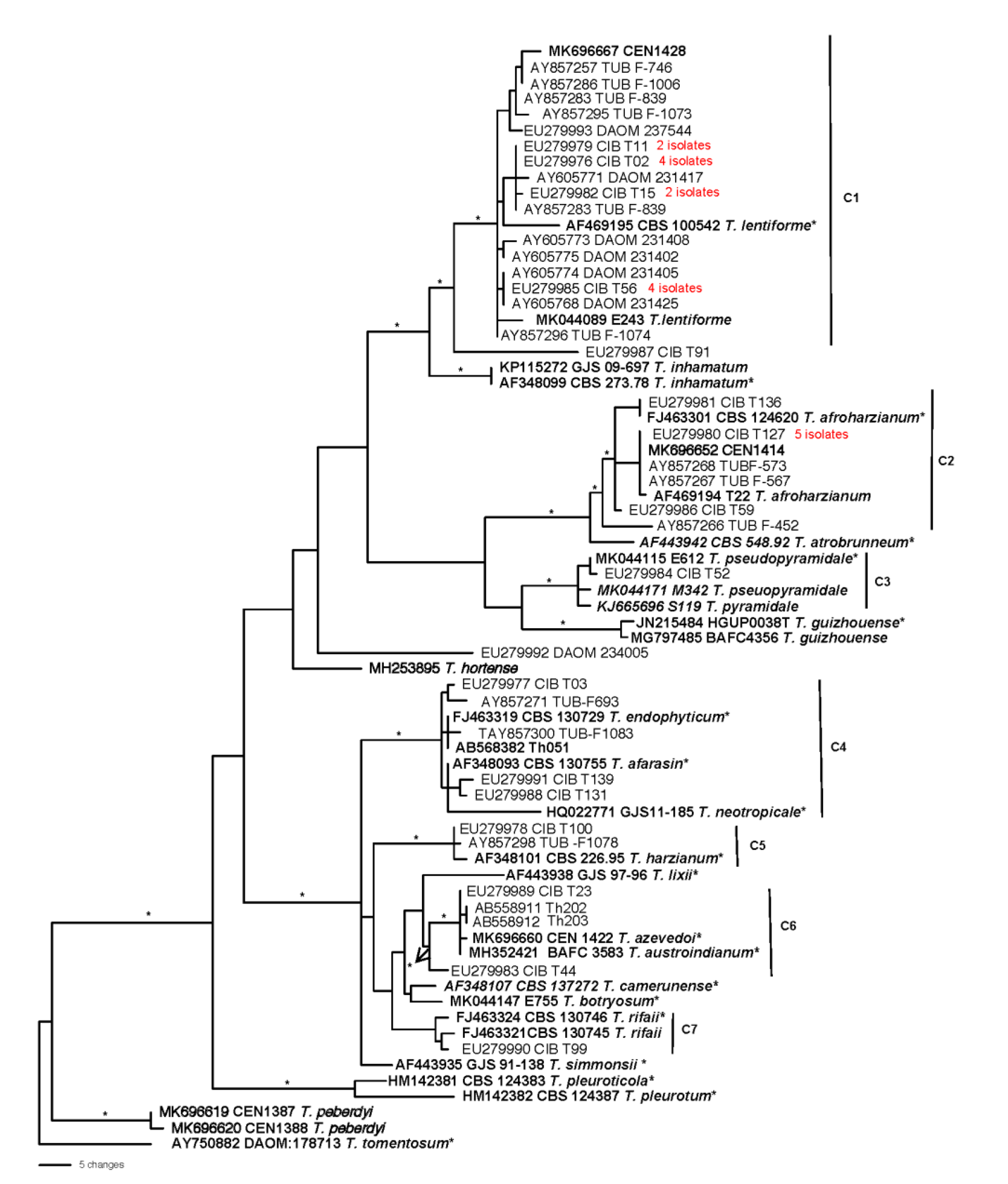
| Trichoderma species | origin | Strain # | Tef1α | ITS | rpb2 | No. of strains |
|---|---|---|---|---|---|---|
| T. lentiformea | Brazil | CEN1428 | MK666796 | MK714909 | MK696827 | |
| Brazil | TUB F-746 | AY857257 | AY857216 | X | ||
| Colombia | CIB T11 | EU279976 | EU280079 | X | 2b | |
| Colombia | CIB T02 | EU279976 | EU280079 | X | 4 | |
| Mexico | DAOM 231417 | AY605771 | AY605728 | X | ||
| Colombia | CIB T15 | EU279982 | EU280079 | X | 2 | |
| Mexico | DAOM 231408 | AY605773 | AY605730 | X | ||
| Mexico | DAOM 231402 | AY605775 | AY605732 | X | ||
| Mexico | DAOM 231439 | EU279994 | AY605728 | X | ||
| Colombia | CIB T91 | EU279987 | EU280079 | X | ||
| Mexico | DAOM 231405 | AY605774 | AY605731 | X | ||
| Colombia | CIB T56 | EU279985 | EU280079 | X | 4 | |
| Peru | DAOM 237544 | EU279993 | EU280133 | X | ||
| Mexico | DAOM 231425 | AY605768 | AY605725 | X | ||
| T. lentiforme | Cameroon | E243 | MK044089 | X | MK044182 | |
| Mexico | TUB F-839 | AY857283 | AY857231 | X | ||
| Brazil | TUB F-1073 | AY857295 | AY857247 | X | ||
| Brazil | TUB F-1006 | AY857286 | AY857235 | X | ||
| T. lentiforme* | French Guiana | CBS 100542 | AF469195 | AF469189 | X | |
| Argentina | TUB F-1074 | AY857296 | AY857248 | |||
| T. inhamatum | Peru | G.J.S. 09-697 | KP115272 | X | X | |
| T. inhamatum* | Colombia | CBS 273.78 | AF348099 | FJ442680 | FJ442725 | |
| Peru | DAOM 234005 | EU279992 | EU280091 | X | ||
| T. hortense* | Argentina | BAFC_cult_4291 | MH253895 | X | X | |
| Colombia | CIB T136 | EU279981 | EU280078 | X | ||
| Colombia | CIB T127 | EU279980 | EU280078 | X | 5 | |
| T. afroharzianum | Colombia | T22 | AF469194 | AF469188 | X | |
| T. afroharzianum | Brazil | CEN1414 | MK696652 | MK714894 | MK696813 | |
| T. afroharzianum* | Peru | CBS 124620 | FJ463301 | FJ442265 | FJ442691 | |
| Colombia | CIB T59 | EU279986 | EU280078 | |||
| T. pseudopyramidale* | COAD 2420 | MK044115 | X | MK044208 | ||
| T. pseudopyramidale | COAD 2439 | MK044171 | X | MK044264 | ||
| T. pyramidale | S119 | KJ665696 | X | X | ||
| T. atrobrunneum* | France | CBS 548.92 | AF443942 | AF443924 | X | |
| Peru | CIB T52 | EU279984 | EU280077 | |||
| T. guizhouense* | China | CBS 131803 | JN215484 | JN191311 | JQ901400 | |
| T. guizhouense | Argentina | BAFC 4356 | MG797485 | X | X | |
| Peru | TUB F-567 | AY857267 | AY857208 | X | ||
| Peru | TUB F-452 | AY857266 | AY857206 | X | ||
| Peru | TUB F-573 | AY857268 | AY857209 | X | ||
| Colombia | CIB T03 | EU279977 | EU280079 | X | ||
| T. endophyticum* | Ecuador | CBS 130729 | FJ463319 | FJ442243 | X | |
