1. Introduction
The global recognition of the need for rapid diagnostic measures in pathogen identification and antimicrobial susceptibility testing is driven by the limited availability of therapeutic options [
1,
2]. The effectiveness of antimicrobials against bacteria in infections has quickly evolved, primarily because of the rise in bacterial resistance and the simultaneous production of resistance genes. This situation particularly limits treatment options for patients suffering from sepsis [
3,
4,
5,
6,
7]. In the treatment of sepsis, it is crucial to begin with empirical, rapid, and broad-spectrum antibiotic therapy. Once culture results and antimicrobial susceptibility testing are obtained, the therapy should be reassessed to consider de-escalation. This means narrowing the antimicrobial spectrum to reduce the selection pressure for multidrug-resistant bacteria [
8]. However, improper or delayed prescription significantly raises patient morbidity and mortality rates. A retrospective study conducted in the United States between 2011 and 2014 found that delays in administering appropriate therapy were linked to poorer clinical outcomes. These included a 70% increase in the length of hospital stays, a 65% rise in total hospital costs, and a 20% higher risk of mortality [
9], demonstrating how rapid and accurate diagnosis is essential for proper treatment.
For many years, the identification of microorganisms in clinical samples relied on determining phenotypic growth characteristics through biochemical tests. However, since the 1970s, the introduction of commercial tests combined with automated instruments has been used in routine laboratory work. These advancements, which include additional biochemical tests and standardized test readings, have enhanced identification accuracy and shortened the time required to release results. With the development of molecular tests, particularly Polymerase Chain Reaction (PCR), regarded as the gold standard for microorganism identification, and the recent availability of mass spectrometry tests, the time to obtain results has become even shorter, significantly impacting patient morbidity and mortality [
10,
11,
12,
13]. However, the swift interaction between the laboratory and the clinician must be effective. Many clinicians, despite receiving laboratory information, often lack the expertise to make the best antimicrobial decisions due to the complexity involved in choosing the right antimicrobials. This is where antimicrobial stewardship becomes crucial.
mHealth, or mobile health, refers to the practice of medicine and public health supported by mobile devices such as smartphones, tablet computers, personal digital assistants, and other wireless devices [
14]. It includes various applications such as health information delivery, patient monitoring, telemedicine, health education, disease surveillance, and health management, utilizing technologies like mobile apps, SMS messaging, and wearable devices. mHealth seeks to enhance healthcare services, patient outcomes, and access to health information and services, particularly targeting underserved populations in remote or resource-limited areas. eHealth, or electronic health, broadly refers to the use of information and communication technologies in healthcare. This encompasses a wide range of processes and services, including electronic health records, telehealth, telemedicine, health information systems, digital health interventions, patient portals, and the use of the internet and mobile devices for health education and information. The aim of eHealth is to improve the quality, efficiency, and accessibility of healthcare services, facilitate better communication between healthcare providers and patients, enhance the management of medical records and data, and support public health initiatives through more effective and efficient use of health information and technologies [
15].
mHealth and eHealth have significant potential within antimicrobial stewardship that needs to be better utilized. In this review, we will compile some examples of the application of mHealth and eHealth in the routine management of antimicrobials, including tele-stewardship. After the brief narrative review, the aim of this study was to perform a systematic review with meta-analysis to evaluate the antimicrobial therapy adequacy on mortality.
2. The Importance of the Antimicrobial Stewardship
The appropriate prescribing of antimicrobial (ATM) in hospitals is crucial for both individual therapeutic success and patient safety. On a broader scale, it is vital for controlling multidrug-resistant microorganisms and managing supply costs [
16,
17,
18]. Selecting the most suitable ATM for treating bacterial and fungal infections, particularly healthcare-associated infections, is a complex process that involves multiple variables. When scaled to a hospital level, this process demands considerable time and a well-trained multidisciplinary team.
In both empirical and targeted treatment phases, there are significant challenges in managing infections. Empirical therapy relies on the clinician's diagnostic expertise to identify the infection site and potential pathogens, utilizing local epidemiological data and understanding of microbial resistance to enhance predictive accuracy regarding the causative agents. Conversely, targeted treatment encounters difficulties related to the collection of samples and the need for rapid and reliable microbiological diagnosis, which are essential for promptly narrowing the antibiotic spectrum [
19]. Additionally, in both phases of treatment, it is essential to consider various factors related to the patient, such as age, pregnancy and lactation status, allergies, and renal and hepatic function. Furthermore, drug-specific factors must be evaluated, including pharmacokinetics, tissue distribution, and the availability of medications within the local therapeutic arsenal [
20,
21].
Considering these challenges, antimicrobial stewardship programs have been implemented in hospitals in developed countries and have gained traction in other regions over the past decade. These programs aim to optimize the use of antimicrobials to improve patient outcomes and combat resistance [
22]. These teams, typically consisting of an infectious disease physician, clinical pharmacist, and clinical microbiologist, implement structured initiatives to prevent and treat infections while auditing antimicrobial prescriptions. Their objective is to tailor antimicrobial therapy to individual patient needs. Numerous studies assessing the effectiveness of antimicrobial stewardship programs interventions in both hospital and outpatient settings have been published, demonstrating predominantly positive outcomes in clinical, economic, and epidemiological domains [
23,
24,
25,
26,
27].
3. Artificial Intelligence and Antimicrobial Stewardship
The initial application of artificial intelligence in healthcare emerged in the field of infectious diseases. Long before "antimicrobial stewardship" was formally defined in the scientific literature, an auxiliary system was developed to aid in diagnosing infections and optimizing antimicrobial therapy prescriptions. Known as MYCIN, this expert AI system was created in 1975 and utilized predefined rules established by programmers to mimic clinical reasoning. By analyzing patient records and microbiological laboratory data, MYCIN was able to assess the likelihood of specific microorganisms and suggest appropriate antimicrobial treatment options [
28]. Despite demonstrating superior performance compared to physicians in tests and effectively reducing unnecessary antimicrobial use, MYCIN was ahead of its time. Technological limitations, such as the time required to input data into the system and the lack of personal computers, alongside ethical considerations, hindered its adoption in clinical practice [
29].
With the advancement and widespread use of personal computers and mobile devices in recent years, machine learning has become a part of our daily lives, evident in streaming services and targeted advertisements based on user preferences. In the medical field, the easy capture and digitization of data have opened up numerous opportunities for leveraging this technology [
30].
AI refers to the ability of a computerized system to mimic human decision-making processes. In healthcare, it exists on a spectrum: at one end are systems with fixed architectures, relying on clear rules and manually programmed formulas, such as evidence-based flowcharts or professional knowledge, exemplified by Expert Systems like MYCIN, which used approximately 600 pre-defined rules to simulate the decision-making of infectious disease specialists. Conversely, at the other end are machine learning systems, including deep learning, which autonomously develop algorithms based on patterns in data, often without programmer intervention but requiring large datasets. For instance, Google's Diabetic Retinopathy project utilized a database of retinography images to create an algorithm capable of diagnosing diabetic retinopathy with sensitivity and specificity comparable to that of ophthalmologists [
31,
32]. Thus, artificial intelligence tools operate along a spectrum of varying dependence on human input, with the complexity of the generated algorithms being inversely related to this dependence. Deep learning algorithms, in particular, often become so intricate that they can be challenging to interpret, leading to what is referred to as a "black box" phenomenon in which the decision-making process is not easily understandable [
30].
By 2019, in the field of infectious diseases, 60 CDSS (Clinical Decision Support Systems) were reported in the scientific literature. Most of these tools were made focusing on bacterial diseases (63%), for use in ICUs (40%), and as infectious disease consultancy (25%), with the goal of diagnosing and predicting sepsis (66%), in high-income countries (90%). To date, only seven systems have been published in low- and middle-income countries, with six focused on HIV and tuberculosis. Evidence regarding the impact of CDSS on clinical practice remains limited. Of these tools, only three have undergone clinical trials, while the others were evaluated solely for their performance metrics [
31].
A randomized clinical trial evaluating a machine learning-based CDSS for sepsis diagnosis found that care and treatment were initiated significantly earlier in the machine learning-assisted group compared to the standard electronic system. Specifically, the administration of antimicrobial therapy and blood culture collection occurred, on average, 2.76 and 2.79 hours earlier, respectively. Consequently, the trial reported a decrease in patient hospital stay from 13.0 to 10.7 days, a reduction of 6.3 hours in average ICU stay, and a 12.4% decrease in hospital mortality within the machine-learning group [
33].
In a multicenter study utilizing predictive models for selecting antimicrobial therapy for the empirical treatment of bacteremia caused by Gram-negative bacteria, demographic and laboratory data were analyzed. Prediction models were developed based on the progression of microbiological examination results. The study reported an area under the curve performance ranging from 0.63 to 0.85 when treatment was guided solely by the Gram stain and from 0.64 to 0.95 when guided by the identified pathogen, suggesting moderate to high performance. Furthermore, the findings indicated that even when compared to culture-guided ATM selection, the predictive model based on the pathogen led to the use of narrow-spectrum antibiotics [
34,
35].
In another randomized clinical trial comparing ATM choices for empirical treatment guided by an antimicrobial stewardship program with a clinical decision support system and clinical protocols versus those selected by the attending physician, several clinical outcomes were assessed. These included average hospital stay, readmission rates, and 30-day mortality, as well as outcomes related to multidrug-resistant organisms, such as
Clostridioides difficile infection and multidrug-resistant Gram-negative bacteria, along with cost implications of ATM. The results indicated no significant differences in outcomes between using the CDSS and not using it. However, when analyzing specific infection types, the intervention by the antimicrobial stewardship program led to a reduction in hospital stay by 0.53 days for cellulitis and a decreased mortality risk (OR 0.58) for community-acquired pneumonia [
36].
