Submitted:
21 July 2024
Posted:
24 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
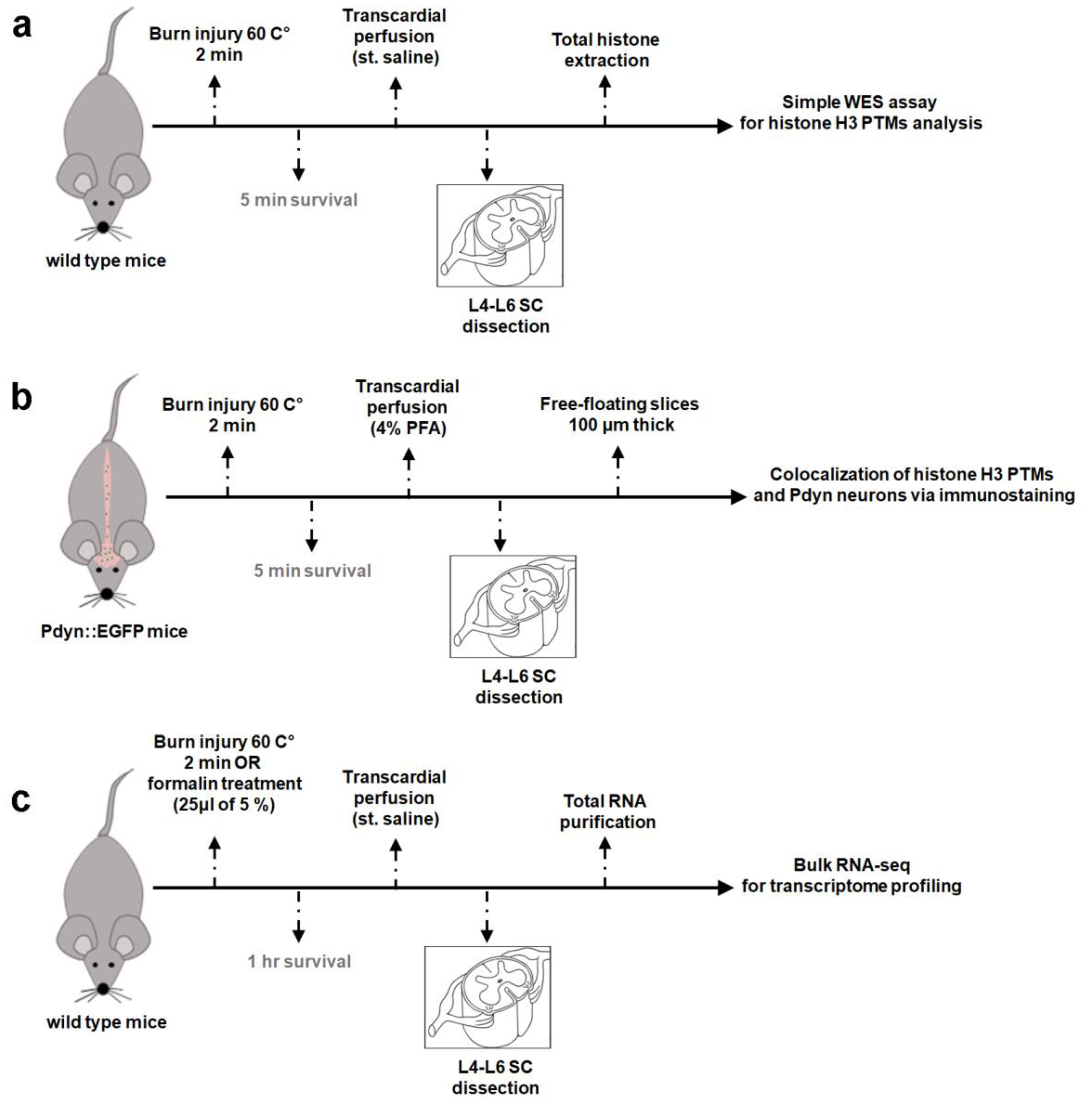
2.1. Burn injury Elevates Methylation Levels Of Histone H3 at Lysine 4 (H3K4) In The Spinal Cord Of Mice As Determined By An Automated Capillary-Based Size Sorting Instrument (WES)
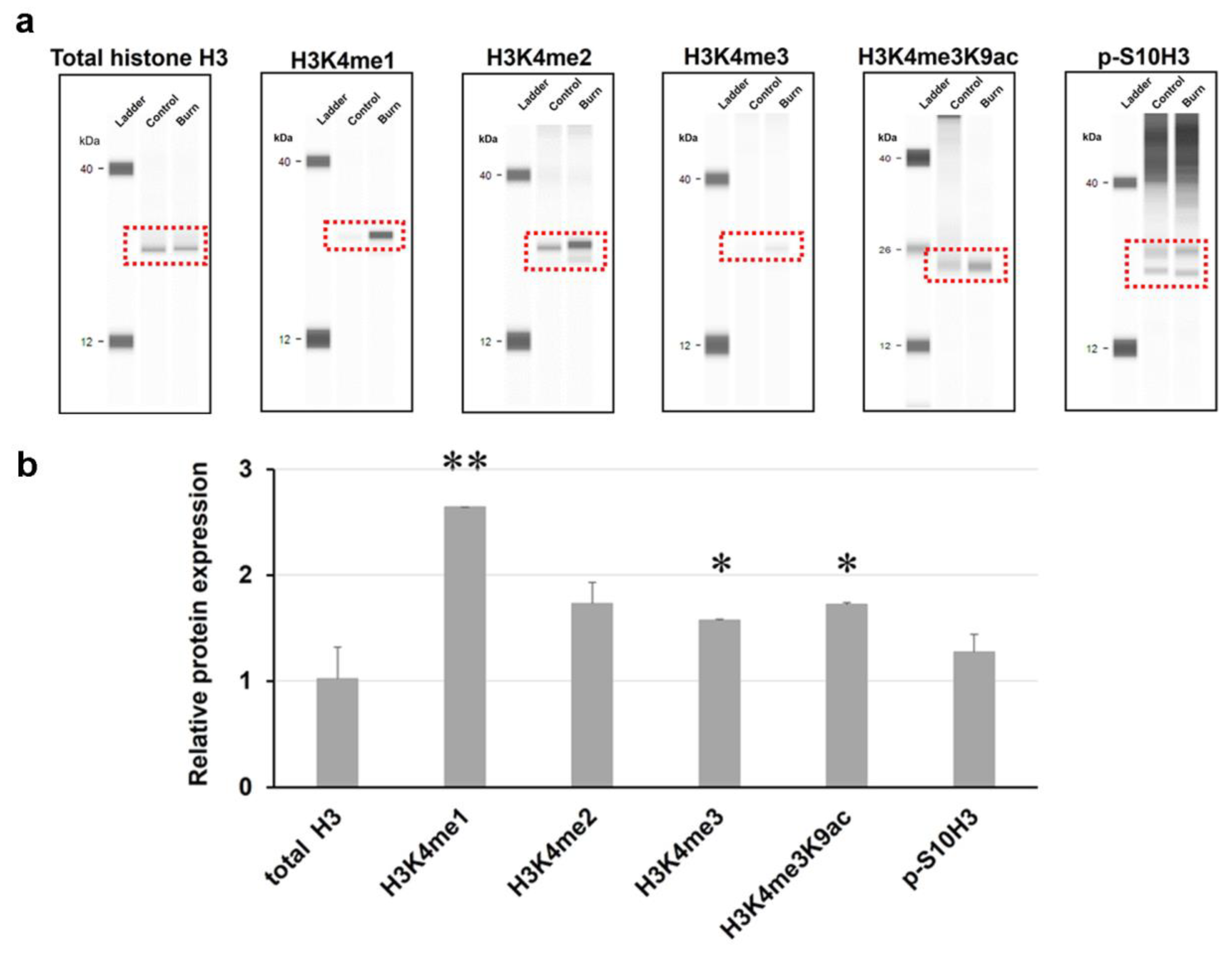
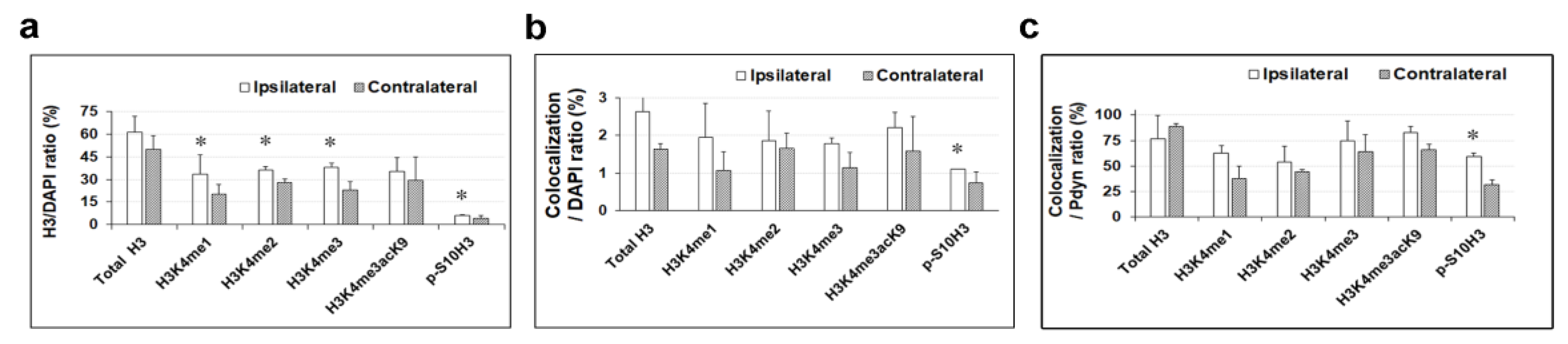
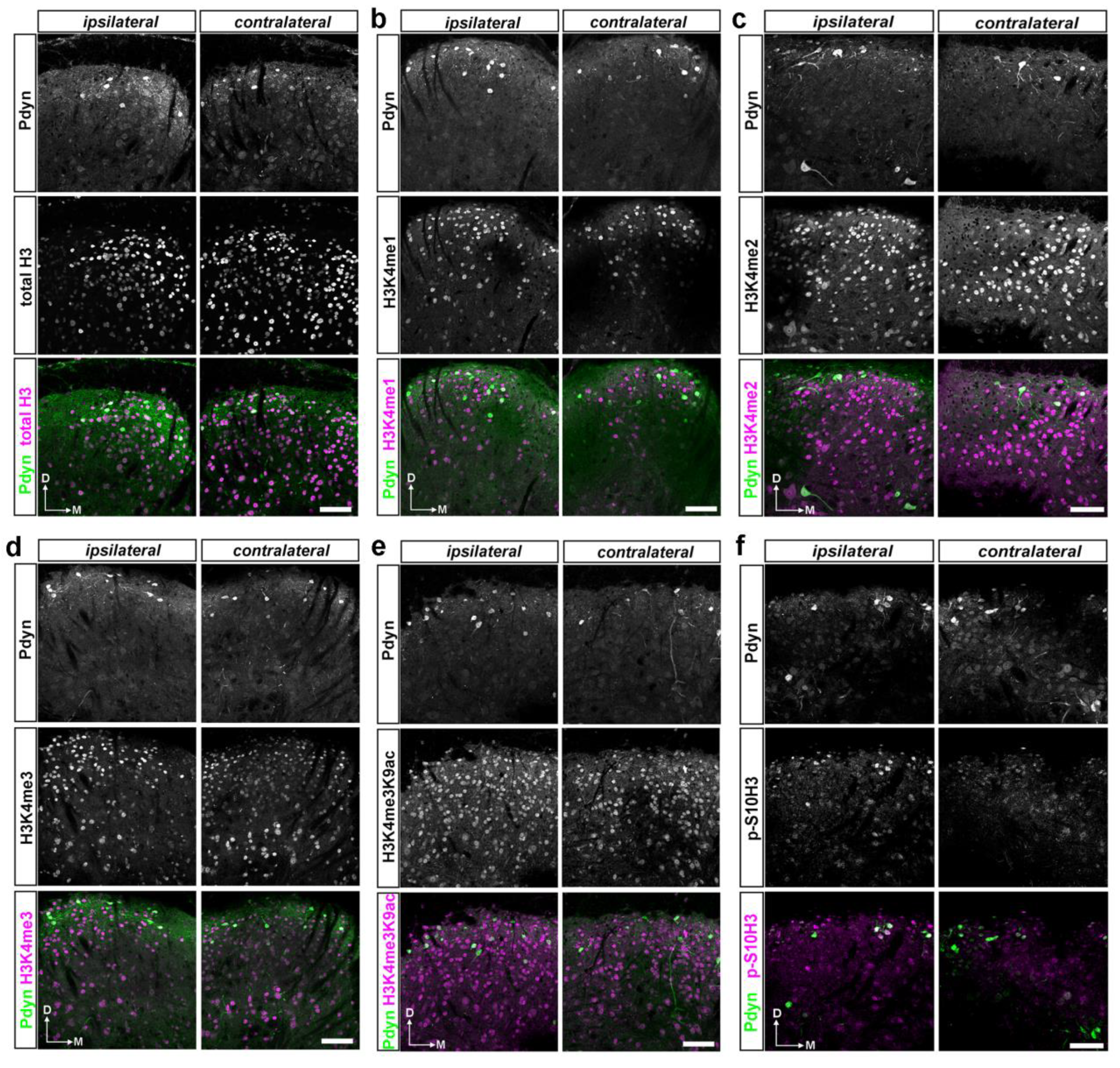
2.2. A dual Immunofluorescence Assay Revealed A Notable Increase In The Proportion Of Cells Containing Methylated Tags Of H3K4 and p-S10H3 After Burn Injury
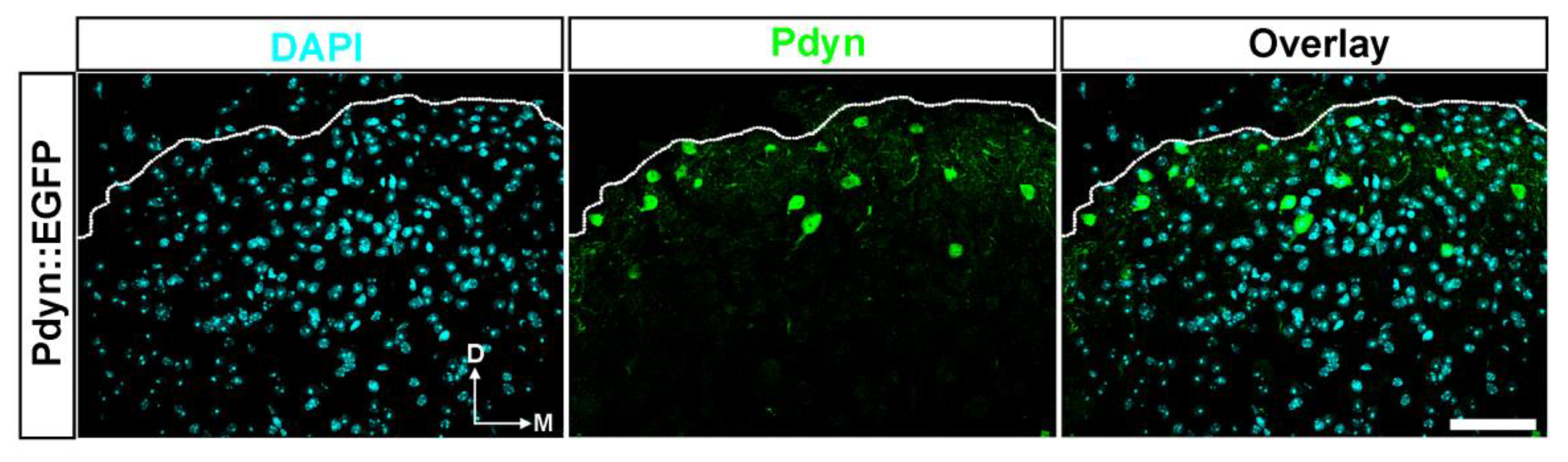
2.3.~3%. of Cells in the SDH of Mice Were Identified as Pdyn Neurons
2.4. A Double Immunofluorescence Assay Confirmed That Spinal Pdyn Neurons Contribute To Burn Injury-Induced Central Sensitization via the p-S10H3 Pathway
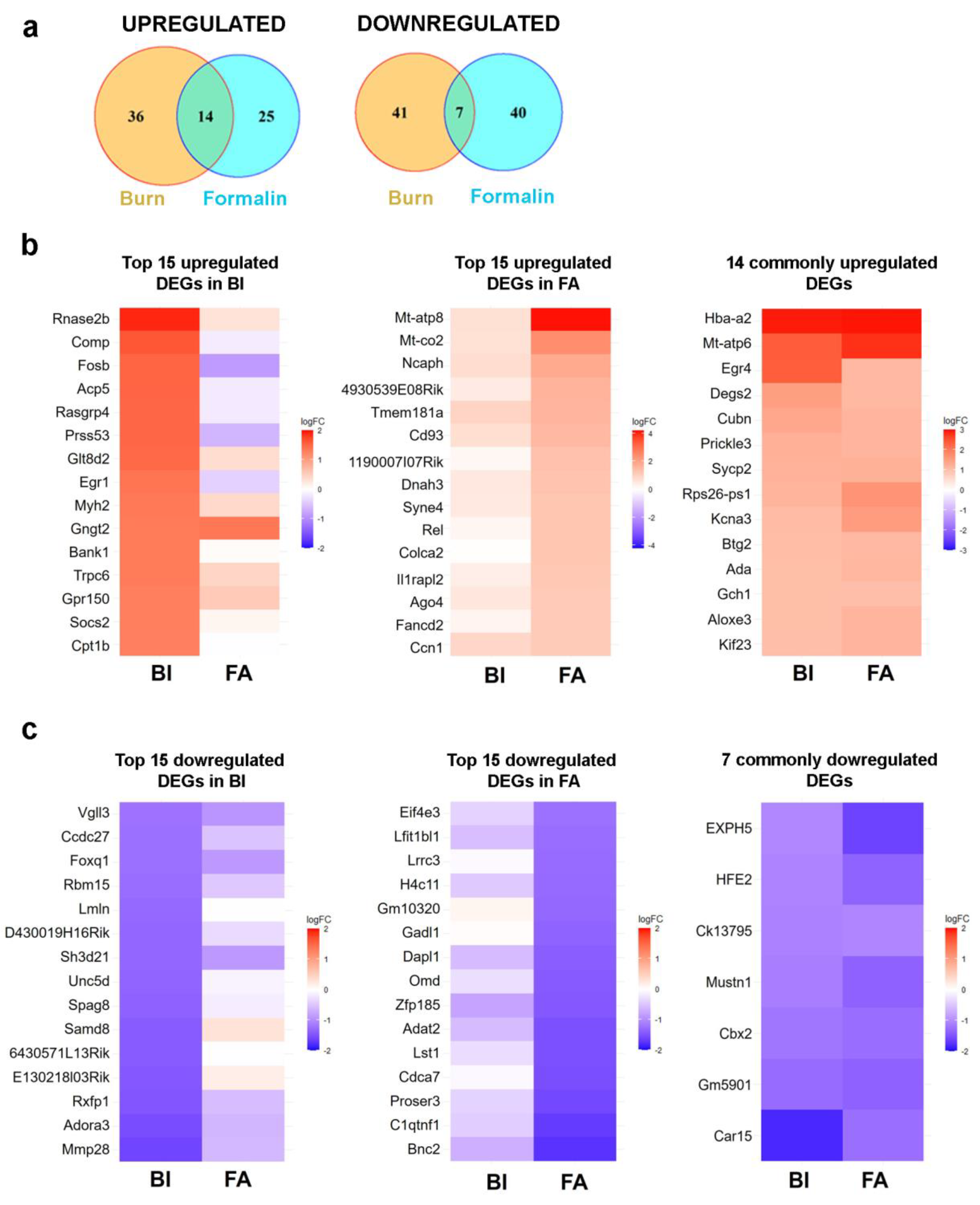
2.5. There is Limited Overlap In Differentially Expressed Genes (Degs) Between Burn Injury And Inflammatory Pain Models As Assessed By Transcriptome Analysis
2.5.1. Burn Injury-Induced Differential Gene Expression in The Spinal Cord
2.5.2. Formalin-Induced Differential Gene Expression in The Spinal Cord
2.5.3. Common Transcriptional Responses to Burn And Inflammatory Pain in Mouse Spinal Cord
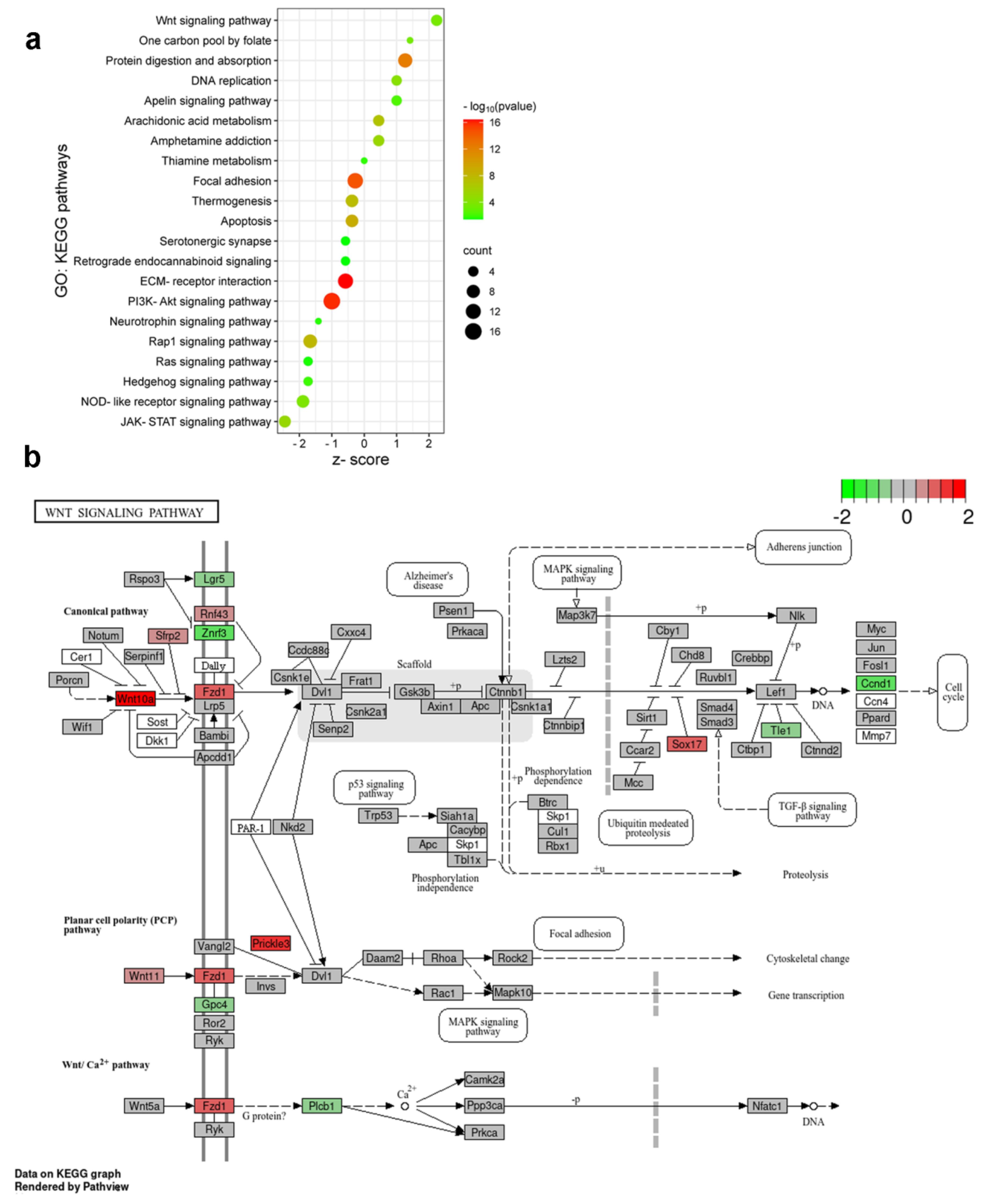
2.6. Enrichment Analysis Revealed The Activation Of Distinct Molecular Pathways In Burn Injury And Inflammatory Pain Models
2.7. Initial Components of the Wnt Signaling Cascade Are Involved In Burn Injury-Induced Nociception in the Spinal Cord of Mice
3. Discussion
4. Materials and Methods
4.1. Animals and Ethical Considerations
4.2. Establishment of Severe Scalding-Type Burn Injury Model
4.3. Establishment of Formalin-Induced Inflammatory Pain Model
4.4. Use of Capillary Western Immunoassay (Wes) for Quantification of Histone H3 PTMs Levels
4.5. Double Immunofluorescent Staining
4.6. Total RNA Isolation and RNA-seq Analysis
4.7. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sneddon, L.U. Comparative Physiology of Nociception and Pain. Physiol. Bethesda Md 2018, 33, 63–73. [Google Scholar] [CrossRef]
- Buchheit, T.; de Ven, T.V.; Shaw, A. Epigenetics and the Transition from Acute to Chronic Pain. Pain Med. Malden Mass 2012, 13, 1474–1490. [Google Scholar] [CrossRef]
- Descalzi, G.; Ikegami, D.; Ushijima, T.; Nestler, E.J.; Zachariou, V.; Narita, M. Epigenetic Mechanisms of Chronic Pain. Trends Neurosci. 2015, 38, 237–246. [Google Scholar] [CrossRef]
