Submitted:
24 July 2024
Posted:
25 July 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. Cell Annotation
2.3. Tumor Marker Analysis
2.4. Workflows of Copy Number Variation (CNV) Analysis
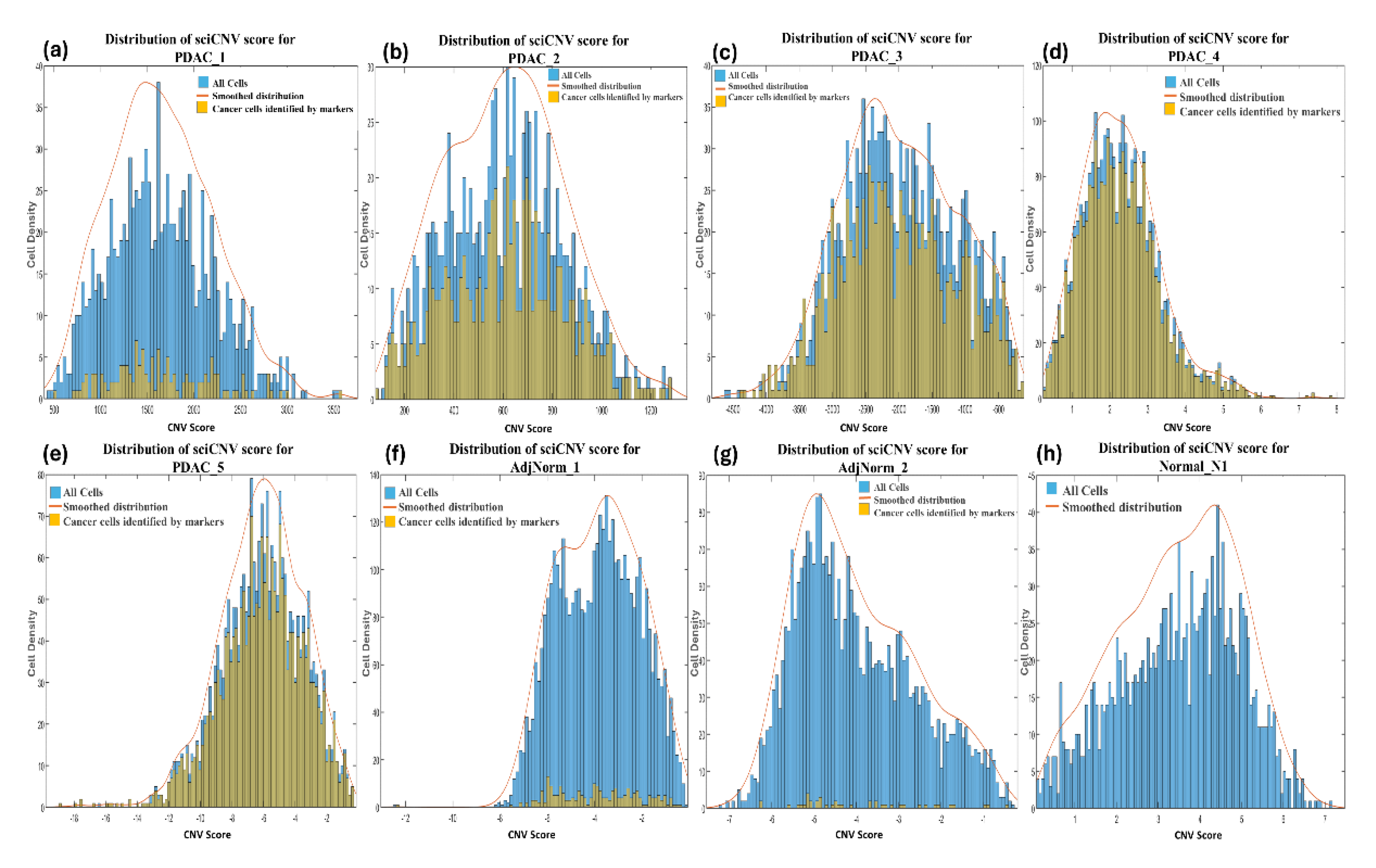
2.4.1. SciCNV (Single-Cell Inferred Chromosomal CNV)
2.4.2. InferCNV
2.4.3. CopyKAT (Copynumber Karyotyping of Aneuploid Tumors)
2.4.4. SCEVAN (Single CELL Variational ANeuploidy analysis)
2.5. Technical Considerations
3. Results
3.1. Cell Composition of Primary and Metastatic Tumor Tissues and Normal Pancreas Controls