4. Diagnostic Stewardship
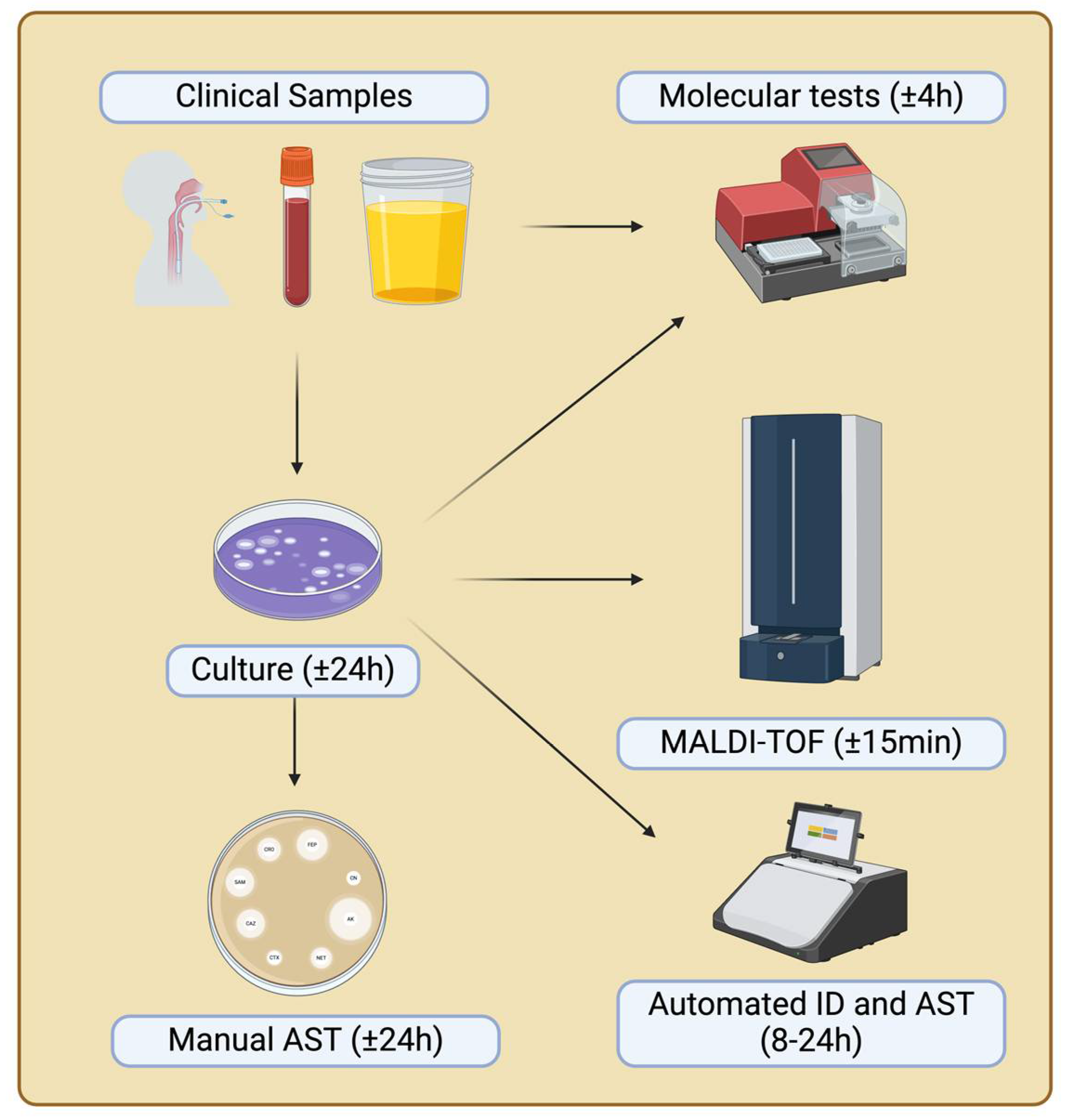
Technological advancements have allowed clinical microbiology laboratories to adopt rapid microorganism identification methods, particularly through mass spectrometry techniques such as matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) (see
Figure 1). The effectiveness of this technology is due to its ability to identify a wide range of organisms, the rapid turnaround time for results (within minutes), the precision and reproducibility of the method, and its cost-effectiveness [
37]. However, advances in molecular methods have introduced equipment capable of identifying different microorganisms present in a sample using the PCR multiplex technique [
38,
39,
40,
41,
42]. Not only clinical laboratories but also public health reference laboratories can investigate hard-to-identify microorganisms using molecular technologies, such as sequencing the 16S rDNA gene, which is representative of prokaryotic organisms [
43].
Like identification, antimicrobial susceptibility tests is essential for the clinical management of patients with infectious diseases, as well as in epidemiology, public health, new antimicrobial development, and clinical microbiology. However, many antimicrobial susceptibility tests currently in use were developed over 50 years ago and rely on phenotypic tests based on microbial growth, which typically require 48 to 72 hours from the time of culture collection [
2]. While various methodologies are being described and tested, the current gold standard for antimicrobial susceptibility test remains manual broth microdilution, as recommended by several standardization bodies. The primary methods for conducting antimicrobial susceptibility test include disk diffusion, concentration gradient diffusion, and automated testing systems.
Advancements in microbiological methodologies directly affect the operational costs of performing tests. Nevertheless, all these tools are designed to optimize and enhance infection management, ultimately improving patient outcomes and supporting diagnostic and antimicrobial administration programs [
44]. The use of these tools is closely linked to the concept of Diagnostic Stewardship, which involves modifying the medical process for requesting microbiological examinations, executing tests, and reporting results. This approach aims to enhance the treatment of infections and other medical conditions [
45]. In the context of Diagnostic Stewardship, implementing measures to preserve antimicrobial use and optimize empirical therapy can be facilitated by utilizing the Cumulative Antibiogram. This tool is designed to guide initial empirical therapy decisions for treating infections in patients who do not yet have microbiological test results available to inform treatment [
46]. The Cumulative Antibiogram is a report that accumulates combined data from microbiological examinations performed over a specified period, describing the percentage of microorganisms sensitive to selected antimicrobials [
47], as well as the possibility of obtaining the minimum inhibitory concentration (MIC) to inhibit the growth of 50% and 90% of the organisms, MIC 50 and MIC 90, respectively [
46].
Beyond empirical therapy, cumulative data are essential for monitoring resistance trends over time, conducting surveillance of antimicrobial-resistant organisms, and identifying areas for intervention through hospital infection prevention programs and antimicrobial stewardship. This approach is not restricted to a specific clinical isolate or hospital area, making it applicable to both individual institutions and broader regions [
47,
48,
49,
50]. The Clinical and Laboratory Standards Institute's M39 document provides guidelines for calculating and compiling cumulative antibiogram data, including establishing confidence intervals, determining statistical significance, estimating percentiles, and calculating MIC 50 and MIC 90. Currently, LabPro software (Beckman Coulter Inc., Sacramento, USA) facilitates the automated generation of cumulative antibiograms, displaying percentages of growth inhibition across various concentrations of available antibiotics by panels, while also allowing for the assessment of MIC 90 and intermediate MICs
5. The choice of ATM
The choice of antibiotics is a crucial aspect of treating bacterial infections and requires careful consideration to ensure therapeutic efficacy while minimizing the development of antimicrobial resistance. Several essential steps are necessary for the appropriate selection of antibiotics. The first step in selecting an antibiotic is making an accurate diagnosis of the infection. This involves identifying the infection site and, whenever possible, the specific pathogen responsible. Diagnostic methods may include clinical evaluations, bacterial cultures, imaging studies, and molecular tests. Once the pathogen is identified, it is essential to assess its sensitivity to antibiotics. Antimicrobial susceptibility tests, such as the disk diffusion method or broth microdilution, offer valuable insights into which antibiotics are effective against the specific pathogen. The selection of an antibiotic should consider its spectrum of activity, which refers to the range of organisms it can effectively target. For infections caused by identified specific pathogens, a narrow-spectrum antibiotic is preferred to minimize disruption to the body’s normal microbiota and reduce the risk of resistance development. In cases of severe infections or when the pathogen is unknown, a broad-spectrum antibiotic may be necessary initially.
It is essential to consider the pharmacokinetics of the antibiotic, including how it is distributed, metabolized, and eliminated by the body, as well as its mechanism of action and the relationship between drug concentration and bactericidal effect (pharmacodynamics). Factors such as the antibiotic's ability to achieve therapeutic concentrations at the site of infection and potential drug interactions are also critical in the selection process. The selection of an antibiotic should also consider individual patient characteristics, such as allergies to antibiotics, comorbidities, renal and hepatic function, age, pregnancy, and lactation. Additionally, factors such as treatment cost and the ease of administration (e.g., oral versus intravenous) may also influence the choice of therapy.
Adherence to the principles of antimicrobial stewardship is essential throughout the antibiotic selection process to promote responsible use of these drugs, maximize therapeutic efficacy, protect public health, and minimize the development of antimicrobial resistance
[51]. This is clearly translated in the meta-analysis we conducted evaluating the literature on the % of error in empirical antibiotic therapy.
6. Systematic Review with Meta-Analysis
6.1. Search Strategy
We searched EMBASE, the Cochrane Library, Web of Science, and MEDLINE for relevant English/Spanish/Portuguese/French/Deutsch-language articles published between 2012 and 2024. To find additional relevant research, we examined the sources cited in the reviews (systematic reviews with or without meta-analysis). The search criteria included terms from the following Medical Subject Headings: (antibiotic OR antimicrobial OR antibacterial) AND (adequate OR inadequate OR appropriate OR inappropriate OR incorrect OR correct). The complete query search is described in the supplementary material.
6.2. Selection Process
Specify the methods used to decide whether a study met the inclusion criteria of the review, including how many reviewers screened each record and each report retrieved, whether they worked independently, and if applicable, details of automation tools used in the process.
6.3. Data Collection Process
Two authors conducted a full-text analysis on the publications that passed the first abstract screening. We included studies with all the following: a) adult hospitalized patients; b) diagnosis of any infections; c) evaluation adequacy of empirical therapy according with following microbiological culture of the site infection; d) evaluation of global mortality. Included were observational cohort studies, randomized controlled trials, and analyses of prospectively obtained data on the optimal timing of antibiotic administration. Not considered were meta-analyses, animal research, opinion pieces, small-scale studies, and editorial letters. Studies without site infection definition were excluded too.
6.4. Data Items
We documented the participant selection procedure, inclusion criteria, study duration and time, study site (emergency room, intensive care unit, ward), study type, and sample size for each infection site. Data on the baseline and the time intervals of empirical antibiotic and adequacy (or not) according with culture were not evaluated due to extensively variability among each patient and study. When microbiological documentation of infection was available, these data were also considered.
6.5. Subgroups Evaluation
Subgroup analyses were conducted to determine whether antimicrobial adequacy was different among different site infections, including bloodstream infection, respiratory tract infection, abdominal infection, urinary tract infection, and sepsis. In the case of sepsis, the site of infection was no included in the analysis. distinct outcomes.
6.6. Outcomes Assessment
All-cause mortality was the primary outcome measure. The mortality was variable, which includes 30 days or during hospitalization (global). The infection-associated mortality was not considered once there are not criteria for this outcome.
6.7. Statistical Analysis
All statistical analyses were performed with Review Manager Version 5.3. Dichotomous data are presented as odds ratios (ORs) with 95% confidence intervals (CIs). Statistical heterogeneity among studies was assessed via a χ2 test (chi-squared, where p < 0.10 indicates significant heterogeneity) and the I2 (degree of heterogeneity) statistic. Publication bias was assessed via visual inspection of the funnel plot.
