1. Introduction
Osteoporosis (OP) is characterized by reduced bone mass and strength and altered bone architecture, predisposing individuals to fragility fractures [
1]. With a prevalence of 20.5% in Central Europe (Romania), this disease emerges as a pressing public health concern, particularly among the elderly population due to the progressive deterioration of metabolic health, including the emergence of obesity, type 2 diabetes mellitus (T2D) and metabolic syndrome (MetS) [
2,
3]. The interplay between obesity and OP is complex since obese women, through mechanical loading or estrogen production by the adipose tissue, often exhibit higher bone mineral density (BMD) with a potential protective role. The increased BMD does not necessarily translate to reduced fracture risk, a phenomenon known as the "obesity paradox" [
4]. Among numerous factors that might contribute to elevated risk of fragility fractures, the genetic predisposition plays a determining role. A bivariate meta-analysis of a large-scale genome-wide association study (GWAS) indicated three loci (2p23.2, 16q12.2, and 18q21.32) with pleiotropic effect on both obesity and OP, corresponding to TRNA Methyltransferase 61B (
TRMT61B), Fat Mass and Obesity-Associated (
FTO), and Melanocortin 4 Receptor (
MC4R) genes, respectively [
5]. Following the initial discovery of
FTO's implication in human obesity, this gene has garnered significant attention in other metabolic diseases such as T2D, non-alcohol fatty liver disease (NAFLD), hypertension, cardiovascular diseases, and OP [
6,
7].
FTO encodes Fe(II) and 2-oxoglutarate-dependent oxygenases that play a crucial role in epigenetic regulation, functioning as a demethylase of mRNA, specifically targeting N(6)-methyladenosine (m6A), the most prevalent RNA modification [
8,
9]. Studies involving cell cultures and animal models have demonstrated
FTO's involvement in adipogenesis, adipocyte apoptosis, and the osteogenic differentiation of bone marrow mesenchymal stem cells (BMSCs) into adipocytes or osteoblasts [
9]. Apart from its expression in adipose tissue, brain, muscle, and heart,
FTO is also expressed in bone marrow, rendering it a promising candidate for genetic predisposition to osteoporosis [
10].
Although investigations into
FTO's role in osteoporosis in humans, particularly postmenopausal women, have been limited, previous studies have identified intriguing associations. For instance, Guo et al. (2011) identified six single nucleotide variations (SNVs) in intron 8 linked to increased hip BMD in Chinese populations, although this finding was not replicated in a Caucasian sample [
11]. Similarly, recent studies have reported several SNVs in intron 1 of
FTO associated with hip fractures, albeit without impacting BMD or bone loss rate [
12,
13].
The objective of this study was to shed light on the potential involvement of the FTO gene in postmenopausal OP by analyzing five common SNVs in intron 1. Our analysis of unphased DNA revealed associations between SNVs and both OP and severe OP with fractures. Fine-scale haplotype mapping using phased DNA unveiled stronger association signals with OP and fragility fractures carried by specific haplotype combinations, aligning with the observed decrease in BMD at various skeletal sites.
2. Materials and Methods
2.1. Population
A number of 188 postmenopausal women were recruited at the
C.I. Parhon National Institute of Endocrinology (Bucharest,
Romania) during the period from May 28, 2020, to April 1, 2022 [
14]. The inclusion criteria were as follows: (1) women aged 50–75 years; (2) time from menopause ≥ 1 year; (3) Caucasian (
Romanian) origin. Excluded were all forms of secondary osteoporosis after hormonal examination, as well as severe chronic diseases (except for T2D). The Institutional Ethical Committee approved the research protocol, and signed informed consent was obtained from each patient in accordance with the Helsinki Declaration [
15].
Association was performed comparing non-OP subjects (controls) and OP subjects (cases) classified according to
American Association of Clinical Endocrinologists [
16]. Criteria in postmenopausal women were based on any of the following: 1) T-score −2.5 or below in the lumbar spine, femoral neck, total proximal femur, or 1/3 radius; 2) Low-trauma spine or hip fracture, regardless of BMD; 3) T-score between −1.0 and −2.5 and fragility fractures of proximal humerus, pelvis, or distal forearm; 4) T-score between −1.0 and −2.5 and high FRAX fracture probability based on country-specific thresholds.
A comprehensive series of clinical, biochemical and hormonal tests were performed (
Table S1) as described [
14]. All patients were examined by Dual X-ray absorptiometry analysis (DEXA) investigated for skeletal alterations and muscular performance (
Table S2). FRAX PLUS (TBS) score for the evaluation of a 10-year risk for low energy fractures was computed on the country-specific website (
https://www.fraxplus.org/). In addition, severe OP was diagnosed based on WHO criteria, namely the association of T score equal to or less than −2.5 SD (e.g., < −3) and fragility fractures [
17,
18,
19].