| T. afarasin* | Cameroon | CBS 130755 | AF348093 | AY027784 | X | |
| Guatemala | TUB F-693 | AY857271 | AY857211 | X | ||
| Mexico | TUB F-1083 | AY857300 | AY857253 | X | ||
| Colombia | CIB T139 | EU279991 | EU280075 | X | ||
| Colombia | Th051 | AB568382 | X | AB568476 | ||
| Colombia | CIB T131 | EU279988 | EU280075 | X | ||
| Colombia | CIB T100 | EU279978 | EU280079 | X | ||
| T. harzianum* | U.K. | CBS 226.95 | AF348101 | AJ222720 | AF545549 | |
| Mexico | TUB F-1078 | AY857298 | AY857250 | X | ||
| T. lixii** | Thailand | CBS 110080 | AF443938 | AF443920 | X | |
| T. camerunense* | Cameroon | CBS 137272 | AF348107 | AY027780 | X | |
| T. botryosum | Ethiopia | COAD 2526 | MK044147 | X | MK044240 | |
| Colombia | CIB T23 | EU279989 | EU280077 | X | ||
| Colombia | Th202 | AB558911 | X | AB558921 | ||
| Colombia | Th203 | AB558912 | X | AB558922 | ||
| T. austroindianum | Argentina | BAFC 3583 | MH352421 | X | X | |
| Colombia | CIB T44 | EU279983 | EU280077 | X | ||
| T. azevedoi | Brazil | CEN1422 | MK696660 | MK714901 | MK696821 | |
| T. rifaii* | Ecuador | CBS 130746 | FJ463324 | FJ442663 | X | |
| T. rifaii | Panama | CBS 130745 | FJ463321 | FJ442621 | FJ442720 | |
| Colombia | CIB T99 | EU279990 | EU280103 | X | ||
| T. simmonsii* | USA, MD | CBS 130431 | AF443935 | AF443917 | FJ442757 | |
| T. neotropicale* | Peru | G.J.S. 11-185 | HQ022771 | HQ022407 | X | |
| T. aggressivum | Northern Ireland | CBS 433.95 | AF348097 | FJ442605 | FJ442704 | |
| T. aggressivum | U.K. | CBS 100525 | AF348095 | AF057600 | AF545541 | |
| T. pleuroticola* | South Korea | CBS 124383 | HM142381 | HM142362 | HM142371 | |
| T. pleuroti* | South Korea | CBS 124387 | HM142382 | HM142363 | HM142372 | |
| T. peberdyi | Brazil | CEN1387 | MK696619 | MK714861 | MK696781 | |
| T. peberdyi | Brazil | CEN1388 | MK696620 | MK714862 | MK696782 | |
| T. tomentusom* | Canada | DAOM 178713a | AY750882 | EU330958 | AF545557 |
| Species | Strain number | Number of isolates | tef1α accession number | Country | Ref. |
|---|---|---|---|---|---|
| T. afroharzianum | CIB T136 | 1 | EU279981 | Colombia | 19 |
| CIB T07, CIB T63, CIB T61, CIB T53, CIB T127 | 5 | EU279980* | Colombia | 19 | |
| CIB 59 | 1 | EU279986 | Colombia | 19 | |
| CEN, 1410, CEN1414, CEN1417 | 3 | MK696648* | Brazil | 22 | |
| TUB F-567, TUB F-573, TUB F-452 | 3 | AY857267, AY857268, AY857266 | Peru | 21 | |
| BAFC 4374, BAF 4392, for the rest see the reference | 35 | MH395411, MH395415 | Argentina | 23 | |
| Total | 48 | ||||
| T. lentiforme | DAOM 237544 | 1 | EU279993 | Peru | 19 |