3.2. PDAC Tumor Cell Identification Using a Marker-Based Method
3.3. PDAC Tumor Cell Identification using Inference of CNVs
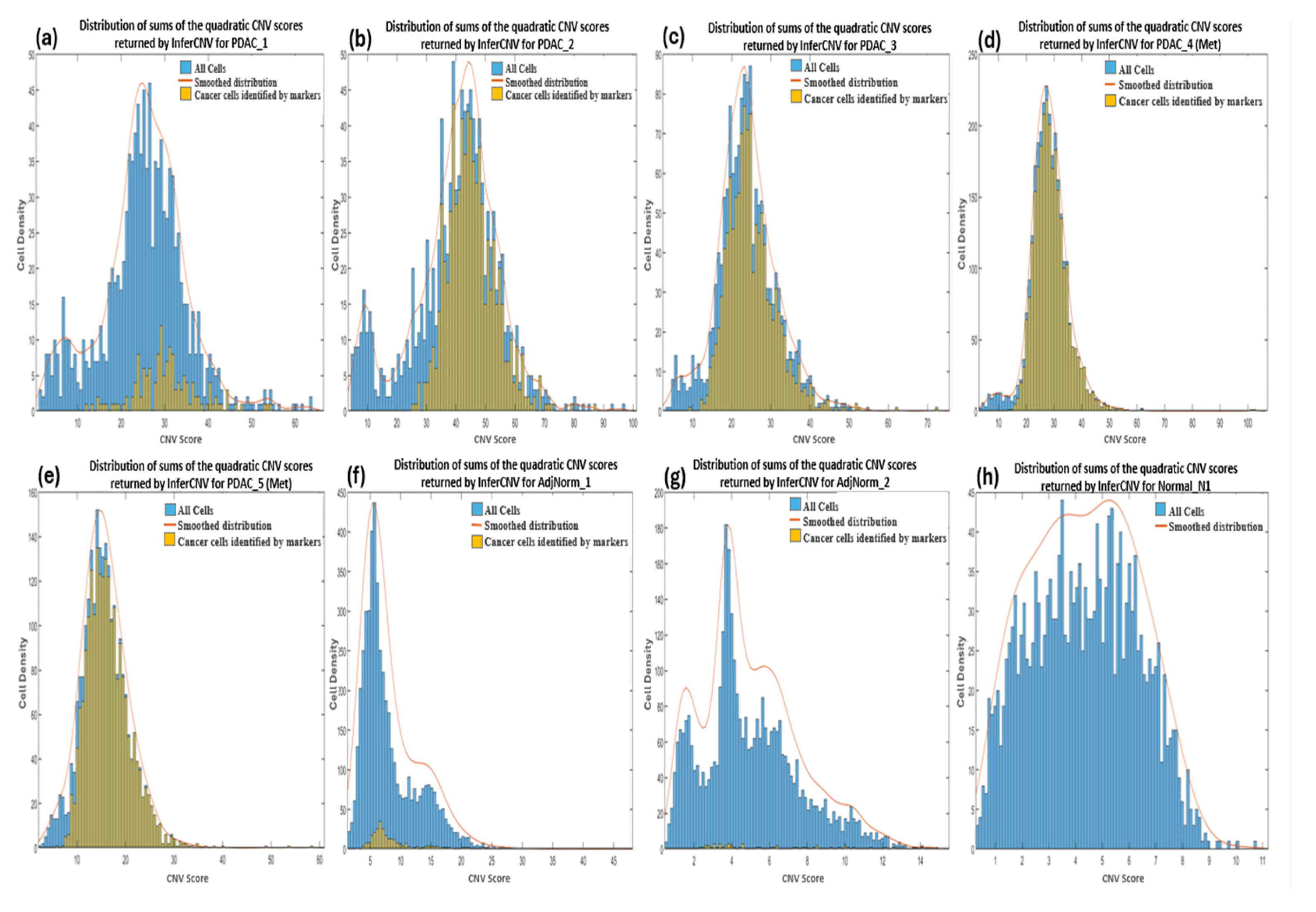
3.3.2. InferCNV
3.3.3. CopyKAT
3.3.4. SCEVAN
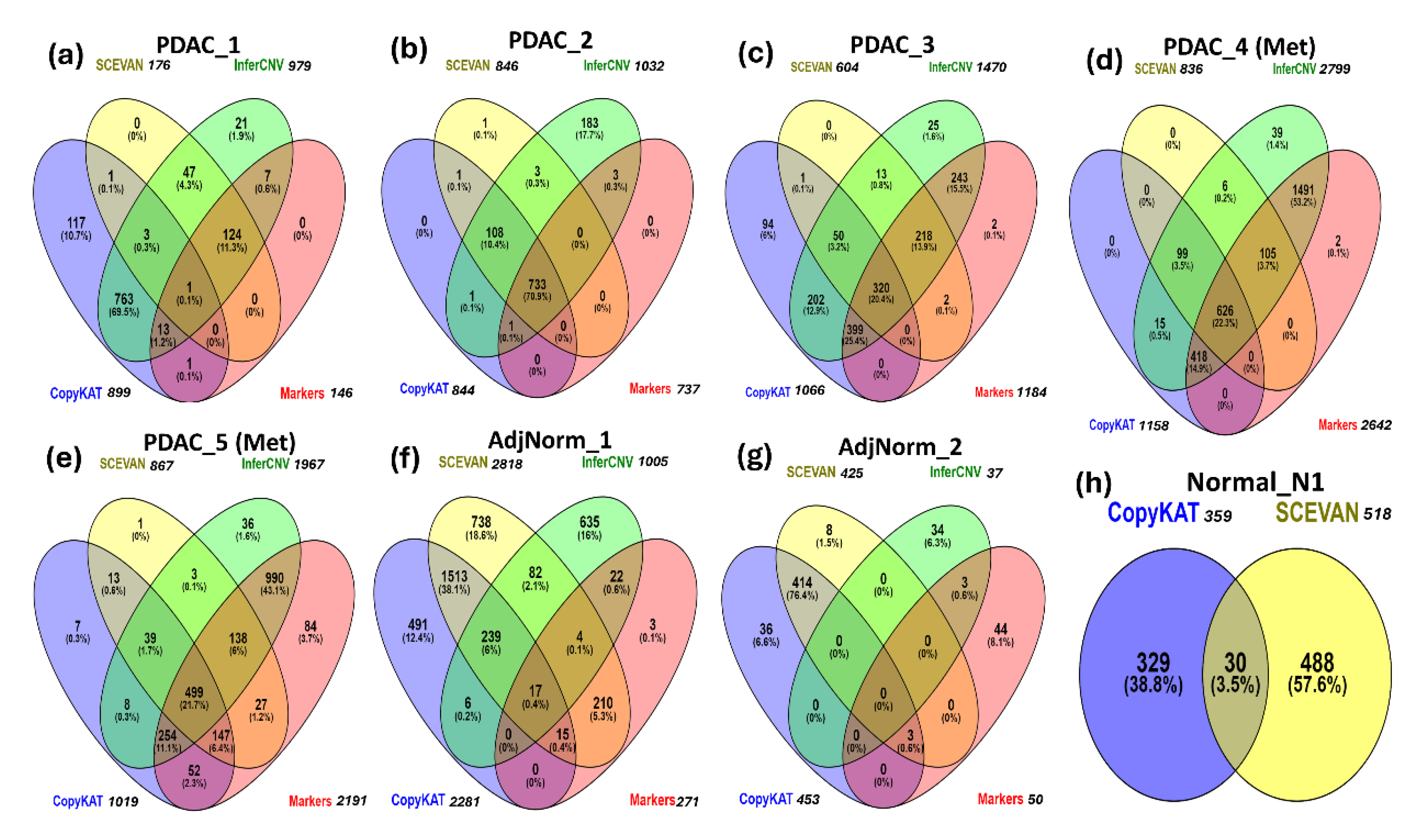
3.4. Comparisons Among the Methods of Inferring CNVs
3.5. Comparison between Tumor Cells Predicted by InferCNV and Markers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Adamska, A.; Domenichini, A.; Falasca, M., Pancreatic Ductal Adenocarcinoma: Current and Evolving Therapies. Int J Mol Sci 2017, 18, (7). [CrossRef]
- Mortezaee, K. Enriched cancer stem cells, dense stroma, and cold immunity: Interrelated events in pancreatic cancer. J Biochem Mol Toxicol 2021, (4), e22708. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R. L.; Giaquinto, A. N.; Jemal, A. Cancer statistics, 2024. CA Cancer J Clin 2024, (1), 12–49. [Google Scholar] [CrossRef] [PubMed]
- Muraro, M. J.; Dharmadhikari, G.; Grun, D.; Groen, N.; Dielen, T.; Jansen, E.; van Gurp, L.; Engelse, M. A.; Carlotti, F.; de Koning, E. J.; van Oudenaarden, A., A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst 2016, 3, (4), 385-394 e3.