Metabolic syndrome (MetS) diagnosis was based on the presence of at least 3 of the harmonized criteria of the National Cholesterol Education Program (NCEP) and Adult Treatment Panel-III (ATP-III), which include: (1) abdominal obesity based on WC ≥ 88 cm, (2) high TG level ≥ 1.7 mmol/L, (3) low HDL-C < 1.03 mmol/L, (4) Hight Blood pressure (HBP) with systolic blood pressure (SBP) ≥ 130 mmHg and/or diastolic blood pressure (DBP) ≥ 85 mmHg, and (5) high fasting glucose levels ≥ 5.36 mmol/L, or current treatment with antihyperlipidemic, antihypertensive or hypoglycemic agents, respectively [
17]. Insulin resistance was assessed using HOMA-IR or as nominative variable defined as having HOMA-IR values above the cutoff of 1.92, which was calculated from fasting insulin levels of lean patients without OP + 2 SEM, as previously described [
21]. To resume the evaluation of muscular strength and physical performance, we composed a statistical instrument (SUM
stat) considering none, 1, 2, 3, 4, or all 5 muscular tests outside the normal values and used as binary (0/1) parameter (
Table S2).
2.3. Genotyping
Genomic DNA was extracted from whole blood using the Wizard Genomic DNA Purification Kit (Promega, Madison, USA) as described [
14]. Five SNVs in intron 1 of the
FTO gene (rs8057044, rs8050136, rs9939609, rs62033406, rs9930506) were selected based on our previous studies [
22,
23,
24,
25]. We also genotyped SNV rs3736228 in the
LRP5 gene identified from GWAS data. Their position was referenced for GRCh37/hg19 and nomenclature was validated using VariantValidation site (
https://variantvalidator.org/). Genotyping was performed by allele discrimination assay (KASPar technique from LCG Genomics, Teddington, UK). For phased DNA, haplotypes were reconstructed in the population using the PHASE 2.1 program [
26] and visualized for linkage disequilibrium (LD) in HAPLOVIEW 3.1 [
27], while predictions of transcriptional activity were examined in HaploReg v.4.1 (
http://archive.broadinstitute.org/mammals/haploreg).
2.4. Statistics and Computation
Statistical analysis was performed StatView 5.0 program (Abacus Concepts, Berkeley, CA) and the study was powered at 0.85 using PBAT, as described [
15]. Numerical variables (mean ± SEM) were tested by non-parametric Kruskal-Wallis and Mann-Whitney tests. In ANOVA the interaction factor α was set at 5%. Nominal variables were analyzed using the χ² test and logistic regression was performed with the descent method to obtain P-values, odds ratios (ORs), and 95% confidence intervals (CIs). Significance was considered at P < 0.05. Bonferroni corrections for genetic association of SNVs were performed in R v3.2.1 program. LD among SNVs was calculated in the NIH database (
https://ldlink.nih.gov) and in HAPLOVIEW 3.1. Genotype-phenotype correlation was performed in OP population and results for haplotypes pairs were correlated with those from independent SNVs on unphased DNA. For BMD all anatomical sites were tested and when indicated, values were adjusted for BMI.
3. Results
3.1. SNP Association
The phenotypic features of OP and controls were previously described in detail [
14]. Briefly, women with OP were on average 66.4 ± 0.7 years old, with BMI of 26.2 ± 0.4 kg/m
2, mean ± SEM), 20.8% being classified as obese and 32% showing insulin resistance. MetS was diagnosed in 48.3% of cases. Fragility fractures were detected in 48.3% of OP cases. Control women were more obese (BMI of 30.6 ± 0.6 kg/m
2), with a higher proportion of MetS (51.5%) but with a comparable level of insulin resistance (35.3%) based on the HOMA-IR index.
Five SNVs in the
FTO gene (rs8057044, rs8050136, rs9939609, rs62033406, and rs9930506) and one SNV in the
LRP5 gene (rs3736228) had comparable minor allele frequency (MAF) to Europeans and were in Hardy-Weinberg equilibrium. Using an over-dominant model in logistic regression, rs9930506 (GA) was associated with OP with a high OR, while the association of rs8057044 (GA) and rs9939609 (TA) appeared as a trend (
Table 1). Association of rs9930506 was supported by the Bonferroni correction (P < 0.0175). No association was detected for rs3736228 in the LRP5 gene, which was no further studied. All five SNVs were significantly associated with severe OP with fractures, among which rs9939609 had a protective effect. Genuine associations with severe OP were sustained by Bonferroni correction for rs8057044 (P < 0.013) and again for rs9930506 (P < 0.001). Conditional analysis showed that rs9930506 and rs9939609 had independent associations (P < 0.0001 and 0.01, respectively) in the corresponding LD block. Of note, the SNVs rs8057044 and rs9930506 were however in reduced LD with r
2 = 0.70 (
https://ldlink.nih.gov). Therefore, for further correlations with biological parameters we considered rs8057044, rs9939609 and rs9930506 as lead SNVs.