7. Results of the Meta-Analysis
7.1. Study Selection
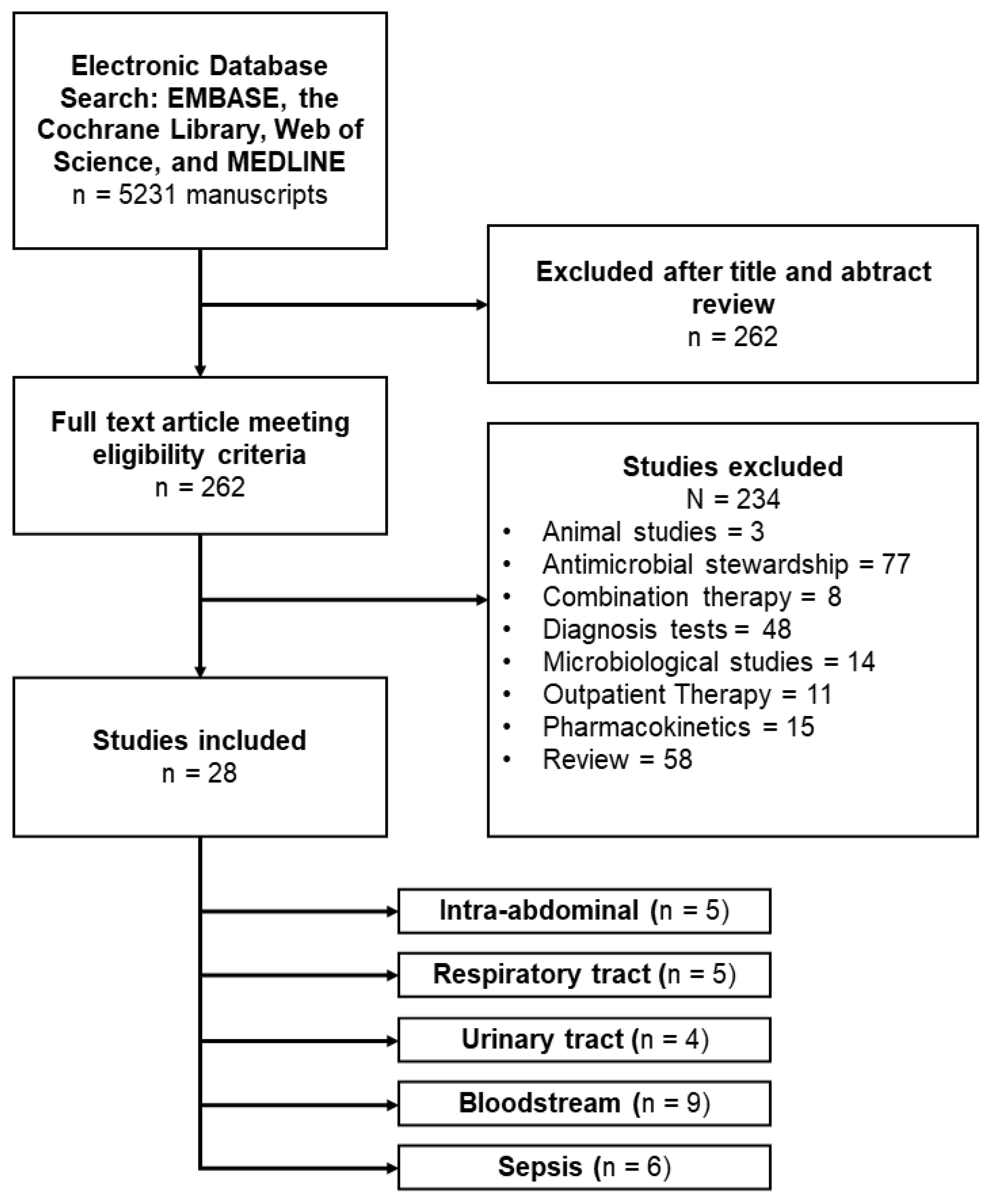
Twenty-eight included studies contained a total of 46.885 participants, with a median of 345 participants per study, a minimum of 28 participants, and a maximum of 35,529 participants [
52,
53,
54,
55,
56,
57,
58,
59,
60,
61,
62,
63,
64,
65,
66,
67,
68,
69,
70,
71,
72,
73,
74,
75,
76,
77].On Cohen's kappa scale, there was a 0.92 agreement between the two reviewers. After conducting a full-text search, we included 28 papers.
Figure 2 depicts reporting items for systematic reviews and meta-analyses (PRISMA) study selection strategy.
7.2. Study Characteristics
Twenty-eight included studies contained a total of 46.885 participants, with a median of 345 participants per study, a minimum of 28 participants, and a maximum of 35,529 participants. Among the sites of infection, the most studied were bloodstream infections (n=9), followed by respiratory tract infections (n=5), intra-abdominal infections (n=5) and 3 studies evaluated urinary tract infections. Studies that exclusively included patients with sepsis and septic shock were evaluated separately due to severity, although primary sites of sepsis were not included in studies that evaluated site-specific infection (
Figure 3).
7.3. Results of All Site Infections
Among the sites of infection, the most studied were bloodstream infections (n=9), followed by respiratory tract infections (n=5), intra-abdominal infections (n=5) and 3 studies evaluated urinary tract infections. Studies that exclusively included patients with sepsis and septic shock were evaluated separately due to severity, although primary sites of sepsis were not included in studies that evaluated site-specific infection.
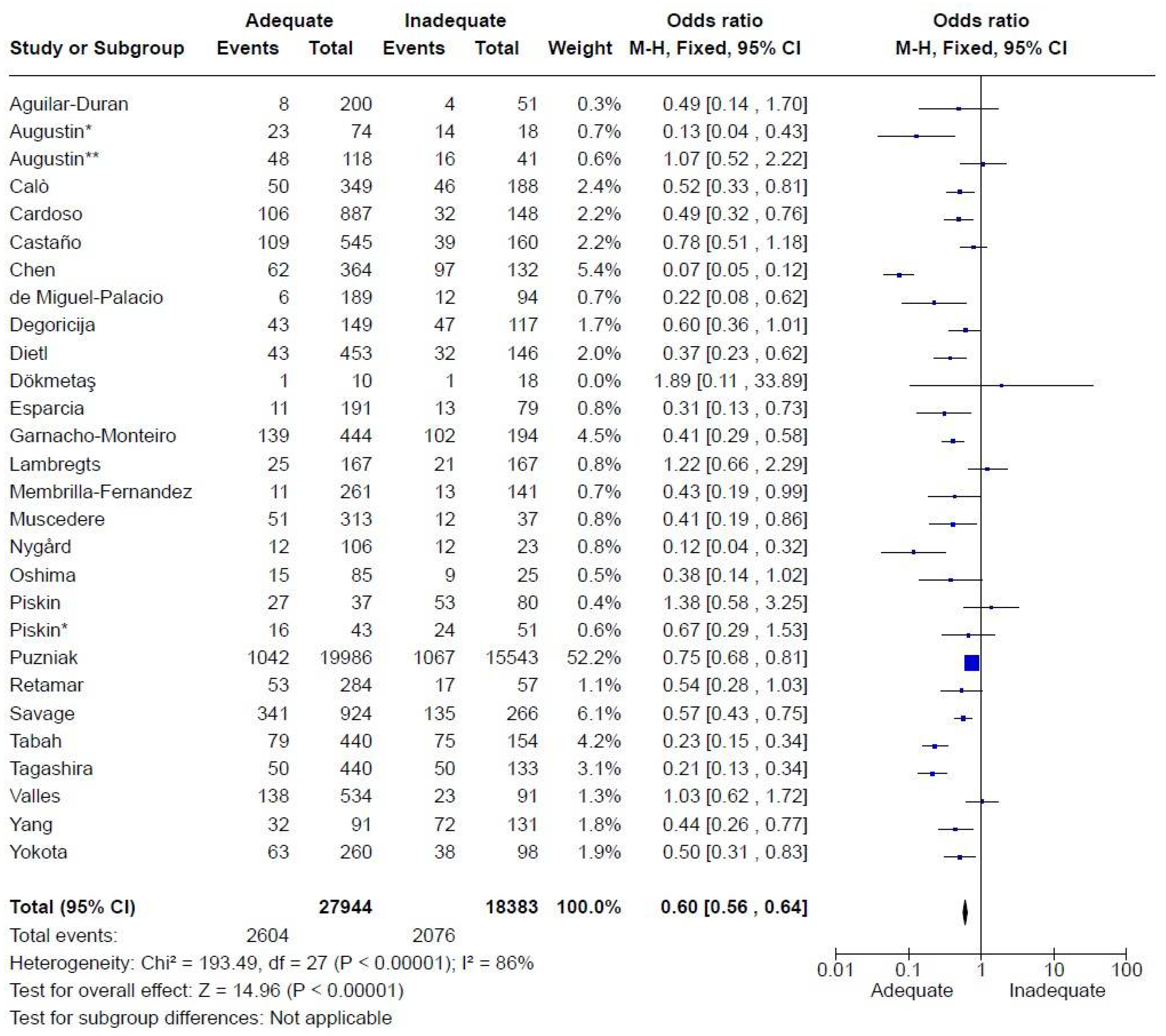
All studies were retrospective, involving adult patients with mortality as the primary outcome. Mortality endpoints varied across studies, with some using 30-day mortality (n=19) and others assessing in-hospital mortality. Preference was given to the analysis of 30-day mortality when both endpoints were available to mitigate the effects of delayed mortality due to causes other than infection, although infection may have contributed to prolonged hospitalization.
Four studies were conducted exclusively in emergency rooms, seven in ICUs, three in general wards, and the remainder in a combination of these settings. While all patients were hospitalized at the time of antibiotic prescription, some infections were defined as community-acquired (eight studies) while the majority were healthcare-associated infections. The number of patients included was 46,327. There were 4,680 deaths, resulting in a mortality rate of 10.1%. Adequate therapy was associated with reduced mortality, with an odds ratio (OR) of 0.60 [0.56-0.64] (
Figure 3).