- Roth, T.L.; Roth, E.D.; Sweatt, J.D. Epigenetic Regulation of Genes in Learning and Memory. Essays Biochem. 2010, 48, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Sultan, F.A.; Day, J.J. Epigenetic Mechanisms in Memory and Synaptic Function. Epigenomics 2011, 3, 157–181. [Google Scholar] [CrossRef] [PubMed]
- Géranton, S.M.; Tochiki, K.K. Regulation of Gene Expression and Pain States by Epigenetic Mechanisms. Prog. Mol. Biol. Transl. Sci. 2015, 131, 147–183. [Google Scholar] [CrossRef]
- Torres-Perez, J.V.; Irfan, J.; Febrianto, M.R.; Giovanni, S.D.; Nagy, I. Histone Post-Translational Modifications as Potential Therapeutic Targets for Pain Management. Trends Pharmacol. Sci. 2021, 42, 897–911. [Google Scholar] [CrossRef] [PubMed]
- Tochiki, K.K.; Maiarú, M.; Norris, C.; Hunt, S.P.; Géranton, S.M. The Mitogen and Stress-Activated Protein Kinase 1 Regulates the Rapid Epigenetic Tagging of Dorsal Horn Neurons and Nocifensive Behaviour. Pain 2016, 157, 2594–2604. [Google Scholar] [CrossRef]
- Torres-Pérez, J.V.; Sántha, P.; Varga, A.; Szucs, P.; Sousa-Valente, J.; Gaal, B.; Sivadó, M.; Andreou, A.P.; Beattie, S.; Nagy, B.; et al. Phosphorylated Histone 3 at Serine 10 Identifies Activated Spinal Neurons and Contributes to the Development of Tissue Injury-Associated Pain. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Varga, A.; Mészár, Z.; Sivadó, M.; Bácskai, T.; Végh, B.; Kókai, É.; Nagy, I.; Szücs, P. Spinal Excitatory Dynorphinergic Interneurons Contribute to Burn Injury-Induced Nociception Mediated by Phosphorylated Histone 3 at Serine 10 in Rodents. Int. J. Mol. Sci. 2021, 22, 1–28. [Google Scholar] [CrossRef]
- Mészár, Z.; Kókai, É.; Varga, R.; Ducza, L.; Papp, T.; Béresová, M.; Nagy, M.; Szücs, P.; Varga, A. CRISPR/Cas9-Based Mutagenesis of Histone H3.1 in Spinal Dynorphinergic Neurons Attenuates Thermal Sensitivity in Mice. Int. J. Mol. Sci. 2022, 23. [Google Scholar] [CrossRef] [PubMed]
- Lo, W.S.; Trievel, R.C.; Rojas, J.R.; Duggan, L.; Hsu, J.Y.; Allis, C.D.; Marmorstein, R.; Berger, S.L. Phosphorylation of Serine 10 in Histone H3 Is Functionally Linked in Vitro and in Vivo to Gcn5-Mediated Acetylation at Lysine 14. Mol. Cell 2000, 5, 917–926. [Google Scholar] [CrossRef]
- Tossetta, G.; Piani, F.; Borghi, C.; Marzioni, D. Role of CD93 in Health and Disease. Cells 2023, 12. [Google Scholar] [CrossRef]
- Tang, Y.; Chen, Y.; Liu, R.; Li, W.; Hua, B.; Bao, Y. Wnt Signaling Pathways: A Role in Pain Processing. Neuromolecular Med. 2022, 24, 233–249. [Google Scholar] [CrossRef] [PubMed]