- Tang, F.; Barbacioru, C.; Wang, Y.; Nordman, E.; Lee, C.; Xu, N.; Wang, X.; Bodeau, J.; Tuch, B. B.; Siddiqui, A.; Lao, K.; Surani, M. A. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 2009, (5), 377–82. [Google Scholar] [CrossRef] [PubMed]
- Hwang, B.; Lee, J. H.; Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med 2018, (8), 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Noel, P.; Borazanci, E. H.; Lee, J.; Amini, A.; Han, I. W.; Heo, J. S.; Jameson, G. S.; Fraser, C.; Steinbach, M.; Woo, Y.; Fong, Y.; Cridebring, D.; Von Hoff, D. D.; Park, J. O.; Han, H. Single-cell transcriptome analysis of tumor and stromal compartments of pancreatic ductal adenocarcinoma primary tumors and metastatic lesions. Genome Med 2020, (1), 80. [Google Scholar] [CrossRef] [PubMed]
- Elyada, E.; Bolisetty, M.; Laise, P.; Flynn, W. F.; Courtois, E. T.; Burkhart, R. A.; Teinor, J. A.; Belleau, P.; Biffi, G.; Lucito, M. S.; Sivajothi, S.; Armstrong, T. D.; Engle, D. D.; Yu, K. H.; Hao, Y.; Wolfgang, C. L.; Park, Y.; Preall, J.; Jaffee, E. M.; Califano, A.; Robson, P.; Tuveson, D. A. Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts. Cancer Discov 2019, (8), 1102–1123. [Google Scholar] [CrossRef] [PubMed]
- Ohlund, D.; Handly-Santana, A.; Biffi, G.; Elyada, E.; Almeida, A. S.; Ponz-Sarvise, M.; Corbo, V.; Oni, T. E.; Hearn, S. A.; Lee, E. J.; Chio, II; Hwang, C. I.; Tiriac, H.; Baker, L. A.; Engle, D. D.; Feig, C.; Kultti, A.; Egeblad, M.; Fearon, D. T.; Crawford, J. M.; Clevers, H.; Park, Y.; Tuveson, D. A., Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med 2017, 214, (3), 579-596.
- Pasquini, G.; Rojo Arias, J. E.; Schafer, P.; Busskamp, V. Automated methods for cell type annotation on scRNA-seq data. Comput Struct Biotechnol J 2021, 19, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Dohmen, J.; Baranovskii, A.; Ronen, J.; Uyar, B.; Franke, V.; Akalin, A. Identifying tumor cells at the single-cell level using machine learning. Genome Biol 2022, (1), 123. [Google Scholar] [CrossRef]
- Patel, A. P.; Tirosh, I.; Trombetta, J. J.; Shalek, A. K.; Gillespie, S. M.; Wakimoto, H.; Cahill, D. P.; Nahed, B. V.; Curry, W. T.; Martuza, R. L.; Louis, D. N.; Rozenblatt-Rosen, O.; Suva, M. L.; Regev, A.; Bernstein, B. E. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, (6190), 1396–401. [Google Scholar] [CrossRef]
- Gao, R.; Bai, S.; Henderson, Y. C.; Lin, Y.; Schalck, A.; Yan, Y.; Kumar, T.; Hu, M.; Sei, E.; Davis, A.; Wang, F.; Shaitelman, S. F.; Wang, J. R.; Chen, K.; Moulder, S.; Lai, S. Y.; Navin, N. E. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat Biotechnol 2021, (5), 599–608. [Google Scholar] [CrossRef] [PubMed]
- Loging, W. T.; Lal, A.; Siu, I. M.; Loney, T. L.; Wikstrand, C. J.; Marra, M. A.; Prange, C.; Bigner, D. D.; Strausberg, R. L.; Riggins, G. J. Identifying potential tumor markers and antigens by database mining and rapid expression screening. Genome Res 2000, (9), 1393–402. [Google Scholar] [CrossRef] [PubMed]
- Wei, R.; Liu, S.; Zhang, S.; Min, L.; Zhu, S. Cellular and Extracellular Components in Tumor Microenvironment and Their Application in Early Diagnosis of Cancers. Anal Cell Pathol (Amst) 2020, 2020, 6283796. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Abbas-Aghababazadeh, F.; Chen, Y. A.; Fridley, B. L. , Statistical and Bioinformatics Analysis of Data from Bulk and Single-Cell RNA Sequencing Experiments. Methods Mol Biol 2021, 2194, 143–175. [Google Scholar] [PubMed]