3.2. Haplotype Mapping
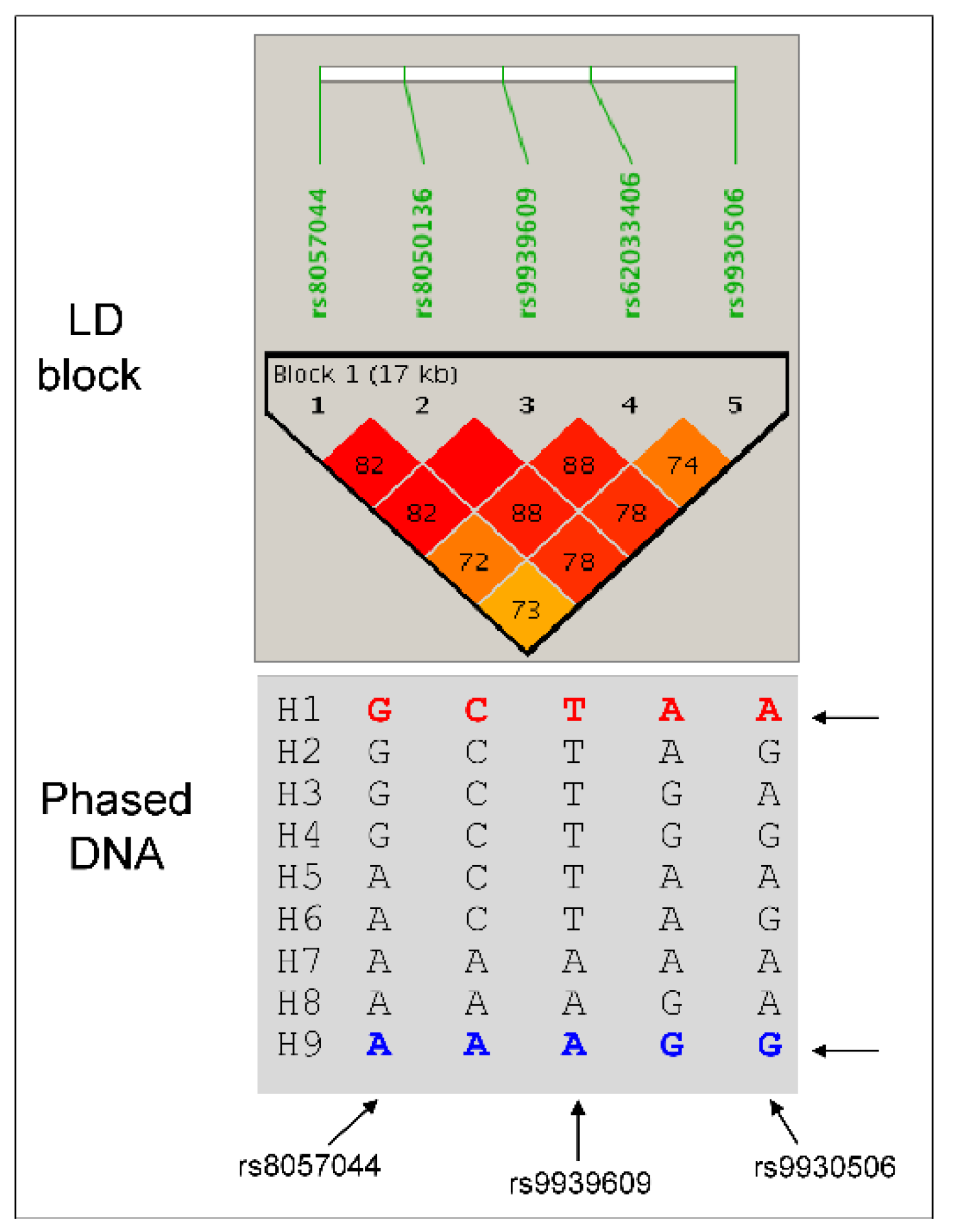
To further understand the biological effects of SNVs, we performed haplotype mapping using the PHASE program. A total of 9 haplotypes were reconstructed in the population, which were then assigned to dizygotic individuals as haplotype pairs (
Figure 1).
Two haplotypes (H1 and H9) were more prevalent (> 40%), while others (H2 to H8) were rare or very rare. Haplotype H4 was absent in controls, while haplotypes H6 and H7 were absent in severe OP, which displayed only haplotype H8. There were 17 haplotype pairs in the population, among which three pairs were frequent: H1/H1 (24.5%), H1/H9 (35.6%), and H9/H9 (20.2%). All other pairs were less than 5%. When tested independently, only one rare haplotype (H6) showed a trend association with OP (P < 0.0557), being protective.
A completely different picture emerged from the analysis of haplotype pairs. While the H1/H1 pair remained non-significant, the H9/H9 was associated with severe OP with a protective effect. By contrast, the heterozygous H1/H9 pair was associated with both OP and severe OP with high OR (
Table 1). The H1/H9 association was stronger in insulin-resistant individuals with an OR of 3.92, 95% CI [1.48–10.367], P < 0.0029. To understand metabolic consequences, variations in OR were examined as a function of the presence or absence of MetS or its components. When the population was stratified as with and without MetS, a significant association was detected for H1/H9 in the absence of MetS (P < 0.03, OR 1.9, 95% CI [1.043–3.626]), the absence of low HDL levels (P < 0.0057, OR 2.3, 95% CI [1.249–4.242]), in individuals with high blood pressure (HBP) with P < 0.0093, OR 2.5, 95% CI [1.228–5.148], and particularly in women with high triglycerides (TG) levels (P < 0.0079, OR 4.58, 95% CI [1.35–15.98]). The same picture was obtained for the H1/H9 association in the sub-population of OP with fractures, the OR for H1/H9 pair being increased up to 9.1 (P < 0.0067) in individuals with high TG levels. These data concordantly indicated the H1/H9 haplotype pair as pathogenic for OP and fractures, contingent upon the presence of insulin resistance with high TG levels and HBP rather than obesity, low HDL levels, hyperglycemia, or the presence of MetS.