- Gräff, J.; Mansuy, I.M. Epigenetic Dysregulation in Cognitive Disorders. Eur. J. Neurosci. 2009, 30, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Fagiolini, M.; Jensen, C.L.; Champagne, F.A. Epigenetic Influences on Brain Development and Plasticity. Curr. Opin. Neurobiol. 2009, 19, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Cai, Y.Q.; Zou, F.; Bie, B.; Pan, Z.Z. Epigenetic Suppression of GAD65 Expression Mediates Persistent Pain. Nat. Med. 2011, 17, 1448–1455. [Google Scholar] [CrossRef] [PubMed]
- Rando, O.J. Combinatorial Complexity in Chromatin Structure and Function: Revisiting the Histone Code. Curr. Opin. Genet. Dev. 2012, 22, 148–155. [Google Scholar] [CrossRef]
- Taylor, B.C.; Young, N.L. Combinations of Histone Post-Translational Modifications. Biochem. J. 2021, 478, 511–532. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The Language of Covalent Histone Modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef]
- Jiang, Y.; Langley, B.; Lubin, F.D.; Renthal, W.; Wood, M.A.; Yasui, D.H.; Kumar, A.; Nestler, E.J.; Akbarian, S.; Beckel-Mitchener, A.C. Mini-Symposium Epigenetics in the Nervous System. 2008. [Google Scholar] [CrossRef]
- Zhang, W.; Shi, Y.; Peng, Y.; Zhong, L.; Zhu, S.; Zhang, W.; Tang, S.J. Neuron Activity-Induced Wnt Signaling up-Regulates Expression of Brain-Derived Neurotrophic Factor in the Pain Neural Circuit. J. Biol. Chem. 2018, 293, 15641–15651. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Tao, Y.X. Expression of Acetyl-Histone H3 and Acetyl-Histone H4 in Dorsal Root Ganglion and Spinal Dorsal Horn in Rat Chronic Pain Models. Life Sci. 2018, 211, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Rolim, L.S.A.; Nascente, P.D.S.; Castilho, R.M.; Squarize, C.H. Feeling the Heat. Mapping the Epigenetic Modifications of Histone during Burn Wound Healing. J. Burn Care Res. Off. Publ. Am. Burn Assoc. 2024, 45, 499–507. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Wang, J.-F.; Yang, C.-X.; Wu, L.; Yin, Q.; Liu, H.; Fu, Z.-J. Intrathecal Injection of Resveratrol Attenuates Burn Injury Pain by Activating Spinal Sirtuin 1. Pharmacogn. Mag. 2016, 12, 201–201. [Google Scholar] [CrossRef]
- Luo, H.M.; Hu, S.; Bai, H.Y.; Wang, H.B.; Du, M.H.; Lin, Z.L.; Ma, L.; Wang, H.; Lv, Y.; Sheng, Z.Y. Valproic Acid Treatment Attenuates Caspase-3 Activation and Improves Survival after Lethal Burn Injury in a Rodent Model. J. Burn Care Res. Off. Publ. Am. Burn Assoc. 2014, 35. [Google Scholar] [CrossRef] [PubMed]