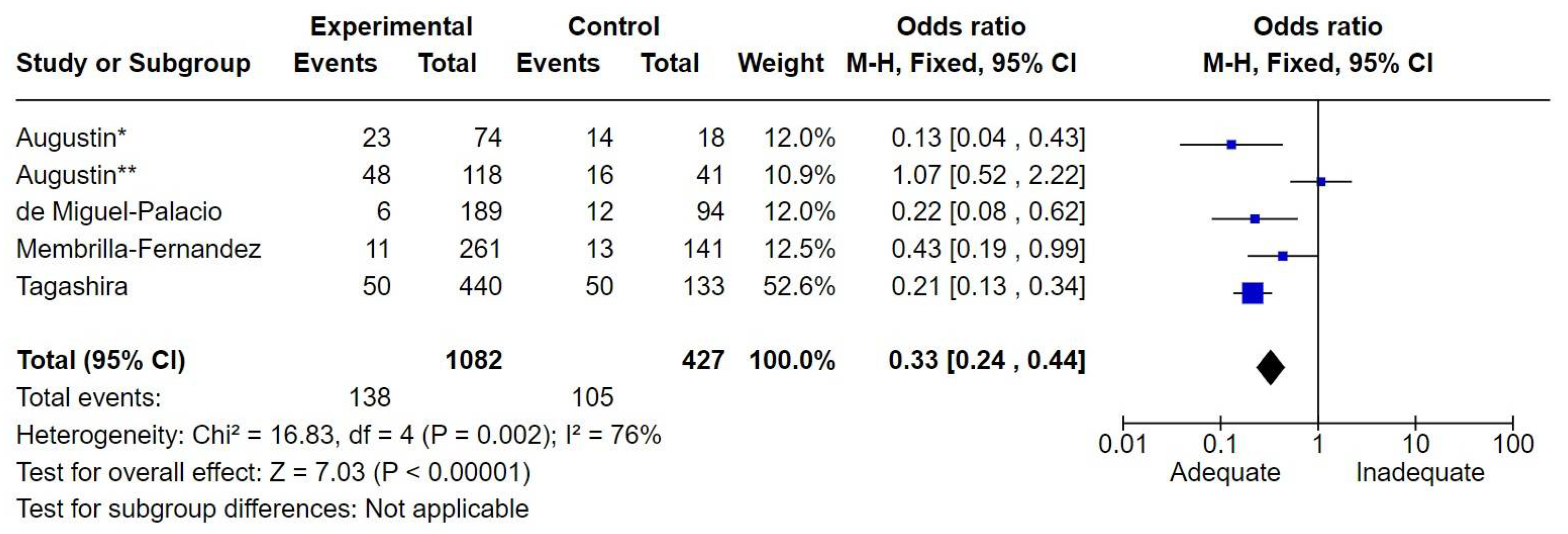
7.4. Abdominal Infections
The number of patients with abdominal infections was 1,509. The nature of the infection varied from secondary peritonitis to cholecystitis, with subgroup classification by infection type and severity not feasible. There were 243 deaths, corresponding to a mortality rate of 16.1%. Adequate therapy was associated with reduced mortality, with an odds ratio (OR) of 0.33 [0.24-0.44] (
Figure 4).
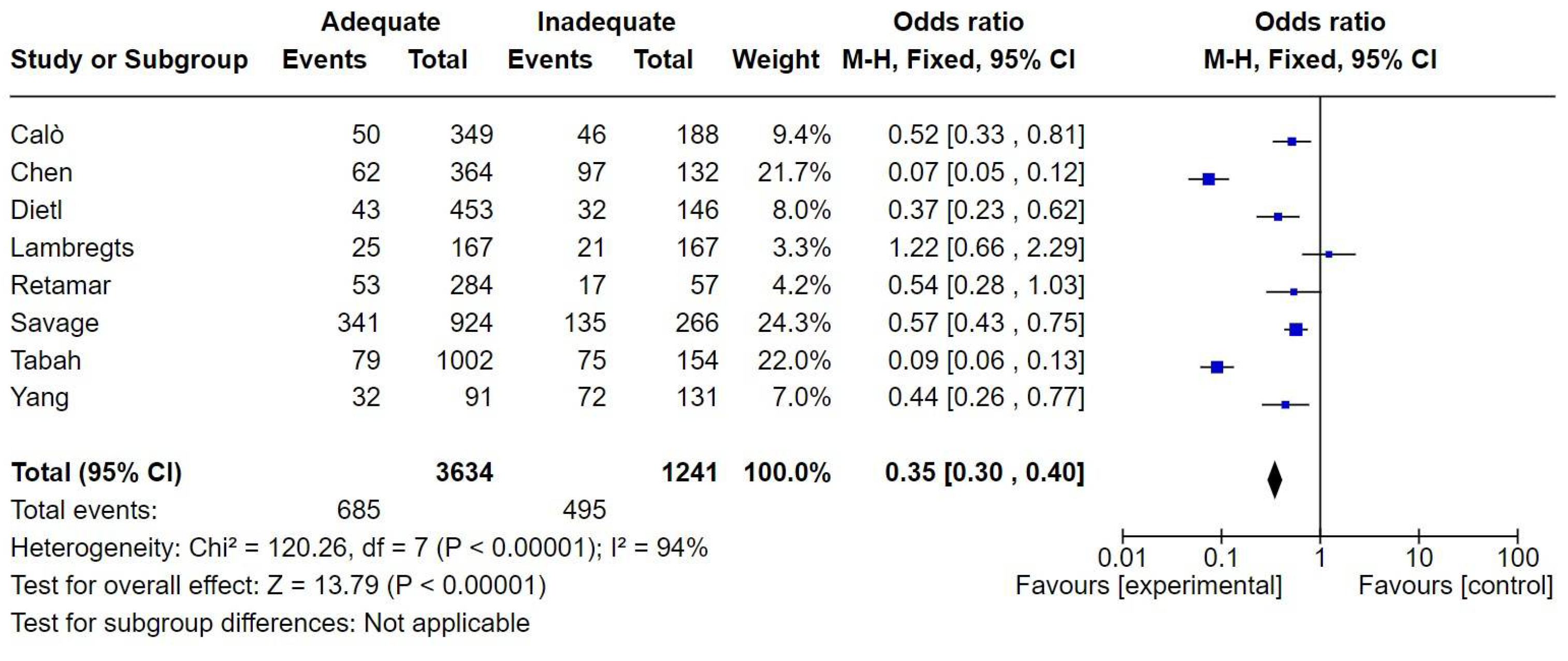
7.5. Bloodstream Infections
The definition of bloodstream infections (BSI) was consistent across the studies, with most studies categorizing infections based on mandatory microorganism identification criteria as per CDC guidelines. The microbiological profile was based on studies with non-species specific and also to target pathogens like
Pseudomonas aeruginosa and ESBL-producing
Enterobacterales. The number of patients with BSI was 4,875. There were 1,180 deaths, resulting in a mortality rate of 24.2%. Adequate therapy was associated with reduced mortality, with an odds ratio (OR) of 0.35 [0.30-0.40] (
Figure 5).
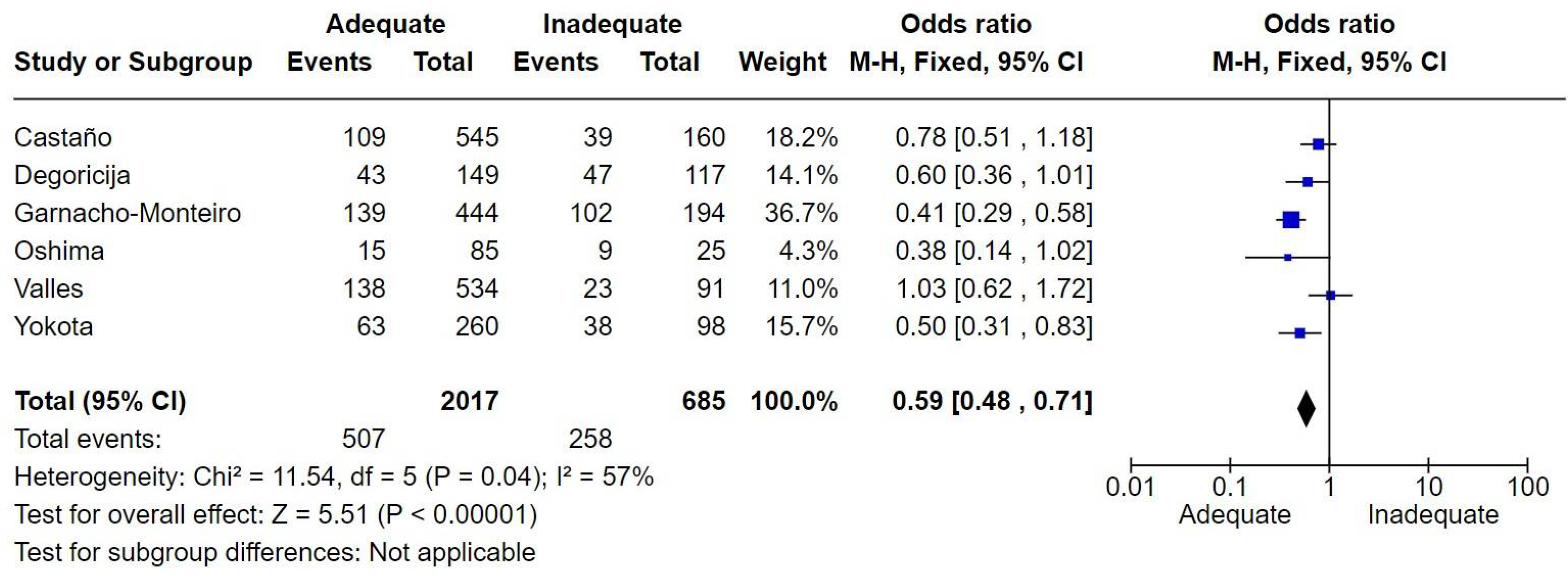
7.6. Respiratory Tract Infections
The definition of respiratory tract infections (RTI) was generally consistent across the studies. However, included community acquired as well as hospital acquired, including ventilator associated pneumonia. The number of patients with RTI was 690. There were 207 deaths, resulting in a mortality rate of 30.0%. Adequate therapy was associated with reduced mortality, with an odds ratio (OR) of 0.53 [0.35-0.80] (
Figure 6).
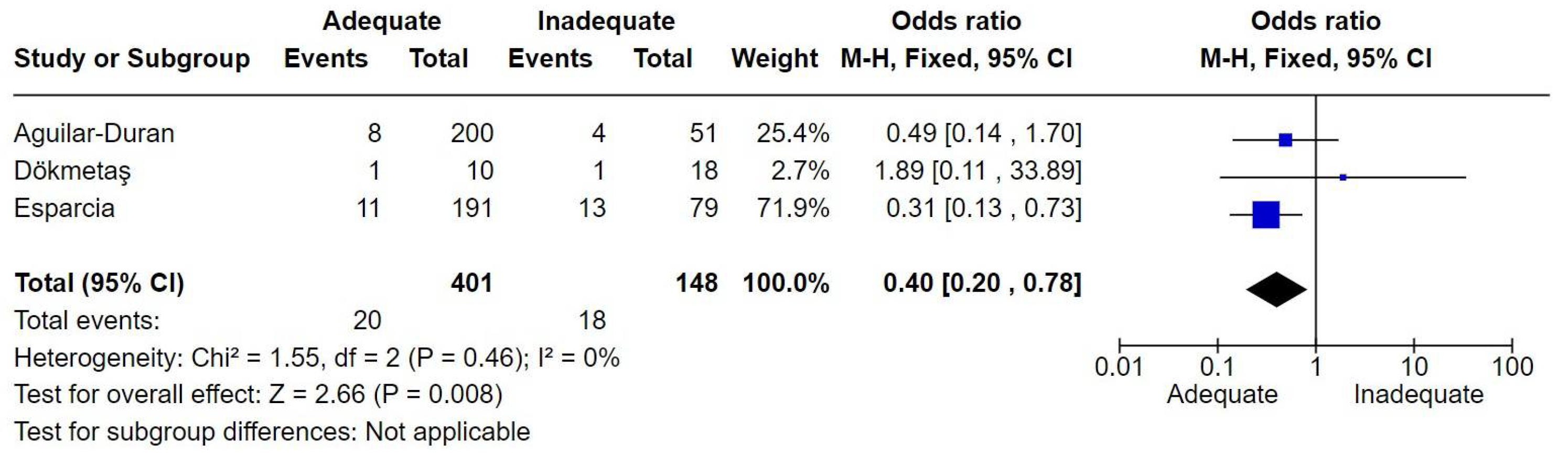
7.7. Urinary Tract Infections
The definition of urinary tract infections was consistent with pyelonephritis and catheter associated infections. The number of patients with RTI was 549. There were 38 deaths, resulting in a mortality rate of 6.9%. Adequate therapy was associated with reduced mortality, with an odds ratio (OR) of 0.40 [0.20-0.78] (
Figure 7).