3.3. Genotype Phenotype Correlation
To search for metabolic consequences and bone alterations, we examined the genotype-phenotype correlation in the OP population by investigating the impact of the three most frequent haplotype pairs and correlated results with those from independent unphased DNA lead SNVs rs8057044, rs9939609 and rs9930506 (
Table S3, S4, S5).
Carriers of H1/H9 haplotypes exhibited leaner phenotype, attributed to the heterozygous GA, TA, or GA genotypes of rs8057044, rs9939609, and rs9930506, respectively (
Table 2).
Despite their leaner phenotype (12.2% obesity), this SNV combination was found to be pathogenic for bone, being associated with 53% fractures and the highest prevalence of muscular alterations (51%). FRAX scores were also the highest. By contrast, homozygous H9/H9 carriers with AA, AA, and GG alleles of three lead SNVs, respectively, were more obese (30.2% obesity) with the highest waist circumference (97.2 cm), central obesity (86%), and SBP. In unphased DNA, these SNVs display higher BMI and prevalence of obesity, larger waist circumference, and central obesity, features that may stem from the effect of the homozygous AA allele of rs9939609, extensively studied in human obesity [
28]. Finally, H1/H1 homozygous carriers of GG, TT and AA alleles of lead SNVs, exhibited an intermediate phenotype combining GG alleles of rs8057044 (influential in obesity) as well as the TT and AA alleles of rs9939609 and rs9930506, respectively (associated with a leaner phenotype). No effects of haplotype pairs were found on HOMA-IR index, insulin resistance as a nominative variable, fasting glycemia, or MetS, although in H1/H1 carriers, there was a trend towards a higher prevalence (39.3%) of MetS and low levels of HDL (46.4%, P < 0.0003). These latter effects may be driven by the dominant effect of the rs9930506 AA genotype.
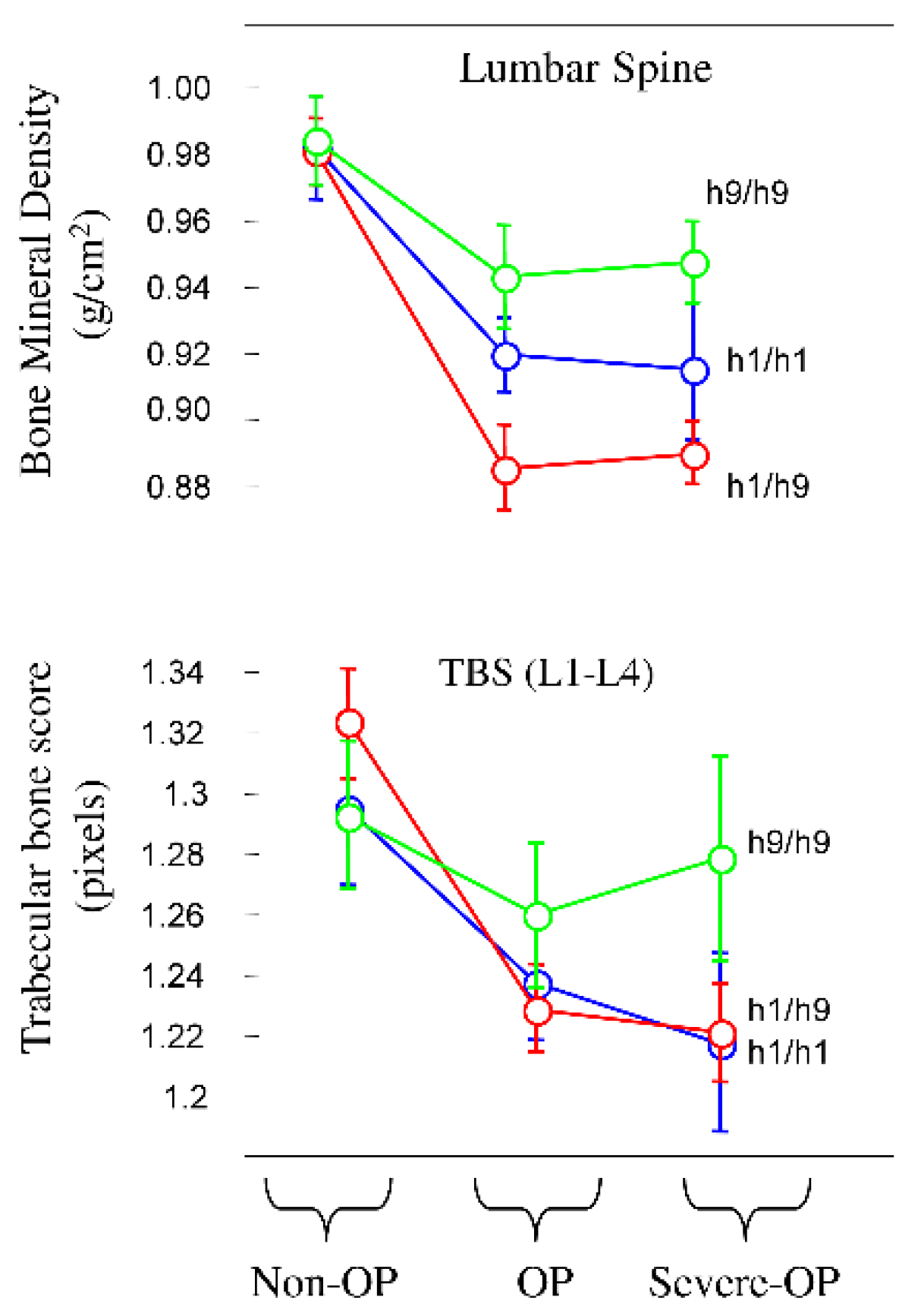
Next, our focus shifted to bone metabolism, particularly BMD, trabecular bone score (TBS) and bone turnover markers (
Table 2). Carriers of the pathogenic pair H1/H9 exhibited lower BMD values at all anatomical sites while H9/H9 revealed higher values, as shown in
Figure 2 for lumbar spine.