- Duan, B.; Cheng, L.; Bourane, S.; Britz, O.; Padilla, C.; Garcia-Campmany, L.; Krashes, M.; Knowlton, W.; Velasquez, T.; Ren, X.; et al. Identification of Spinal Circuits Transmitting and Gating Mechanical Pain. Cell 2014, 159, 1417–1432. [Google Scholar] [CrossRef]
- Brewer, C.L.; Styczynski, L.M.; Serafin, E.K.; Baccei, M.L. Postnatal Maturation of Spinal Dynorphin Circuits and Their Role in Somatosensation. Pain 2020, 161, 1906–1924. [Google Scholar] [CrossRef]
- Huang, K.; Hu, D.-D.; Bai, D.; Wu, Z.-Y.; Chen, Y.-Y.; Zhang, Y.-J.; Lv, X.; Wang, Q.-X.; Zhang, L. Persistent Extracellular Signal-Regulated Kinase Activation by the Histamine H4 Receptor in Spinal Neurons Underlies Chronic Itch. J. Invest. Dermatol. 2018, 138, 1843–1850. [Google Scholar] [CrossRef]
- Kardon, A.P.; Polgár, E.; Hachisuka, J.; Snyder, L.M.; Cameron, D.; Savage, S.; Cai, X.; Karnup, S.; Fan, C.R.; Hemenway, G.M.; et al. Dynorphin Acts as a Neuromodulator to Inhibit Itch in the Dorsal Horn of the Spinal Cord. Neuron 2014, 82, 573–586. [Google Scholar] [CrossRef]
- Boyle, K.A.; Gutierrez-Mecinas, M.; Polgár, E.; Mooney, N.; O’Connor, E.; Furuta, T.; Watanabe, M.; Todd, A.J. A Quantitative Study of Neurochemically Defined Populations of Inhibitory Interneurons in the Superficial Dorsal Horn of the Mouse Spinal Cord. Neuroscience 2017, 363, 120–133. [Google Scholar] [CrossRef] [PubMed]
- Hozumi, T.; Sawai, S.; Jitsuishi, T.; Kitajo, K.; Inage, K.; Eguchi, Y.; Shiga, Y.; Narita, M.; Orita, S.; Ohtori, S.; et al. Gene Expression Profiling of the Spinal Cord at the Chronic Pain Phase Identified CDKL5 as a Candidate Gene for Neural Remodeling. Neurosci. Lett. 2021, 749. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Wang, J.; Nie, H.; Zeng, D.; Yin, C.; Li, Y.; Wei, H.; Liu, B.; Tai, Y.; Hu, Q.; et al. Genome-Wide Expression Profiling by RNA-Sequencing in Spinal Cord Dorsal Horn of a Rat Chronic Postsurgical Pain Model to Explore Potential Mechanisms Involved in Chronic Pain. J. Pain Res. 2022, 15, 985–1001. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Shi, J.; Wang, M.; An, S.; Guo, X.; Wang, Z. Analyses of Gene Expression Profiles in the Rat Dorsal Horn of the Spinal Cord Using RNA Sequencing in Chronic Constriction Injury Rats. J. Neuroinflammation 2018, 15. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Zhang, F.; Wang, Y.; Huang, Y.; He, Y.; Ren, J.; Deng, Y.T.; Gao, Y.; Li, X.; Yu, L.; et al. Spinal CBX2 Contributes to Neuropathic Pain by Activating ERK Signaling Pathway in Male Mice. Neurosci. Lett. 2023, 812. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Yin, C.; Hu, Q.; Liu, B.; Tai, Y.; Zheng, X.; Li, Y.; Fang, J.; Liu, B. Expression Profiling of Spinal Cord Dorsal Horn in a Rat Model of Complex Regional Pain Syndrome Type-I Uncovers Potential Mechanisms Mediating Pain and Neuroinflammation Responses. J. Neuroinflammation 2020, 17. [Google Scholar] [CrossRef]
- Huang, J.; Liu, P.; Wang, G. Regulation of Mitochondrion-Associated Cytosolic Ribosomes by Mammalian Mitochondrial Ribonuclease T2 (RNASET2). J. Biol. Chem. 2018, 293, 19633–19644. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, Z.Y.; Gerner, P.; Woolf, C.J.; Ji, R.R. ERK Is Sequentially Activated in Neurons, Microglia, and Astrocytes by Spinal Nerve Ligation and Contributes to Mechanical Allodynia in This Neuropathic Pain Model. Pain 2005, 114, 149–159. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Xu, Z.Z.; Wang, X.; Park, J.Y.; Zhuang, Z.Y.; Tan, P.H.; Gao, Y.J.; Roy, K.; Corfas, G.; Lo, E.H.; et al. Distinct Roles of Matrix Metalloproteases in the Early- and Late-Phase Development of Neuropathic Pain. Nat. Med. 2008, 14, 331–336. [Google Scholar] [CrossRef]
- Hannocks, M.J.; Zhang, X.; Gerwien, H.; Chashchina, A.; Burmeister, M.; Korpos, E.; Song, J.; Sorokin, L. The Gelatinases, MMP-2 and MMP-9, as Fine Tuners of Neuroinflammatory Processes. Matrix Biol. J. Int. Soc. Matrix Biol. 2019, 75–76, 102–113. [Google Scholar] [CrossRef]
- Tansley, S.; Gu, N.; Guzmán, A.U.; Cai, W.; Wong, C.; Lister, K.C.; Muñoz-Pino, E.; Yousefpour, N.; Roome, R.B.; Heal, J.; et al. Microglia-Mediated Degradation of Perineuronal Nets Promotes Pain. Science 2022, 377, 80–86. [Google Scholar] [CrossRef]
- Ichiro Hiraga, S.; Itokazu, T.; Nishibe, M.; Yamashita, T. Neuroplasticity Related to Chronic Pain and Its Modulation by Microglia. Inflamm. Regen. 2022, 42. [Google Scholar] [CrossRef]
- Rodgers, U.R.; Kevorkian, L.; Surridge, A.K.; Waters, J.G.; Swingler, T.E.; Culley, K.; Illman, S.; Lohi, J.; Parker, A.E.; Clark, I.M. Expression and Function of Matrix Metalloproteinase (MMP)-28. Matrix Biol. J. Int. Soc. Matrix Biol. 2009, 28, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Abboud, C.; Brochoire, L.; Drouet, A.; Hossain, M.A.; Hleihel, W.; Gundlach, A.L.; Landry, M. Analgesic Effect of Central Relaxin Receptor Activation on Persistent Inflammatory Pain in Mice: Behavioral and Neurochemical Data. PAIN Rep. 2021, 6, e937. [Google Scholar] [CrossRef] [PubMed]