- Rosenberg, A. Z.; Armani, M. D.; Fetsch, P. A.; Xi, L.; Pham, T. T.; Raffeld, M.; Chen, Y.; O’Flaherty, N.; Stussman, R.; Blackler, A. R.; Du, Q.; Hanson, J. C.; Roth, M. J.; Filie, A. C.; Roh, M. H.; Emmert-Buck, M. R.; Hipp, J. D.; Tangrea, M. A. High-Throughput Microdissection for Next-Generation Sequencing. PLoS One 2016, (3), e0151775. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.; Quesnel-Vallieres, M.; Wang, D.; Thomas-Tikhonenko, A.; Lynch, K. W.; Barash, Y. Identifying common transcriptome signatures of cancer by interpreting deep learning models. Genome Biol 2022, (1), 117. [Google Scholar] [CrossRef] [PubMed]
- Nevins, J. R.; Potti, A. Mining gene expression profiles: expression signatures as cancer phenotypes. Nat Rev Genet 2007, (8), 601–9. [Google Scholar] [CrossRef] [PubMed]
- Nofech-Mozes, I.; Soave, D.; Awadalla, P.; Abelson, S. Pan-cancer classification of single cells in the tumour microenvironment. Nat Commun 2023, (1), 1615. [Google Scholar] [CrossRef] [PubMed]
- McShane, L. M.; Hayes, D. F. Publication of tumor marker research results: the necessity for complete and transparent reporting. J Clin Oncol 2012, (34), 4223–32. [Google Scholar] [CrossRef]
- Munz, M.; Baeuerle, P. A.; Gires, O. The emerging role of EpCAM in cancer and stem cell signaling. Cancer Res 2009, (14), 5627–9. [Google Scholar] [CrossRef] [PubMed]
- Spizzo, G.; Fong, D.; Wurm, M.; Ensinger, C.; Obrist, P.; Hofer, C.; Mazzoleni, G.; Gastl, G.; Went, P. EpCAM expression in primary tumour tissues and metastases: an immunohistochemical analysis. J Clin Pathol 2011, (5), 415–20. [Google Scholar] [CrossRef] [PubMed]
- Fong, D.; Steurer, M.; Obrist, P.; Barbieri, V.; Margreiter, R.; Amberger, A.; Laimer, K.; Gastl, G.; Tzankov, A.; Spizzo, G. Ep-CAM expression in pancreatic and ampullary carcinomas: frequency and prognostic relevance. J Clin Pathol 2008, (1), 31–5. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Gu, W.; Hurles, M. E.; Lupski, J. R. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 2009, 10, 451–81. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, G.; Petreaca, R. C. Distribution of copy number variations and rearrangement endpoints in human cancers with a review of literature. Mutat Res 2022, 824, 111773. [Google Scholar] [CrossRef] [PubMed]
- Shlien, A.; Malkin, D. Copy number variations and cancer. Genome Med 2009, (6), 62. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.; Zhao, X.; Meyerson, M. Somatic alterations in the human cancer genome. Cancer Cell 2004, (5), 433–8. [Google Scholar] [CrossRef] [PubMed]
- Shao, X.; Lv, N.; Liao, J.; Long, J.; Xue, R.; Ai, N.; Xu, D.; Fan, X. Copy number variation is highly correlated with differential gene expression: a pan-cancer study. BMC Med Genet 2019, (1), 175. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Niu, L.; Wu, B.; He, C.; Deng, L.; Chen, C.; Lan, Z.; Lin, C.; Kuang, W.; Lin, H.; Zou, J.; Zhang, W.; Luo, Z. Copy number variants landscape of multiple cancers and clinical applications based on NGS gene panel. Ann Med 2023, (2), 2280708. [Google Scholar] [CrossRef] [PubMed]
- Oketch, D. J. A.; Giulietti, M.; Piva, F., Copy Number Variations in Pancreatic Cancer: From Biological Significance to Clinical Utility. Int J Mol Sci 2023, 25, (1).