Among anatomical sites the radius demonstrated the most significant difference (P < 0.0001). Although TBS values were lower compared to controls, they did not reach statistical significance (P < 0.0501). However, significance was obtained for GA of rs8057044 (P = 0.0192), AA of rs9939609 (P = 0.0135), and GG for rs9930506 (P = 0.0453) when independent SNVs were considered in unphased DNA. For bone turnover markers, only Beta-crosslaps showed higher values in the H9/H9 pair. Additionally, there was a trend towards a higher prevalence of muscular alterations (51.0%) in H1/H9 carriers compared to H1/H1 and H9/H9 (32.1% and 46.3%, respectively), the significance being obtained only for reduced grip strength (right hand). Carriers of the H1/H9 pair exhibited higher values for FRAX PLUS major and hip fracture risks (9.8 and 3.2, respectively), with significance between H1/H9 and H9/H9 carriers (
Table 2 and
Table S6).
4. Discussion
In this paper, we present evidence for the association of SNVs in intron 1 of the FTO gene with OP and severe OP with fragility fractures, designating this gene as a promising candidate for OP. Association was observed using unphased independent SNVs and, in addition, haplotype mapping indicated stronger association signals and correlated well with biological parameters of bone metabolism, particularly BMD. These data reinforce previous studies on the FTO gene and, by haplotype mapping, provide new data useful in the description of potential biomarkers for the genetic predisposition for OP in postmenopausal women.
We studied five SNVs in intron 1 of
FTO gene selected from our previous investigations in human obesity and MetS in French, Romanian and North African populations and extensively studied in human obesity [
16,
17,
18]. The rs8057044 is reported here for the first time as being associated with OP and fracture risk. This SNV displayed low LD with the other four SNVs in intron 1 suggesting that it might contain a distinct signal. Except for rs8057044, the remaining four SNVs were already reported in relation with hip fractures but without correlation with BMD [
11,
12,
13]. The rs9939609 was extensively studied as marker for human obesity. One article reported rs9939609 associated with spine BMD (P = 0.037) in Chinese population, although protective or pathogenenic role was not indicated, the paper being focused on SNVs in intron 8 [
11]. Two other studies in Australian
Dubbo Osteoporosis Epidemiology Study (DOES) collection investigated SNVs in intron 1 and found association with hip fractures, but again without effect on BMD [
12,
13]. In these Australian studies, the rs9930506 was the best associated with hip fracture (HR of 2.19, P < 0.01), result which is concordant to our best association for the same SNV in the Romanian population. The well-known obesity associated rs9939609 was non-significant (P < 0.21) in Australian samples in association with hip fractures [
12] while in our sample, the same SNV showed a rather protective effect regarding OP, being linked with obesity. Finally, another SNV (rs17817712) which is located between rs9939609 and rs62033406 was identified by meta-analysis of large-scale GWAS data as being associated with both obesity and OP. Data were confirmed in UK Biobank, indicating the pleiotropic effect of
FTO on both obesity and OP [
5]. From all these data of independent SNVs in unphased DNA, including this study, we conclude that intron 1 contains indeed a strong associated signal for OP and fracture risk, although we cannot exclude the possibility that intron 1 contains distinct signals, at least between rs8057044 and rs9930506 (pathogenic) and rs9939609 (protective) as function of the concomitant association with obesity or lack thereof.
Our study further analyzed intron 1 in phased DNA by fine-scale haplotype mapping and obtained better associations for OP and severe OP with fractures. Data were concordant with the decrease in BMD at all anatomical sites, including the lumbar spine, femoral neck, hip, and radius. These results were expected due to the detailed resolution of this genomic region [
23,
24,
25]. Indeed, 9 haplotypes obtained from 5 SNVs indicate good resolution compared to the potential 2
5 theoretical combinations of SNVs. While independent haplotypes remained not significant, haplotype pairs reached the highest OR for association. We focused on three major haplotype pairs (H1/H1, H9/H9, and H1/H9), among which H1/H9 was pathogenic in lean subjects. Effects on biological parameters were concordant with those drawn from independent SNVs in unphased DNA. The pair H9/H9 was by contrast associated with high BMI and obesity, containing the pathogenic AA alleles of rs9939609, while H1/H9 with GA, TA, and GA alleles of lead SNVs were associated with both leaner and obese phenotype. These results suggest that there would be a variability in OP phenotypes as a function of combinations of SNVs with synergistic or antagonistic effects. This might explain why the association of OP with rs9939609 was not always found in human studies as for instance in GWAS for OP.