- Cioato, S.G.; Medeiros, L.F.; Lopes, B.C.; de Souza, A.; Medeiros, H.R.; Assumpção, J.A.F.; Caumo, W.; Roesler, R.; Torres, I.L.S. Antinociceptive and Neurochemical Effects of a Single Dose of IB-MECA in Chronic Pain Rat Models. Purinergic Signal. 2020, 16, 573–584. [Google Scholar] [CrossRef]
- Wang, X.Q.; Liu, Z.H.; Xue, L.; Lu, L.; Gao, J.; Shen, Y.; Yang, K.; Chen, Q.J.; Zhang, R.Y.; Shen, W.F. C1q/TNF-Related Protein 1 Links Macrophage Lipid Metabolism to Inflammation and Atherosclerosis. Atherosclerosis 2016, 250, 38–45. [Google Scholar] [CrossRef]
- Sugeno, A.; Piao, W.; Yamazaki, M.; Takahashi, K.; Arikawa, K.; Matsunaga, H.; Hosokawa, M.; Tominaga, D.; Goshima, Y.; Takeyama, H.; et al. Cortical Transcriptome Analysis after Spinal Cord Injury Reveals the Regenerative Mechanism of Central Nervous System in CRMP2 Knock-in Mice. Neural Regen. Res. 2021, 16, 1258–1265. [Google Scholar] [CrossRef]
- Richter, F.; Meurers, B.H.; Zhu, C.; Medvedeva, V.P.; Chesselet, M.F. Neurons Express Hemoglobin Alpha- and Beta-Chains in Rat and Human Brains. J. Comp. Neurol. 2009, 515, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Niu, R.N.; Shang, X.P.; Teng, J.F. Overexpression of Egr2 and Egr4 Protects Rat Brains against Ischemic Stroke by Downregulating JNK Signaling Pathway. Biochimie 2018, 149, 62–70. [Google Scholar] [CrossRef]
- Gaudio, N.D.; Costanzo, A.D.; Liu, N.Q.; Conte, L.; Dell’Aversana, C.; Bove, G.; Benedetti, R.; Montella, L.; Ciardiello, F.; Carafa, V.; et al. CBX2 Shapes Chromatin Accessibility Promoting AML via P38 MAPK Signaling Pathway. Mol. Cancer 2022, 21. [Google Scholar] [CrossRef]
- Wang, K.; Ju, Z.; Yong, Y.; Chen, T.; Song, J.; Zhou, J. The Effects of Electroacupuncture on the Apelin/APJ System in the Spinal Cord of Rats With Inflammatory Pain. Anesth. Analg. 2016, 123, 1603–1610. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhao, Q.; Chen, L.; Li, Z. Apelin/APJ System: An Emerging Therapeutic Target for Neurological Diseases. Mol. Biol. Rep. 2023, 50, 1639–1653. [Google Scholar] [CrossRef]
- Yam, M.F.; Loh, Y.C.; Tan, C.S.; Adam, S.K.; Manan, N.A.; Basir, R. General Pathways of Pain Sensation and the Major Neurotransmitters Involved in Pain Regulation. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.; Kim, M.; Hwang, S.W. Molecular Mechanisms Underlying the Actions of Arachidonic Acid-Derived Prostaglandins on Peripheral Nociception. J. Neuroinflammation 2020, 17. [Google Scholar] [CrossRef]