- Dancey, J. E.; Bedard, P. L.; Onetto, N.; Hudson, T. J. The genetic basis for cancer treatment decisions. Cell 2012, (3), 409–20. [Google Scholar] [CrossRef] [PubMed]
- Mahdipour-Shirayeh, A.; Erdmann, N.; Leung-Hagesteijn, C.; Tiedemann, R. E., sciCNV: high-throughput paired profiling of transcriptomes and DNA copy number variations at single-cell resolution. Brief Bioinform 2022, 23, (1).
- De Falco, A.; Caruso, F.; Su, X. D.; Iavarone, A.; Ceccarelli, M. A variational algorithm to detect the clonal copy number substructure of tumors from scRNA-seq data. Nat Commun 2023, (1), 1074. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, H.; Lyu, S.; Zhai, J.; Ji, Z.; Zhang, Z.; Zhang, X.; Liu, Z.; Wang, H.; Xu, J.; Fan, H.; Kou, J.; Li, L.; Lang, R.; He, Q. Single-cell transcriptomics reveals heterogeneous progression and EGFR activation in pancreatic adenosquamous carcinoma. Int J Biol Sci 2021, (10), 2590–2605. [Google Scholar] [CrossRef]
- Steele, N. G.; Carpenter, E. S.; Kemp, S. B.; Sirihorachai, V. R.; The, S.; Delrosario, L.; Lazarus, J.; Amir, E. D.; Gunchick, V.; Espinoza, C.; et al. Multimodal Mapping of the Tumor and Peripheral Blood Immune Landscape in Human Pancreatic Cancer. Nat Cancer 2020, (11), 1097–1112. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Looney, A. P.; Liu, L.; Wu, E.; Fong, V.; Hsu, A.; Chak, S.; Naikawadi, R. P.; Wolters, P. J.; Abate, A. R.; Butte, A. J.; Bhattacharya, M. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019, (2), 163–172. [Google Scholar] [CrossRef] [PubMed]
- Mabbott, N. A.; Baillie, J. K.; Brown, H.; Freeman, T. C.; Hume, D. A. An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genomics 2013, 14, 632. [Google Scholar] [CrossRef] [PubMed]
- Brown M, G. C., Haas B infercnvApp: An R shiny app to Infer Copy Number Variation from Single-Cell RNA-Seq Data https://github.com/broadinstitute/infercnvApp/wiki.
- Li, C.; Heidt, D. G.; Dalerba, P.; Burant, C. F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M. F.; Simeone, D. M. Identification of pancreatic cancer stem cells. Cancer Res 2007, (3), 1030–7. [Google Scholar] [CrossRef] [PubMed]
- Hermann, P. C.; Huber, S. L.; Herrler, T.; Aicher, A.; Ellwart, J. W.; Guba, M.; Bruns, C. J.; Heeschen, C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell 2007, (3), 313–23. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Li, T.; Wang, H.; Wu, J.; Yi, C.; Shi, J.; Wang, P.; Song, C.; Dai, L.; Jiang, G.; Huang, Y.; Yu, Y.; Li, J. TSPAN1, TMPRSS4, SDR16C5, and CTSE as Novel Panel for Pancreatic Cancer: A Bioinformatics Analysis and Experiments Validation. Front Immunol 2021, 12, 649551. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, H.; Chen, X.; Wang, Y.; Huang, S.; Chen, B.; Zhang, C.; Hou, B. Identification of LIPH as an unfavorable biomarkers correlated with immune suppression or evasion in pancreatic cancer based on RNA-seq. Cancer Immunol Immunother 2022, (3), 601–612. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Quan, G.; Huang, W.; Jiang, J. VSIG2 promotes malignant progression of pancreatic ductal adenocarcinoma by enhancing LAMTOR2-mediated mTOR activation. Cell Commun Signal 2023, (1), 223. [Google Scholar] [CrossRef]
- Zhang, B.; Xiao, J.; Cheng, X.; Liu, T. MAL2 interacts with IQGAP1 to promote pancreatic cancer progression by increasing ERK1/2 phosphorylation. Biochem Biophys Res Commun 2021, 554, 63–70. [Google Scholar] [CrossRef]
- Du, Y.; Hou, S.; Chen, Z.; Li, W.; Li, X.; Zhou, W., Comprehensive Analysis Identifies PKP3 Overexpression in Pancreatic Cancer Related to Unfavorable Prognosis. Biomedicines 2023, 11, (9).