All these data corroborated with new results from literature indicate that the
FTO gene, operating as demethylase, is a promising candidate in the pathogenesis of OP. The mechanism is not completely understood. The
FTO gene is expressed in numerous tissues, including brain, adipose tissue and bone marrow. Very likely, the demethylase activity involves a shift from differentiation to osteoblasts towards bone marrow adipocytes. Recently it was shown that methylated m6A RNA level was up-regulated in bone marrow in patients with OP. Moreover, there is experimental evidence that
FTO overexpression in normal BMSC cells would compromise osteogenic potential, by decreasing the methylated m6A and the level of runt related trascriptional factor 2 (Runx2) mRNA [
29]. Although the SNVs studied are involved in demethylation process, the proteins bound are different: Nanog and Pou5f1 (rs9939609), P300 (rs8050136), HMG-IY (rs62033406). The rs8057044 involves Pax-5 and Rad21, while rs9930506 involves IRX proteins. We cannot exclude the possibility that the best associated SNV rs9930506 operates in a similar mode as another upstream SNV (rs1421085), by binding to the AT-rich interaction domain 5B (ARID5B) repressor protein and the enhancer region of Iroquois Homeobox (IRX) 3 and 5, known to suppress the expression of
IRX3 and
5 [
30]. The rs1421085 was not investigated in this study, but we previously showed its role in simple and morbid obesity and its association with insulin resistance and MetS in Romanian subjects [
23,
24,
25].
We were unable to find a direct relationship between SNVs and insulin resistance as measured by the HOMA-IR index, nor with altered glycemic levels. This result was unexpected since the OR of association was found to be higher when population was stratified by insulin resistance. This might be explained by the multiple determinants of HOMA-IR values, including immunological alterations such as those involved in MetS [
3]. Indeed, in the same population, we previously showed that HOMA-IR was proportional to the cumulative criteria for MetS and not simply to BMI [
14].
The relationship between obesity and OP remains complex, involving not only systemic insulin resistance, but also other factors such as lifestyle (physical activity, smoking, alcohol consumption) and metabolic complications in the elderly population with OP. In the same population, we previously reported that the relationship between BMI (expressed as percentiles) and BMD was not linear, and the decrease in BMD in OP occurred beyond an inflection point of 27.2 kg/m² of BMI [
14]. A similar inflection point was found in other populations and correlated to the proportion of fat versus lean mass [
31]. These recent observations suggest that some variability in FTO studies might be explained by the clinical classification of individuals as lean, overweight, or obese, as well as different definition of MetS in ethnic populations.
5. Conclusions
In conclusion, the strength of this paper is the identification of SNVs in intron 1 of the FTO gene robustly associated with primary OP and severe OP with fragility fractures and in concordance with the decrease in BMD at different anatomical sites. The study performed in well-characterized postmenopausal women, albeit in a small size sample, indicates the FTO as a promising gene candidate for OP, in which haplotype mapping in phased DNA offers supplementary new insights into the multifaceted association signals of FTO gene with OP.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Table S1 : Clinical and biochemical assessments of osteoporosis ; Table S2 : Skeletal assessment, muscular strength and physical performance ; Table S3 : Genotype phenotype correlation of SNVs rs8057044 (G/A) in osteoporosis ; Table S4: Genotype phenotype correlation of SNV rs9939609 (T/A) in osteoporotic patients; Table S5: Genotype phenotype correlation of SNVs rs9930506 (G/A) in osteoporosis; Table S6: Genotype phenotype correlation of haplotype pairs of the FTO gene in osteoporosis.
Author Contributions
Conceptualization: D.G., C.P., F.G.; Data curation: F.G., S.H., C.L.; Formal analysis: D.G., S.H., F.G., C.L.; Investigation: D.M., D.G., G.V.; Methodology: D.G., D.M., F.G., C.L.; Resources: C.P., G.V.; Software: F.G.; Writing – original draft: D.G., S.H., F.G., and C.P.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board (or Ethics Committee) of CI. Pahon National Institute of Endocrinology (protocol number 13 on 27/06/2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Full data may be obtained from the author opon requenst.
Acknowledgments
Publication of this paper was supported by the C.I. Parhon National Institute of Endocrinology, Bucharest (Romania). FG is honorary professor at Carol Davila University of Medicine and Pharmacy, Bucharest (Romania).
Conflicts of Interest
The authors declare that they have no conflict of interest.