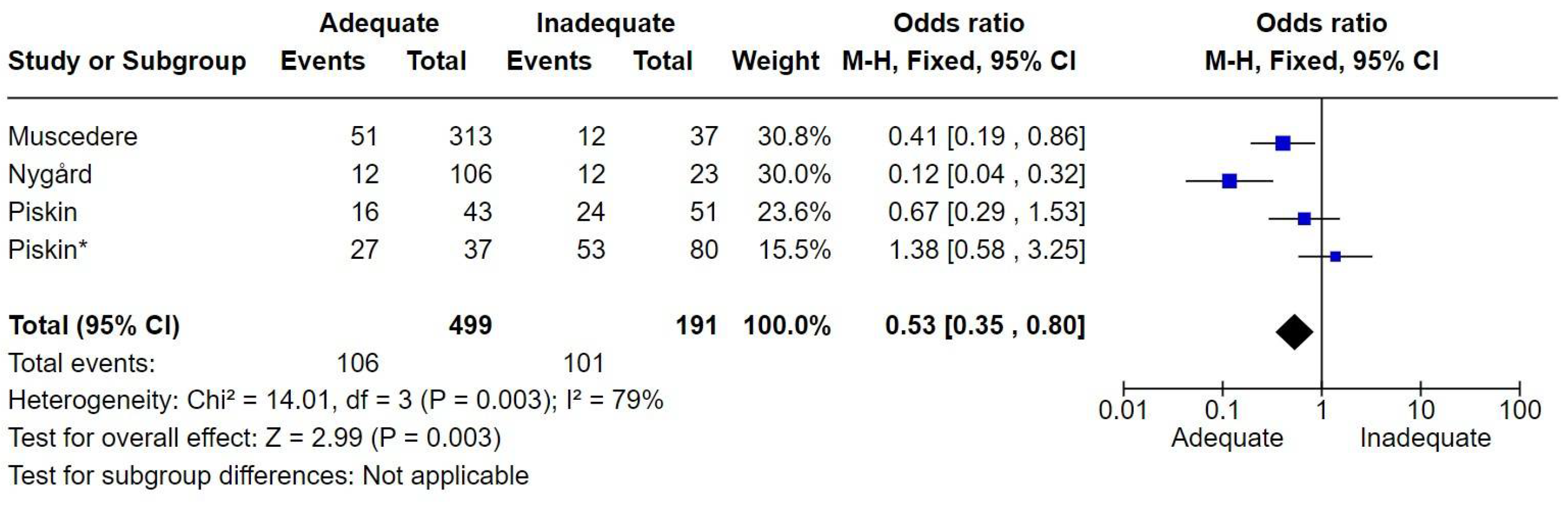
7.8. Sepsis and Shock Septic
The number of patients with sepsis or septic shock was 2,702. There were 765 deaths, corresponding to a mortality rate of 28.3%. Adequate therapy was associated with reduced mortality, with an odds ratio (OR) of 0.59 [0.48-0.71] (
Figure 8).
7.9. Results of Syntheses
All studies were retrospective, as it would be ethically impossible to propose inadequate therapy or to maintain it after pathogen identification. Among the biases encountered, the primary issue was an imbalance in cases, favoring a larger number of patients from the Puzniak et al. study. The etiologies of infection varied across studies, and it was not feasible to separate them by pathogens, knowing that the severity of each infection varies depending on the pathogens involved.
8. Discussion
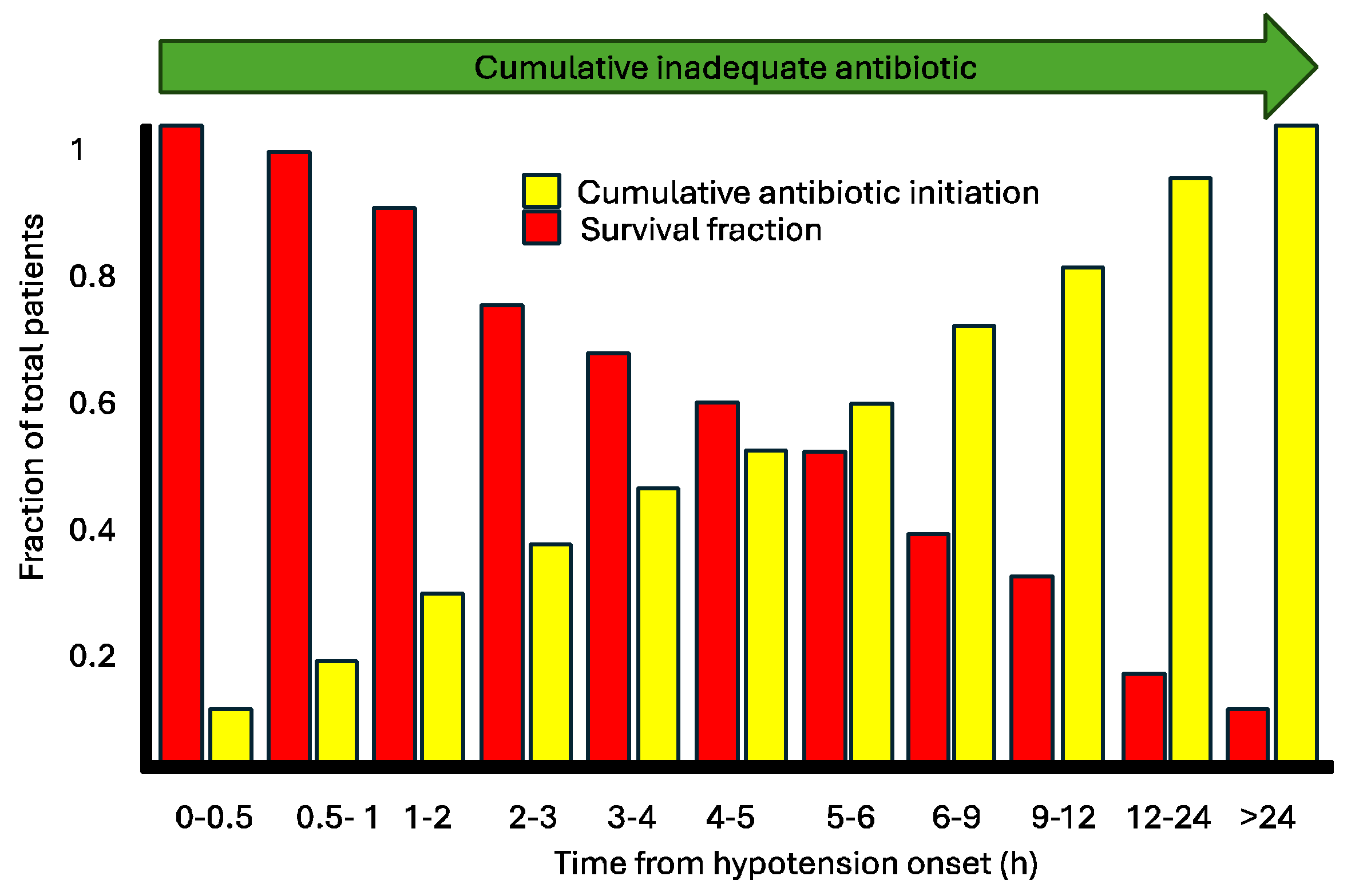
The mortality increases significantly when empirical antibiotics are not adequate. The risk of death increases 1.7 (70%), showing the importance of rapid identification of bacteria or correct empirical antimicrobial therapy based on concrete epidemiological data. The correct empirical therapy is the most effective approach to reduce mortality, emphasizing the importance of initiating treatment as early as possible [
78]. One of the seminal works illustrating the association between timing and appropriate therapy was published by Kumar et al. in 2006, focusing on patients with infection and associated hypotension, which would now be categorized as sepsis and septic shock [
79]. According to this study, each hour of delay in appropriate therapy corresponded to an increase in mortality proportionally. We consider delayed appropriate therapy in the same interpretive framework as Kumar et al.'s study (
Figure 9). However, we lack studies specifically evaluating the impact of hours of delay in appropriate versus inappropriate therapy. What we can assert is that inappropriate therapy within 24 hours is associated with higher mortality [
80].
This systematic review clearly demonstrates that for all infection sites included, mortality decreases with appropriate therapy, even for infections traditionally considered less severe such as urinary tract infections. Interestingly, while mortality rates in respiratory tract infections were noted, we did not identify a clear reason for the higher mortality in RTIs compared to sepsis, where one might expect higher mortality [
81]. In the studies of RTIs, patients were admitted to the ICU, whereas in another study, they were already in the ICU. In the study including community-acquired pneumonia patients, severity criteria were met but not the sepsis definition, thus excluding them from the sepsis group. Conversely, in studies involving sepsis patients, potentially lower mortality rates were attributed to faster initiation of therapy, as sepsis protocols demanding broad-spectrum therapy within the first hour (bundles) are stringent in this regard [
81]. Additionally, the time from symptom onset to therapy initiation is typically longer for community-acquired pneumonia patients who present from home.
In bloodstream infection studies, there was a predominance of specific pathogen etiology investigations. Consequently, we could not determine superior severity or mortality outcomes among the studies. As previously described, mortality associated with inadequate therapy in hospital-acquired bloodstream infection is higher in pathogens like
Acinetobacter baumannii, extended-spectrum beta-lactamase-producing
Enterobacteriaceae, and carbapenemase-producing KPC types [
80,
82,
83].
Another crucial variable to consider is the antibiotic class used in treating these patients, as pharmacokinetic and pharmacodynamic factors can influence clinical response, despite conflicting views in the literature. Adequate therapy should not solely consider antibiotic spectrum coverage against the pathogen; factors such as tissue penetration, dosage, and adverse events are equally important [
19,
84].
Of particular interest is the potential 50% reduction in mortality risk attributed to appropriate therapy. However, analysis of hospitalization duration and costs could not be included as a sub-analysis in this meta-analysis but could provide additional insights into patient quality of life, acquisition of multidrug-resistant bacteria, and costs associated with prolonged hospital stays, as demonstrated in various studies [
21].