- Parisien, M.; Samoshkin, A.; Tansley, S.N.; Piltonen, M.H.; Martin, L.J.; El-Hachem, N.; Dagostino, C.; Allegri, M.; Mogil, J.S.; Khoutorsky, A.; et al. Genetic Pathway Analysis Reveals a Major Role for Extracellular Matrix Organization in Inflammatory and Neuropathic Pain. Pain 2019, 160, 932–944. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.M.; Lu, Y.C.; Zhang, J. Dexmedetomidine Reduces Diabetic Neuropathy Pain in Rats through the Wnt 10a/ β-Catenin Signaling Pathway. BioMed Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Pan, Y.; Xu, H.; Song, X. Hyperbaric Oxygen Attenuates Neuropathic Pain and Reverses Inflammatory Signaling Likely via the Kindlin-1/Wnt-10a Signaling Pathway in the Chronic Pain Injury Model in Rats. J. Headache Pain 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Yuan, S.; Li, B.; Wang, J.; Carlton, S.M.; Chung, K.; Chung, J.M.; Tang, S.J. Regulation of Wnt Signaling by Nociceptive Input in Animal Models. Mol. Pain 2012, 8. [Google Scholar] [CrossRef]
- Wu, Z.S.; Lo, J.J.; Wu, S.H.; Wang, C.Z.; Chen, R.F.; Lee, S.S.; Chai, C.Y.; Huang, S.H. Early Hyperbaric Oxygen Treatment Attenuates Burn-Induced Neuroinflammation by Inhibiting the Galectin-3-Dependent Toll-Like Receptor-4 Pathway in a Rat Model. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef]
- Imai, S.; Narita, M.; Ikegami, D.; Yamashita, A.; Shimizu, T.; Narita, M.; Niikura, K.; Furuya, M.; Kobayashi, Y.; Miyashita, K.; et al. Epigenetic Transcriptional Activation of Monocyte Chemotactic Protein 3 Contributes to Long-Lasting Neuropathic Pain. Brain J. Neurol. 2013, 136, 828–843. [Google Scholar] [CrossRef]
- Krashes, M.J.; Shah, B.P.; Madara, J.C.; Olson, D.P.; Strochlic, D.E.; Garfield, A.S.; Vong, L.; Pei, H.; Watabe-Uchida, M.; Uchida, N.; et al. An Excitatory Paraventricular Nucleus to AgRP Neuron Circuit That Drives Hunger. Nature 2014, 507, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Platt, R.J.; Chen, S.; Zhou, Y.; Yim, M.J.; Swiech, L.; Kempton, H.R.; Dahlman, J.E.; Parnas, O.; Eisenhaure, T.M.; Jovanovic, M.; et al. CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell 2014, 159, 440–455. [Google Scholar] [CrossRef] [PubMed]