Abbreviations and Acronyms
| AACE |
American Association of Clinical Endocrinologists |
| ALP |
Alkaline phosphatase |
| ATP-III |
Adult Treatment Panel-III |
| BMD |
Bone mineral density |
| BMI |
Body mass index |
| BMSC |
Bone marrow mesenchymal stem cells |
| CRP |
C-reactive protein |
| CST |
Chair stand test |
| DBP |
Diastolic blood pressure |
| DEXA |
Dual X-ray absorptiometry |
| ECLIA |
Electrochemiluminescent immunoassay |
| HBP |
High blood pressure |
| HDL-C |
High-density lipoprotein cholesterol |
| HOMA |
Homeostasis Model Assessment |
| LDL-C |
Low-density lipoprotein cholesterol |
| GWAS |
Genome-wide association study |
| LD |
Linkage disequilibrium |
| MAF |
Minor allele frequency |
| MetS |
Metabolic syndrome |
| m6A |
N6-methyladenosine |
| NCEP |
National Cholesterol Education Program |
| OB |
Obesity |
| OP |
Osteoporosis |
| P1NP |
Procollagen type I N-terminal propeptide |
| PTH |
Parathyroid hormone |
| SBP |
Systolic blood pressure |
| SHBG |
Sex hormone binding globulin |
| SNV |
Single nucleotide variation |
| T2D |
Type 2 diabetes mellitus |
| TBS |
Trabecular Bone Score |
| TG |
Triglycerides |
| TUG |
Timed up and go test |
| WC |
Waist circumference |
| WHO |
World Health Organisation |
References
- Kanis, J. Assessment of osteoporosis at the primary health care level, WHO Scientific Group Technical Report. University of Sheffield, UK; 2007. https://frax.shef.ac.uk/FRAX/pdfs/WHO_Technical_Report.pdf.
- Grigorie, D.; Sucaliuc, A.; Johansson, H.; Kanis, J.A.; McCloskey, E. Incidence of Hip Fracture in Romania and the Development of a Romanian FRAX Model. Calcif. Tissue Int. 2013, 92, 429–436. [Google Scholar] [CrossRef]
- Greere, D. I, Grigorescu F., Manda D., Lautier C., and C. Poianã, Insulin resistance and pathogenesis of postmenopausal osteoporosis. Acta Endocrinologica (Buc)., vol. XIX, (2023) 349-363. 2023. [Google Scholar] [CrossRef]
- Rinonapoli G, Pace V, Ruggiero C, Ceccarini P, Bisaccia M, Meccariello L, et al. Obesity and Bone: A Complex Relationship. International Journal of Molecular Sciences. 1: 2021 Dec 20;22(24), 2021. [CrossRef] [PubMed]
- Pei, Y.; Wei, X.; Feng, G.; Zhang, H.; Yang, X.; Zhang, S.; Fang, C.; Huang, Y.; Tian, Q.; Deng, H.; et al. Bivariate genome-wide association analysis identified three pleiotropic loci underlying osteoporosis and obesity. Clin. Genet. 2020, 97, 785–786. [Google Scholar] [CrossRef] [PubMed]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.B.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Huang, C.; Shen, M.; Zhan, H.; Xu, K. RNA N6-methyladenosine: a promising molecular target in metabolic diseases. Cell Biosci. 2020, 10, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gerken, T. , Girard CA., Tung YC., Webby CJ., Saudek V., Hewitson KS. et al., The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318 (2007) 1469–1472.
- Chen, X.; Hua, W.; Huang, X.; Chen, Y.; Zhang, J.; Li, G. Regulatory Role of RNA N6-Methyladenosine Modification in Bone Biology and Osteoporosis. Front. Endocrinol. 2020, 10, 911. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Riddle, R.C.; Yang, Q.; Rosen, C.R.; Guttridge, D.C.; Dirckx, N.; Faugere, M.-C.; Farber, C.R.; Clemens, T.L. The RNA demethylase FTO is required for maintenance of bone mass and functions to protect osteoblasts from genotoxic damage. Proc. Natl. Acad. Sci. 2019, 116, 17980–17989. [Google Scholar] [CrossRef]
- Guo, Y.; Liu, H.; Yang, T.-L.; Li, S.M.; Li, S.K.; Tian, Q.; Liu, Y.-J.; Deng, H.-W. The Fat Mass and Obesity Associated Gene, FTO, Is Also Associated with Osteoporosis Phenotypes. PLOS ONE 2011, 6, e27312. [Google Scholar] [CrossRef]
- De Dios, K.; Huynh, N.; Tran, T.S.; Center, J.R.; Nguyen, T.V. Association between Fat Mass and Obesity-Related Transcript Polymorphisms and Osteoporosis Phenotypes. J. Bone Metab. 2024, 31, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Tran, B.; Nguyen, N.D.; Center, J.R.; Eisman, J.A.; Nguyen, T.V. Association between fat-mass-and-obesity-associated (FTO) gene and hip fracture susceptibility. Clin. Endocrinol. 2013, 81, 210–217. [Google Scholar] [CrossRef]
- Greere, D.; Grigorescu, F.; Manda, D.; Voicu, G.; Lautier, C.; Nitu, I.; Poiana, C. Relative Contribution of Metabolic Syndrome Components in Relation to Obesity and Insulin Resistance in Postmenopausal Osteoporosis. J. Clin. Med. 2024, 13, 2529. [Google Scholar] [CrossRef]
- World Medical Association. WMA - the World Medical Association-WMA Declaration of Helsinki – Ethical Principles for Medical Research Involving Human Subjects. Wma.net. WMA - the World Medical Association-WMA Declaration of Helsinki – Ethical Principles for Medical Research Involving Human Subjects; 2022. https://www.wma.net/policies-post/wma-declaration-of-helsinki-ethical-principles-for-medical-research-involving-human-subjects.