| CIB T02, CIB T112, CIB T35, JB M10-2 | 4 | EU279976* | Mexico, Colombia | 19 | |
| CIB T15, CIB T41 | 2 | EU279982* | Colombia | 19 | |
| DAOM 231417 | 1 | AY605771 | Mexico | 19 | |
| DAOM 231439 | 1 | EU279994 | Mexico | 19 | |
| DAOM 231408 | 1 | AY605773 | Mexico | 19 | |
| DAOM 231405 | 1 | AY605774 | Mexico | 19 | |
| DAOM 231425 | 1 | AY605768 | Mexico | 19 | |
| CIB T56, CIB T60, CIB T16,DAOM 229985 | 4 | EU279985* | Panama, Colombia | 19 | |
| CIB T91 | 1 | EU279987 | |||
| CIB T11, CIB T102 | 2 | EU279979* | Colombia | 19 | |
| DAOM 231402 | 1 | AY605775 | Mexico | 19 | |
| CEN1412, CEN1415, CEN1416, CEN1428, CEN1429 | 5 | MK696650, MK696653, MK696654, MK696668, MK696667 | Brazil | 22 | |
| TUB F-839, TUB F-1073, TUB F-1006, TUB F-746, TUB F-1074 | 5 | AY857283, AY857295, AY857286, AY857257, AY857296 | México, Brazil, Brazil, Brazil, Argentina | 21 | |
| BAFC 4391, BAFC 4394, for the rest see the reference | 9 | MH036883, MH036885 | Argentina | 23 | |
| Total | 39 | ||||
| T. endophyticuma | CIB T03, CIB T31, CIB T139 | 3 | EU279977, EU279988, EU279991 | Colombia | 19 |
| TUB F-1083, TUB F-693 | 2 | AY857300, AY857271 | Mexico, Guatemala | 21 | |
| Th051 | 1 | AB568382 | Colombia | 20 | |
| BAFC 4358, BAFC 4372, for the rest see the reference | 18 | MH371393, MH371397 | Argentina | 23 | |
| Total | 24 | ||||
| T. azevedoi | CIB T23, CIB T24, CIB T126, CIB T128 | 4 | EU279989* | Colombia | 19 |
| CIB T44 | 1 | EU279983 | Colombia | 19 | |
| CEN1422, CEN1423, CEN1403 | 3 | MK696638* | Brazil | 22 | |
| Th202, Th203 | 2 | AB558911* | Colombia | 20 | |
| BAFC 3583, BAFC 3844, GJS 08-128, GJS 08-181, VAB-T051 | 5 | MH352421, MG822709, MH352423, MH352422, MH352424 | Argentina | 23 | |
| Total | 15 | ||||
| T. harzianum | CIB T100, PER4-2 | 2 | EU279978* | Colombia, Peru | 19 |
| TUB F-1078 | 1 | AY857298 | Mexico | 21 | |
| GJS 08-172, GJS 08-173, VAB-T032, VAB-T052, VAB-T053 | 5 | KT275197, KT275198, KT275199, MH364354, MH364355 | Argentina | 23 | |
| Total | 8 | ||||
| T. guizhouense | BAFC 4356, BAFC 4370, GJS 08-102, GJS 08-121, VAB-T047 | 5 | MG797485, MG797486, MG797484, MG797483, MG797482 | Argentina | 23 |
| T. neotropicale | GJS 08-182, GJS 08-183, VAB-T049 | 3 | MG822718, MG822719, MG822720 | Argentina | 23 |
| T. syagri | BAFC 4357, BAFC 4371 | 2 | MG227714, MG227710 | Argentina | 23 |
| T. pseudopyramidale | CIB T52 | 1 | EU279984 | Peru | 19 |
| T. hortense | GJS 08-116 | 1 | MH253895 | Argentina | 23 |
| T. rifaii | CIB T99 | 1 | EU279990 | Colombia | 19 |
| T. sp. | JB PER6-2 | 1 | EU279992 | Peru | 19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