Presently, healthcare systems' sustainability is increasingly scrutinized, especially in developing countries' hospitals. By conserving financial resources, investments can be made in enhanced diagnostic capabilities to expedite pathogen identification, thereby increasing diagnostic accuracy and reducing mortality. In this context, the application of molecular panels directly from blood or specific infection sites enables rapid pathogen identification, reinforcing the importance of timely, appropriate therapy within 24 hours.
However, the mere fact of rapid identification is not sufficient. Several studies demonstrate that without an effective antimicrobial stewardship team, the implementation of diagnostic technologies may not achieve the desired effect [
85,
86]. In addition to the "aggressive" activity of antimicrobial stewardship programs for adjusting, de-escalating, or even discontinuing unnecessary antibiotics, not all hospitals have the capability to maintain a full team of dedicated professionals for antimicrobial care [
25]. In this regard, the implementation of tele-stewardship is appealing, as it allows for cost reduction in staffing and integrates real-time technologies with diagnostic resources.
This is a meta-analysis, and classical limitations must be considered. Many limitations were described during the discussion, but retrospective studies have a significant impact. However, it would not be possible to conduct prospective studies on antimicrobial error. The doses of antibiotics were not considered, nor were the specific antibiotics used and the duration of treatment. The severity of the patients was not assessable due to the differences between the studies, nor was the matching of groups by age and comorbidities. Although one study had a larger number of patients than the others, the trend of reduced mortality was consistent across the other publications. Thus, we conclude that there was significant heterogeneity. Publication bias is an important factor, as it is uncommon for studies to show increased mortality with appropriate therapy, and few have shown no difference. Among the comparability biases, the mortality outcome varied between 30-day mortality and total hospital mortality. The clinical response variable would be the best way to analyze the antimicrobial response, but this information is rarely used in publications due to its variability in clinical interpretation. A variable not found in these studies was the length of hospital stay in patients with inadequate versus adequate therapy. With this variable, we could also include a cost-benefit analysis of rapid diagnosis.
9. Conclusion
Early appropriate therapy within the first 24 hours is associated with reduced mortality in respiratory, urinary, abdominal, bloodstream infections, and cases of sepsis. In this context, we highlight that healthcare technologies aimed at minimizing therapeutic inadequacies contribute to mortality reduction and, by extension, facilitate shorter hospital stays and decreased healthcare costs. These savings enable investments in antimicrobial stewardship programs and rapid diagnostic laboratory initiatives.
Author Contributions
F.F.T.; K.O.S, M.S.S: writing—original draft preparation, F.F.T.; T.Z. writing—review and editing. All authors have read and agreed to the published version of the manuscript. ChatGPT was used to improve the English language.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available under request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Holmes, A.H.; Moore, L.S.; Sundsfjord, A.; Steinbakk, M.; Regmi, S.; Karkey, A.; Guerin, P.J.; Piddock, L.J. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 2016, 387, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Wenzler, E.; Maximos, M.; Asempa, T.E.; Biehle, L.; Schuetz, A.N.; Hirsch, E.B. Antimicrobial susceptibility testing: An updated primer for clinicians in the era of antimicrobial resistance: Insights from the Society of Infectious Diseases Pharmacists. Pharmacotherapy 2023, 43, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Liu, Y.; Wang, R.; Wang, Q.; Jin, L.; Wang, H. The transferability and evolution of NDM-1 and KPC-2 co-producing Klebsiella pneumoniae from clinical settings. EBioMedicine 2020, 51, 102599. [Google Scholar] [CrossRef] [PubMed]
- Oteo, J.; Ortega, A.; Bartolome, R.; Bou, G.; Conejo, C.; Fernandez-Martinez, M.; Gonzalez-Lopez, J.J.; Martinez-Garcia, L.; Martinez-Martinez, L.; Merino, M.; et al. Prospective multicenter study of carbapenemase-producing Enterobacteriaceae from 83 hospitals in Spain reveals high in vitro susceptibility to colistin and meropenem. Antimicrob Agents Chemother 2015, 59, 3406–3412. [Google Scholar] [CrossRef] [PubMed]
- Quiles, M.G.; Rocchetti, T.T.; Fehlberg, L.C.; Kusano, E.J.; Chebabo, A.; Pereira, R.M.; Gales, A.C.; Pignatari, A.C. Unusual association of NDM-1 with KPC-2 and armA among Brazilian Enterobacteriaceae isolates. Braz J Med Biol Res 2015, 48, 174–177. [Google Scholar] [CrossRef] [PubMed]
- Vazquez-Ucha, J.C.; Seoane-Estevez, A.; Rodino-Janeiro, B.K.; Gonzalez-Bardanca, M.; Conde-Perez, K.; Martinez-Guitian, M.; Alvarez-Fraga, L.; Arca-Suarez, J.; Lasarte-Monterrubio, C.; Gut, M.; et al. Activity of imipenem/relebactam against a Spanish nationwide collection of carbapenemase-producing Enterobacterales. J Antimicrob Chemother 2021, 76, 1498–1510. [Google Scholar] [CrossRef] [PubMed]
- Lasarte-Monterrubio, C.; Guijarro-Sanchez, P.; Vazquez-Ucha, J.C.; Alonso-Garcia, I.; Alvarez-Fraga, L.; Outeda, M.; Martinez-Guitian, M.; Pena-Escolano, A.; Maceiras, R.; Lence, E.; et al. Antimicrobial Activity of Cefiderocol against the Carbapenemase-Producing Enterobacter cloacae Complex and Characterization of Reduced Susceptibility Associated with Metallo-beta-Lactamase VIM-1. Antimicrob Agents Chemother 2023, 67, e0150522. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.M.; Evans, L.E.; Rhodes, A. The Surviving Sepsis Campaign Bundle: 2018 update. Intensive Care Med 2018, 44, 925–928. [Google Scholar] [CrossRef] [PubMed]
- Bonine, N.G.; Berger, A.; Altincatal, A.; Wang, R.; Bhagnani, T.; Gillard, P.; Lodise, T. Impact of Delayed Appropriate Antibiotic Therapy on Patient Outcomes by Antibiotic Resistance Status From Serious Gram-negative Bacterial Infections. Am J Med Sci 2019, 357, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.M.; Newton, D.; Kunapuli, A.; Gandhi, T.N.; Washer, L.L.; Isip, J.; Collins, C.D.; Nagel, J.L. Impact of rapid organism identification via matrix-assisted laser desorption/ionization time-of-flight combined with antimicrobial stewardship team intervention in adult patients with bacteremia and candidemia. Clin Infect Dis 2013, 57, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Kock, R.; Wullenweber, J.; Horn, D.; Lanckohr, C.; Becker, K.; Idelevich, E.A. Implementation of short incubation MALDI-TOF MS identification from positive blood cultures in routine diagnostics and effects on empiric antimicrobial therapy. Antimicrob Resist Infect Control 2017, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Lockwood, A.M.; Perez, K.K.; Musick, W.L.; Ikwuagwu, J.O.; Attia, E.; Fasoranti, O.O.; Cernoch, P.L.; Olsen, R.J.; Musser, J.M. Integrating Rapid Diagnostics and Antimicrobial Stewardship in Two Community Hospitals Improved Process Measures and Antibiotic Adjustment Time. Infect Control Hosp Epidemiol 2016, 37, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Timbrook, T.T.; Morton, J.B.; McConeghy, K.W.; Caffrey, A.R.; Mylonakis, E.; LaPlante, K.L. The Effect of Molecular Rapid Diagnostic Testing on Clinical Outcomes in Bloodstream Infections: A Systematic Review and Meta-analysis. Clin Infect Dis 2017, 64, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Moungui, H.C.; Nana-Djeunga, H.C.; Anyiang, C.F.; Cano, M.; Ruiz Postigo, J.A.; Carrion, C. Dissemination Strategies for mHealth Apps: Systematic Review. JMIR Mhealth Uhealth 2024, 12, e50293. [Google Scholar] [CrossRef]
- Reno, B.; Oliveira, E.M.; Souza, A.D. A Systematic Literature Review on Trustworthiness for Applications Used in eHealth Environments. J Multidiscip Healthc 2023, 16, 3393–3403. [Google Scholar] [CrossRef] [PubMed]
- Nathwani, D.; Lawson, W.; Dryden, M.; Stephens, J.; Corman, S.; Solem, C.; Li, J.; Charbonneau, C.; Baillon-Plot, N.; Haider, S.; et al. Implementing criteria-based early switch/early discharge programmes: a European perspective. Clin Microbiol Infect 2015, 21 Suppl 2, S47–55. [Google Scholar] [CrossRef]
- Nathwani, D. Developments in outpatient parenteral antimicrobial therapy (OPAT) for Gram-positive infections in Europe, and the potential impact of daptomycin. J Antimicrob Chemother 2009, 64, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Karanika, S.; Paudel, S.; Grigoras, C.; Kalbasi, A.; Mylonakis, E. Systematic Review and Meta-analysis of Clinical and Economic Outcomes from the Implementation of Hospital-Based Antimicrobial Stewardship Programs. Antimicrob Agents Chemother 2016, 60, 4840–4852. [Google Scholar] [CrossRef] [PubMed]
- Telles, J.P.; Morales, R., Jr.; Yamada, C.H.; Marins, T.A.; D'Amaro Juodinis, V.; Sztajnbok, J.; Silva, M., Jr.; Bassetti, B.R.; Albiero, J.; Tuon, F.F. Optimization of Antimicrobial Stewardship Programs Using Therapeutic Drug Monitoring and Pharmacokinetics-Pharmacodynamics Protocols: A Cost-Benefit Review. Ther Drug Monit 2023, 45, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Cruz, J.A.W.; da Cunha, M.; de Moraes, T.P.; Marques, S.; Tuon, F.F.; Gomide, A.L.; de Paula Linhares, G. Brazilian private health system: history, scenarios, and trends. BMC Health Serv Res 2022, 22, 49. [Google Scholar] [CrossRef] [PubMed]