- White, J.P.M.; Ko, C.W.; Fidalgo, A.R.; Cibelli, M.; Paule, C.C.; Anderson, P.J.; Cruz, C.; Gomba, S.; Matesz, K.; Veress, G.; et al. Severe Burn Injury Induces a Characteristic Activation of Extracellular Signal-Regulated Kinase 1/2 in Spinal Dorsal Horn Neurons. Eur. J. Pain Lond. Engl. 2011, 15, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Tjølsen, A.; Berge, O.G.; Hunskaar, S.; Rosland, J.H.; Hole, K. The Formalin Test: An Evaluation of the Method. Pain 1992, 51, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Lucera, M.B.; Tilton, C.A.; Mao, H.; Dobrowolski, C.; Tabler, C.O.; Haqqani, A.A.; Karn, J.; Tilton, J.C. The Histone Deacetylase Inhibitor Vorinostat (SAHA) Increases the Susceptibility of Uninfected CD4+ T Cells to HIV by Increasing the Kinetics and Efficiency of Postentry Viral Events. J. Virol. 2014, 88, 10803–10812. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Milasta, S.; Hu, D.; AlTahan, A.M.; Interiano, R.B.; Zhou, J.; Davidson, J.; Low, J.; Lin, W.; Bao, J.; et al. Targeting Histone Demethylases in MYC-Driven Neuroblastomas with Ciclopirox. Cancer Res. 2017, 77, 4626–4638. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinforma. Oxf. Engl. 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-Based Differential Expression Analysis of RNA Sequencing Data Using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A Free Online Platform for Data Visualization and Graphing. PloS One 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
| Primary antibody | Abbreviations | Company Cat. No. |
Dilution for WES | Predicted band (kDa) based on datasheet (WES) |
Observed band (kDa) / peak on chromatogram (WES) | Dilution for IF |
|---|---|---|---|---|---|---|
| Total histone H3 | Total H3 | #ab1791 | 1:100 | 17 | 26.9 | 1:1000 |
| histone H3 mono methyl K4 | H3K4me1 | #ab239402 | 1:10 | 15 | 28.2 | 1:1000 |
| histone H3 di methyl K4 | H3K4me2 | #ab32356 | 1:10 | 17 | 28 | 1:1000 |
| histone H3 tri methyl K4 | H3K4me3 | #ab213224 | 1:10 | 15 | 28.3 | 1:500 |
| histone H3 tri methyl K4 acetyl K9 |
H3K4me3K9ac | #ab272164 | 1:10 | 15 | 28.3 | 1:100 |
| histone H3 phospho-S10 | p-S10H3 | #ab5176 | 1:10 | 15 | 25.0 | 1:300 |
| GFP | GFP | #ab13970 | n.r | n.r | n.r | 1:2000 |
| Compound | Target | Final cc. | Company #Cat. No |
Ref. |
|---|---|---|---|---|
| Vorinostat (SAHA) | histone deacetylase (HDAC) inhibitor |
2 µM | Sigma-Aldrich #SML0061-VAR |
[65] |
| ciclopirox | Pan-histone demethylase inhibitor | 100 µM | Sigma-Aldrich #SML2011 |
[66] |
| Na-fluoride | protein phosphoseryl- and phosphothreonyl phosphatases inhibitor | 0.2% (wt/v) | Sigma-Aldrich #S7920 |
[8] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