- Camacho, P.M.; Petak, S.M.; Binkley, N.; Diab, D.L.; Eldeiry, L.S.; Farooki, A.; Harris, S.T.; Hurley, D.L.; Kelly, J.; Lewiecki, E.M.; et al. American Association of Clinical Endocrinologists/American College of Endocrinology Clinical Practice Guidelines for the Diagnosis and Treatment of Postmenopausal Osteoporosis— 2020 Update Executive Summary. Endocr. Pr. 2020, 26, 564–570. [Google Scholar] [CrossRef] [PubMed]
- Kanis, J.A.; Cooper, C.; Rizzoli, R.; Reginster, J.-Y. European Guidance for the Diagnosis and Management of Osteoporosis in Postmenopausal Women. Osteoporos. Int. 2019, 30, 3–44. [Google Scholar] [CrossRef] [PubMed]
- Kanis, J.A. Assessment of fracture risk and its application to screening for postmenopausal osteoporosis: Synopsis of a WHO report. Osteoporos. Int. 1994, 4, 368–381. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Jentoft AJ, Bahat G, Bauer J, Boirie Y, Bruyère O, Cederholm T, et al. Sarcopenia: revised European consensus on definition and diagnosis. Age and Ageing. 2018 Sep 24;48(1):16–31. [CrossRef] [PubMed]
- Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults[M1]. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). JAMA 2001, 285, 2486–2497. [CrossRef]
- Lautier, C.; El Mkadem, S.A.; Renard, E.; Brun, J.F.; Gris, J.C.; Bringer, J.; Grigorescu, F. Complex haplotypes of IRS2 gene are associated with severe obesity and reveal heterogeneity in the effect of Gly1057Asp mutation. Hum. Genet. 2003, 113, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Haydar, S.; Grigorescu, F.; Vintilă, M.; Cogne, Y.; Lautier, C.; Tutuncu, Y.; Brun, J.F.; Robine, J.M.; Pugeat, M.; Normand, C.; et al. Fine-scale haplotype mapping of MUT, AACS, SLC6A15 and PRKCA genes indicates association with insulin resistance of metabolic syndrome and relationship with branched chain amino acid metabolism or regulation. PLOS ONE 2019, 14, e0214122. [Google Scholar] [CrossRef]
- Attaoua, R.; El Mkadem, S.A.; Radian, S.; Fica, S.; Hanzu, F.; Albu, A.; Gheorghiu, M.; Coculescu, M.; Grigorescu, F. FTO gene associates to metabolic syndrome in women with polycystic ovary syndrome. Biochem. Biophys. Res. Commun. 2008, 373, 230–234. [Google Scholar] [CrossRef] [PubMed]
- Attaoua, R.; El Mkadem, S.A.; Lautier, C.; Kaouache, S.; Renard, E.; Brun, J.-F.; Fedou, C.; Gris, J.-C.; Bringer, J.; Grigorescu, F. Association of the FTO gene with obesity and the metabolic syndrome is independent of the IRS-2 gene in the female population of Southern France. Diabetes Metab. 2009, 35, 476–483. [Google Scholar] [CrossRef] [PubMed]
- Ben Salem, A.; Attaoua, R.; Mtiraoui, N.; Meddeb, S.; Kacem, O.; Ajina, M.; Souissi, M.; Poucheret, P.; Normand, C.; Mahjoub, T.; et al. Haplotyping strategy highlights the specificity of FTO gene association with polycystic ovary syndrome in Tunisian women population. Gene 2015, 565, 166–170. [Google Scholar] [CrossRef]
- Stephens, M.; Smith, N.J.; Donnelly, P. A New Statistical Method for Haplotype Reconstruction from Population Data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Loos, R.J.F.; Yeo, G.S.H. The bigger picture of FTO—The first GWAS-identified obesity gene. Nat. Rev. Endocrinol. 2015, 10, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Fu, Q.; Yang, J.; Liu, J.-L.; Hou, S.-M.; Huang, X.; Cao, J.-S.; Liu, T.-L.; Wang, K.-Z. RNA N6-methyladenosine demethylase FTO promotes osteoporosis through demethylating Runx2 mRNA and inhibiting osteogenic differentiation. Aging 2021, 13, 21134–21141. [Google Scholar] [CrossRef] [PubMed]
- Vámos, A.; Arianti, R.; Vinnai, B. .; Alrifai, R.; Shaw, A.; Póliska, S.; Guba, A.; Csősz,.; Csomós, I.; Mocsár, G.; et al. Human abdominal subcutaneous-derived active beige adipocytes carrying FTO rs1421085 obesity-risk alleles exert lower thermogenic capacity. Front. Cell Dev. Biol. 2023, 11, 1155673. [Google Scholar] [CrossRef]
- Liu, P.-Y.; Ilich, J.Z.; Brummel-Smith, K.; Ghosh, S. New insight into fat, muscle and bone relationship in women: determining the threshold at which body fat assumes negative relationship with bone mineral density. Int J Prev Med. 2014, 5, 1452–63. [Google Scholar]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).