- Loesch, G.H.; Cruz, J.A.W.; Gasparetto, J.; Oliveira, D.D.S.; Telles, J.P.; Tuon, F.F. Cost minimization analysis of outpatient parenteral/oral antibiotic therapy at a trauma hospital: Public health system. Infect Control Hosp Epidemiol 2021, 42, 1445–1450. [Google Scholar] [CrossRef]
- Van Dijck, C.; Vlieghe, E.; Cox, J.A. Antibiotic stewardship interventions in hospitals in low-and middle-income countries: a systematic review. Bull World Health Organ 2018, 96, 266–280. [Google Scholar] [CrossRef] [PubMed]
- Davey, P.; Marwick, C.A.; Scott, C.L.; Charani, E.; McNeil, K.; Brown, E.; Gould, I.M.; Ramsay, C.R.; Michie, S. Interventions to improve antibiotic prescribing practices for hospital inpatients. Cochrane Database Syst Rev 2017, 2, CD003543. [Google Scholar] [CrossRef] [PubMed]
- Zequinao, T.; Telles, J.P.; Gasparetto, J.; Tuon, F.F. Carbapenem stewardship with ertapenem and antimicrobial resistance-a scoping review. Rev Soc Bras Med Trop 2020, 53, e20200413. [Google Scholar] [CrossRef] [PubMed]
- Zequinao, T.; Gasparetto, J.; Oliveira, D.D.S.; Silva, G.T.; Telles, J.P.; Tuon, F.F. A broad-spectrum beta-lactam-sparing stewardship program in a middle-income country public hospital: antibiotic use and expenditure outcomes and antimicrobial susceptibility profiles. Braz J Infect Dis 2020, 24, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Tuon, F.F.; Telles, J.P.; Gasparetto, J.; Zequinao, T. Antibiotic price rise and antibiotic stewardship programs-Stimulus or discouragement? Infect Control Hosp Epidemiol 2020, 41, 994–995. [Google Scholar] [CrossRef] [PubMed]
- Gasparetto, J.; Tuon, F.F.; Dos Santos Oliveira, D.; Zequinao, T.; Pipolo, G.R.; Ribeiro, G.V.; Beninca, P.D.; Cruz, J.A.W.; Moraes, T.P. Intravenous-to-oral antibiotic switch therapy: a cross-sectional study in critical care units. BMC Infect Dis 2019, 19, 650. [Google Scholar] [CrossRef] [PubMed]
- Shortliffe, E.H.; Davis, R.; Axline, S.G.; Buchanan, B.G.; Green, C.C.; Cohen, S.N. Computer-based consultations in clinical therapeutics: explanation and rule acquisition capabilities of the MYCIN system. Comput Biomed Res 1975, 8, 303–320. [Google Scholar] [CrossRef] [PubMed]
- Yu, V.L.; Fagan, L.M.; Wraith, S.M.; Clancey, W.J.; Scott, A.C.; Hannigan, J.; Blum, R.L.; Buchanan, B.G.; Cohen, S.N. Antimicrobial selection by a computer. A blinded evaluation by infectious diseases experts. JAMA 1979, 242, 1279–1282. [Google Scholar] [CrossRef] [PubMed]
- Beam, A.L.; Kohane, I.S. Big Data and Machine Learning in Health Care. JAMA 2018, 319, 1317–1318. [Google Scholar] [CrossRef] [PubMed]
- Peiffer-Smadja, N.; Rawson, T.M.; Ahmad, R.; Buchard, A.; Georgiou, P.; Lescure, F.X.; Birgand, G.; Holmes, A.H. Machine learning for clinical decision support in infectious diseases: a narrative review of current applications. Clin Microbiol Infect 2020, 26, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.C.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef]
- Shimabukuro, D.W.; Barton, C.W.; Feldman, M.D.; Mataraso, S.J.; Das, R. Effect of a machine learning-based severe sepsis prediction algorithm on patient survival and hospital length of stay: a randomised clinical trial. BMJ Open Respir Res 2017, 4, e000234. [Google Scholar] [CrossRef] [PubMed]
- Bucheeri, M.; Elligsen, M.; Lam, P.W.; Daneman, N.; MacFadden, D. A sepsis treatment algorithm to improve early antibiotic de-escalation while maintaining adequacy of coverage (Early-IDEAS): A prospective observational study. PLoS One 2023, 18, e0295908. [Google Scholar] [CrossRef] [PubMed]
- MacFadden, D.R.; Coburn, B.; Shah, N.; Robicsek, A.; Savage, R.; Elligsen, M.; Daneman, N. Decision-support models for empiric antibiotic selection in Gram-negative bloodstream infections. Clin Microbiol Infect 2019, 25, 108 e101–108 e107. [Google Scholar] [CrossRef] [PubMed]
- Ridgway, J.P.; Robicsek, A.; Shah, N.; Smith, B.A.; Singh, K.; Semel, J.; Acree, M.E.; Grant, J.; Ravichandran, U.; Peterson, L.R. A Randomized Controlled Trial of an Electronic Clinical Decision Support Tool for Inpatient Antimicrobial Stewardship. Clin Infect Dis 2021, 72, e265–e271. [Google Scholar] [CrossRef] [PubMed]
- Carreno, J.J.; Eaton, R.; Itro, L.; Babowicz, F.; Falvo, J.; Tobin, E.; Mitchell, C.; George, M. Time to clinical response in sepsis associated with an algorithm for blood-culture pathogen identification using matrix-assisted laser desorption ionization time-of-flight mass spectroscopy. Am J Health Syst Pharm 2019, 76, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Kakizaki, N.; Asai, K.; Kuroda, M.; Watanabe, R.; Kujiraoka, M.; Sekizuka, T.; Katagiri, M.; Moriyama, H.; Watanabe, M.; Saida, Y. Rapid identification of bacteria using a multiplex polymerase chain reaction system for acute abdominal infections. Front Microbiol 2023, 14, 1220651. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, V.S.T.; Cieslinski, J.; Bertol, J.; Schumacher, A.L.; Telles, J.P.; Tuon, F.F. Detection of Microorganisms in Clinical Sonicated Orthopedic Devices Using Conventional Culture and qPCR. Rev Bras Ortop (Sao Paulo) 2022, 57, 689–696. [Google Scholar] [CrossRef] [PubMed]
- Cieslinski, J.; Ribeiro, V.S.T.; Kraft, L.; Suss, P.H.; Rosa, E.; Morello, L.G.; Pillonetto, M.; Tuon, F.F. Direct detection of microorganisms in sonicated orthopedic devices after in vitro biofilm production and different processing conditions. Eur J Orthop Surg Traumatol 2021, 31, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Ai, J.; Wu, J.; Yu, S.; Cui, P.; Gao, Y.; Jin, J.; Weng, X.; Zhang, W. Rapid detection of respiratory organisms with FilmArray respiratory panel and its impact on clinical decisions in Shanghai, China, 2016-2018. Influenza Other Respir Viruses 2020, 14, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Rule, R.; Paruk, F.; Becker, P.; Neuhoff, M.; Chausse, J.; Said, M. Clinical utility of the BioFire FilmArray Blood Culture Identification panel in the adjustment of empiric antimicrobial therapy in the critically ill septic patient. PLoS One 2021, 16, e0254389. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.; Lau, S.K.; Teng, J.L.; Tse, H.; Yuen, K.Y. Then and now: use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect 2008, 14, 908–934. [Google Scholar] [CrossRef] [PubMed]
- Messacar, K.; Parker, S.K.; Todd, J.K.; Dominguez, S.R. Implementation of Rapid Molecular Infectious Disease Diagnostics: the Role of Diagnostic and Antimicrobial Stewardship. J Clin Microbiol 2017, 55, 715–723. [Google Scholar] [CrossRef] [PubMed]
- Claeys, K.C.; Johnson, M.D. Leveraging diagnostic stewardship within antimicrobial stewardship programmes. Drugs Context 2023, 12. [Google Scholar] [CrossRef] [PubMed]
- Hindler, J.F.; Stelling, J. Analysis and presentation of cumulative antibiograms: a new consensus guideline from the Clinical and Laboratory Standards Institute. Clin Infect Dis 2007, 44, 867–873. [Google Scholar] [CrossRef] [PubMed]
- Bielec, F.; Wenecka, M.; Brauncajs, M.; Pastuszak-Lewandoska, D. Analysis of Cumulative Antibiogram Reports in Search for Optimal Empirical Urinary Tract Infection Treatment at the Central Teaching Hospital of the Medical University of Lodz, Poland: Results of a 3-Year Surveillance. J Clin Med 2023, 12. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.; Antosz, K.; Daniels, R.; Gainey, A.B.; Burch, A.K. Providing value to patients and providers via a pediatric statewide antibiogram in South Carolina. Antimicrob Steward Healthc Epidemiol 2023, 3, e78. [Google Scholar] [CrossRef] [PubMed]
- Darboe, S.; Mirasol, R.; Adejuyigbe, B.; Muhammad, A.K.; Nadjm, B.; De St Maurice, A.; Dogan, T.L.; Ceesay, B.; Umukoro, S.; Okomo, U.; et al. Using an Antibiogram Profile to Improve Infection Control and Rational Antimicrobial Therapy in an Urban Hospital in The Gambia, Strategies and Lessons for Low- and Middle-Income Countries. Antibiotics (Basel) 2023, 12. [Google Scholar] [CrossRef] [PubMed]
- Negm, E.M.; Elgharabawy, E.S.; Badran, S.G.; Soliman, A.M.; El Sayed, A.M.; Raafat, A.O.N.; Soliman, S.T.; Mahmoud, H.M.; Tawfik, A.E.; El Hawary, A.T.; et al. Analysis of cumulative antibiogram reports in intensive care units at an Egyptian University Hospital. J Infect Public Health 2023, 16, 1220–1229. [Google Scholar] [CrossRef] [PubMed]
- Dupuis, C.; Timsit, J.F. Antibiotics in the first hour: is there new evidence? Expert Rev Anti Infect Ther 2021, 19, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Degoricija, V.; Sharma, M.; Legac, A.; Gradiser, M.; Sefer, S.; Vucicevic, Z. Survival analysis of 314 episodes of sepsis in medical intensive care unit in university hospital: impact of intensive care unit performance and antimicrobial therapy. Croat Med J 2006, 47, 385–397. [Google Scholar] [PubMed]
- Muscedere, J.G.; Shorr, A.F.; Jiang, X.; Day, A.; Heyland, D.K.; Canadian Critical Care Trials, G. The adequacy of timely empiric antibiotic therapy for ventilator-associated pneumonia: an important determinant of outcome. J Crit Care 2012, 27, 322 e327–314. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Duran, S.; Horcajada, J.P.; Sorlí, L.; Montero, M.; Salvadó, M.; Grau, S.; Gómez, J.; Knobel, H. Community-onset healthcare-related urinary tract infections: comparison with community and hospital-acquired urinary tract infections. J Infect 2012, 64, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Tabah, A.; Koulenti, D.; Laupland, K.; Misset, B.; Valles, J.; Bruzzi de Carvalho, F.; Paiva, J.A.; Cakar, N.; Ma, X.; Eggimann, P.; et al. Characteristics and determinants of outcome of hospital-acquired bloodstream infections in intensive care units: the EUROBACT International Cohort Study. Intensive Care Med 2012, 38, 1930–1945. [Google Scholar] [CrossRef] [PubMed]
- Piskin, N.; Aydemir, H.; Oztoprak, N.; Akduman, D.; Comert, F.; Kokturk, F.; Celebi, G. Inadequate treatment of ventilator-associated and hospital-acquired pneumonia: risk factors and impact on outcomes. BMC Infect Dis 2012, 12, 268. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.C.; Lin, W.L.; Lin, C.C.; Hsieh, W.H.; Hsieh, C.H.; Wu, M.H.; Wu, J.Y.; Lee, C.C. Outcome of inadequate empirical antibiotic therapy in emergency department patients with community-onset bloodstream infections. J Antimicrob Chemother 2013, 68, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, T.; Ribeiro, O.; Aragão, I.; Costa-Pereira, A.; Sarmento, A. The impact of healthcare-associated infection on mortality: failure in clinical recognition is related with inadequate antibiotic therapy. PLoS One 2013, 8, e58418. [Google Scholar] [CrossRef]
- Retamar, P.; Lopez-Prieto, M.D.; Natera, C.; de Cueto, M.; Nuno, E.; Herrero, M.; Fernandez-Sanchez, F.; Munoz, A.; Tellez, F.; Becerril, B.; et al. Reappraisal of the outcome of healthcare-associated and community-acquired bacteramia: a prospective cohort study. BMC Infect Dis 2013, 13, 344. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.J.; Chung, Y.C.; Chen, T.C.; Chang, H.L.; Tsai, Y.M.; Huang, M.S.; Chen, Y.H.; Lu, P.L. The impact of inappropriate antibiotics on bacteremia patients in a community hospital in Taiwan: an emphasis on the impact of referral information for cases from a hospital affiliated nursing home. BMC Infect Dis 2013, 13, 500. [Google Scholar] [CrossRef]
- Nygård, S.T.; Langeland, N.; Flaatten, H.K.; Fanebust, R.; Haugen, O.; Skrede, S. Aetiology, antimicrobial therapy and outcome of patients with community acquired severe sepsis: a prospective study in a Norwegian university hospital. BMC Infect Dis 2014, 14, 121. [Google Scholar] [CrossRef] [PubMed]
- Esparcia, A.; Artero, A.; Eiros, J.M.; Balaguer, M.; Madrazo, M.; Alberola, J.; Nogueira, J.M. Influence of inadequate antimicrobial therapy on prognosis in elderly patients with severe urinary tract infections. Eur J Intern Med 2014, 25, 523–527. [Google Scholar] [CrossRef] [PubMed]
- Yokota, P.K.; Marra, A.R.; Martino, M.D.; Victor, E.S.; Durao, M.S.; Edmond, M.B.; dos Santos, O.F. Impact of appropriate antimicrobial therapy for patients with severe sepsis and septic shock--a quality improvement study. PLoS One 2014, 9, e104475. [Google Scholar] [CrossRef] [PubMed]
- Membrilla-Fernandez, E.; Sancho-Insenser, J.J.; Girvent-Montllor, M.; Alvarez-Lerma, F.; Sitges-Serra, A.; Secondary Peritonitis Spanish Study, G. Effect of initial empiric antibiotic therapy combined with control of the infection focus on the prognosis of patients with secondary peritonitis. Surg Infect (Larchmt) 2014, 15, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Garnacho-Montero, J.; Gutiérrez-Pizarraya, A.; Escoresca-Ortega, A.; Fernández-Delgado, E.; López-Sánchez, J.M. Adequate antibiotic therapy prior to ICU admission in patients with severe sepsis and septic shock reduces hospital mortality. Crit Care 2015, 19, 302. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Kodama, Y.; Takahashi, W.; Hayashi, Y.; Iwase, S.; Kurita, T.; Saito, D.; Yamaji, Y.; Oda, S. Empiric Antibiotic Therapy for Severe Sepsis and Septic Shock. Surg Infect (Larchmt) 2016, 17, 210–216. [Google Scholar] [CrossRef] [PubMed]
- Savage, R.D.; Fowler, R.A.; Rishu, A.H.; Bagshaw, S.M.; Cook, D.; Dodek, P.; Hall, R.; Kumar, A.; Lamontagne, F.; Lauzier, F.; et al. The Effect of Inadequate Initial Empiric Antimicrobial Treatment on Mortality in Critically Ill Patients with Bloodstream Infections: A Multi-Centre Retrospective Cohort Study. PLoS One 2016, 11, e0154944. [Google Scholar] [CrossRef] [PubMed]
- Tagashira, Y.; Sakamoto, N.; Isogai, T.; Hikone, M.; Kosaka, A.; Chino, R.; Higuchi, M.; Uehara, Y.; Honda, H. Impact of inadequate initial antimicrobial therapy on mortality in patients with bacteraemic cholangitis: a retrospective cohort study. Clin Microbiol Infect 2017, 23, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Dokmetas, I.; Hamidi, A.A.; Bulut, M.E.; Cetin, S.; Oncul, A.; Uzun, N. Clinical effect of discordance in empirical treatment of cases with urinary tract infection accompanied by bacteremia. Turk J Urol 2017, 43, 543–548. [Google Scholar] [CrossRef]
- Castaño, P.; Plaza, M.; Molina, F.; Hincapié, C.; Maya, W.; Cataño, J.; González, J.; León, A.; Jaimes, F. Antimicrobial agent prescription: a prospective cohort study in patients with sepsis and septic shock. Trop Med Int Health 2019, 24, 175–184. [Google Scholar] [CrossRef] [PubMed]
- Valles, J.; Fontanals, D.; Oliva, J.C.; Martínez, M.; Navas, A.; Mesquida, J.; Gruartmoner, G.; de Haro, C.; Mestre, J.; Guía, C.; et al. Trends in the incidence and mortality of patients with community-acquired septic shock 2003-2016. J Crit Care 2019, 53, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Augustin, P.; Tanaka, S.; Tran-Dinh, A.; Parenti Ribeiro, L.; Arapis, K.; Grall, N.; Al Qarni, A.; Montravers, P. Outcome and Adequacy of Empirical Antibiotherapy in Post-Operative Peritonitis: A Retrospective Study. Surg Infect (Larchmt) 2020, 21, 284–292. [Google Scholar] [CrossRef] [PubMed]
- Lambregts, M.M.C.; Wijnakker, R.; Bernards, A.T.; Visser, L.G.; Cessie, S.L.; Boer, M.G.J. Mortality after Delay of Adequate Empiric Antimicrobial Treatment of Bloodstream Infection. J Clin Med 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Calo, F.; Retamar, P.; Martinez Perez-Crespo, P.M.; Lanz-Garcia, J.; Sousa, A.; Goikoetxea, J.; Reguera-Iglesias, J.M.; Leon, E.; Arminanzas, C.; Mantecon, M.A.; et al. Catheter-related bloodstream infections: predictive factors for Gram-negative bacteria aetiology and 30 day mortality in a multicentre prospective cohort. J Antimicrob Chemother 2020, 75, 3056–3061. [Google Scholar] [CrossRef] [PubMed]
- Puzniak, L.; Bauer, K.A.; Yu, K.C.; Moise, P.; Finelli, L.; Ye, G.; De Anda, C.; Vankeepuram, L.; Gupta, V. Effect of Inadequate Empiric Antibacterial Therapy on Hospital Outcomes in SARS-CoV-2-Positive and -Negative US Patients With a Positive Bacterial Culture: A Multicenter Evaluation From March to November 2020. Open Forum Infect Dis 2021, 8, ofab232. [Google Scholar] [CrossRef] [PubMed]
- Dietl, B.; Boix-Palop, L.; Gisbert, L.; Mateu, A.; Garreta, G.; Xercavins, M.; Badía, C.; López-Sánchez, M.; Pérez, J.; Calbo, E. Risk factors associated with inappropriate empirical antimicrobial treatment in bloodstream infections. A cohort study. Front Pharmacol 2023, 14, 1132530. [Google Scholar] [CrossRef] [PubMed]
- de Miguel-Palacio, M.; González-Castillo, A.M.; Membrilla-Fernández, E.; Pons-Fragero, M.J.; Pelegrina-Manzano, A.; Grande-Posa, L.; Morera-Casaponsa, R.; Sancho-Insenser, J.J. Impact of empiric antibiotic therapy on the clinical outcome of acute calculous cholecystitis. Langenbecks Arch Surg 2023, 408, 345. [Google Scholar] [CrossRef] [PubMed]
- Bateman, R.M.; Sharpe, M.D.; Jagger, J.E.; Ellis, C.G.; Sole-Violan, J.; Lopez-Rodriguez, M.; Herrera-Ramos, E.; Ruiz-Hernandez, J.; Borderias, L.; Horcajada, J.; et al. 36th International Symposium on Intensive Care and Emergency Medicine : Brussels, Belgium. 15-18 March 2016. Crit Care 2016, 20, 94. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L.; et al. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 2006, 34, 1589–1596. [Google Scholar] [CrossRef] [PubMed]
- Tuon, F.F.; Rymsza, A.M.; Penteado-Filho, S.R.; Pilonetto, M.; Arend, L.N.; Levin, A.S. Should polymyxin be used empirically to treat infections in patients under high risk for carbapenem-resistant Acinetobacter? J Infect 2011, 62, 246–249. [Google Scholar] [CrossRef]
- Orsatti, V.N.; Ribeiro, V.S.T.; de Oliveira Montenegro, C.; Costa, C.J.; Raboni, E.A.; Sampaio, E.R.; Michielin, F.; Gasparetto, J.; Telles, J.P.; Tuon, F.F. Sepsis death risk factor score based on systemic inflammatory response syndrome, quick sequential organ failure assessment, and comorbidities. Med Intensiva (Engl Ed) 2024, 48, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Tuon, F.F.; Kruger, M.; Terreri, M.; Penteado-Filho, S.R.; Gortz, L. Klebsiella ESBL bacteremia-mortality and risk factors. Braz J Infect Dis 2011, 15, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Tuon, F.F.; Bianchet, L.C.; Penteado-Filho, S.R. Epidemiology of extended spectrum beta-lactamase producing Enterobacter bacteremia in a brazilian hospital. Rev Soc Bras Med Trop 2010, 43, 452–454. [Google Scholar] [CrossRef] [PubMed]
- Telles, J.P.; Leme, R.C.P.; Campos, M.L.; Ito, C.; Bail, L.; Nogueira, K.D.S.; Tuon, F.F. Ceftriaxone and methicillin-susceptible staphylococcus aureus: a perspective from pharmacokinetics/pharmacodynamics studies. Expert Opin Drug Metab Toxicol 2021, 17, 1039–1048. [Google Scholar] [CrossRef] [PubMed]
- Reiber, C.; Bodendoerfer, E.; Brugger, S.D.; Eberhard, N.; Hitz, E.; Hofmaenner, D.A.; Herren, S.; Kolesnik-Goldmann, N.; Manicini, S.; Zbinden, R.; et al. Rapid antimicrobial susceptibility testing in patients with bacteraemia due to Enterobacterales: an implementation study. Swiss Med Wkly 2023, 153, 40066. [Google Scholar] [CrossRef] [PubMed]
- Cartuliares, M.B.; Rosenvinge, F.S.; Mogensen, C.B.; Skovsted, T.A.; Andersen, S.L.; Østergaard, C.; Pedersen, A.K.; Skjøt-Arkil, H. Evaluation of point-of-care multiplex polymerase chain reaction in guiding antibiotic treatment of patients acutely admitted with suspected community-acquired pneumonia in Denmark: A multicentre randomised controlled trial. PLoS Med 2023, 20, e1004314. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).