- Ramachandran, V.; Arumugam, T.; Wang, H.; Logsdon, C. D. Anterior gradient 2 is expressed and secreted during the development of pancreatic cancer and promotes cancer cell survival. Cancer Res 2008, (19), 7811–8. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Sun, B. F.; Chen, C. Y.; Zhou, J. Y.; Chen, Y. S.; Chen, H.; Liu, L.; Huang, D.; Jiang, J.; Cui, G. S.; Yang, Y.; Wang, W.; Guo, D.; Dai, M.; Guo, J.; Zhang, T.; Liao, Q.; Liu, Y.; Zhao, Y. L.; Han, D. L.; Zhao, Y.; Yang, Y. G.; Wu, W. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res 2019, (9), 725–738. [Google Scholar] [CrossRef] [PubMed]
- Anand, S.; Khan, M. A.; Khushman, M.; Dasgupta, S.; Singh, S.; Singh, A. P., Comprehensive Analysis of Expression, Clinicopathological Association and Potential Prognostic Significance of RABs in Pancreatic Cancer. Int J Mol Sci 2020, 21, (15).
- Xiong, F.; Wu, G. H.; Wang, B.; Chen, Y. J. Plastin-3 is a diagnostic and prognostic marker for pancreatic adenocarcinoma and distinguishes from diffuse large B-cell lymphoma. Cancer Cell Int 2021, (1), 411. [Google Scholar] [CrossRef]
- Kayed, H.; Kleeff, J.; Kolb, A.; Ketterer, K.; Keleg, S.; Felix, K.; Giese, T.; Penzel, R.; Zentgraf, H.; Buchler, M. W.; Korc, M.; Friess, H. FXYD3 is overexpressed in pancreatic ductal adenocarcinoma and influences pancreatic cancer cell growth. Int J Cancer 2006, (1), 43–54. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y. C.; Ma, J.; Zhang, J.; Wang, J. C. Long non-coding RNA LINC01133 silencing exerts antioncogenic effect in pancreatic cancer through the methylation of DKK1 promoter and the activation of Wnt signaling pathway. Cancer Biol Ther 2019, (3), 368–380. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Liang, X.; Shi, L.; Tang, L.; Chen, D.; Liu, A.; Shao, C. SYT8 promotes pancreatic cancer progression via the TNNI2/ERRalpha/SIRT1 signaling pathway. Cell Death Discov 2021, (1), 390. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Fan, R. Identification and verification of CCNB1 as a potential prognostic biomarker by comprehensive analysis. Sci Rep 2022, (1), 16153. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y. F.; Yin, X. M.; Liu, X. Q. TOP2A induces malignant character of pancreatic cancer through activating beta-catenin signaling pathway. Biochim Biophys Acta Mol Basis Dis 2018, (1), 197–207. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Gao, W.; Li, X.; Yu, L.; Luo, D.; Liu, Y.; Yu, X. S100A14 promotes progression and gemcitabine resistance in pancreatic cancer. Pancreatology 2021, (3), 589–598. [Google Scholar] [CrossRef] [PubMed]
- Song, N.; Cui, K.; Zeng, L.; Fan, Y.; Wang, Z.; Shi, P.; Su, W.; Wang, H. Calpain 8 as a potential biomarker regulates the progression of pancreatic cancer via EMT and AKT/ERK pathway. J Proteomics 2024, 301, 105182. [Google Scholar] [CrossRef] [PubMed]
- Carrasco-Garcia, E.; Lopez, L.; Moncho-Amor, V.; Carazo, F.; Aldaz, P.; Collado, M.; Bell, D.; Gaafar, A.; Karamitopoulou, E.; Tzankov, A.; Hidalgo, M.; Rubio, A.; Serrano, M.; Lawrie, C. H.; Lovell-Badge, R.; Matheu, A., SOX9 Triggers Different Epithelial to Mesenchymal Transition States to Promote Pancreatic Cancer Progression. Cancers (Basel) 2022, 14, (4).
- Li, J.; Chen, X.; Zhu, L.; Lao, Z.; Zhou, T.; Zang, L.; Ge, W.; Jiang, M.; Xu, J.; Cao, Y.; Du, S.; Yu, Y.; Fan, G.; Wang, H. SOX9 is a critical regulator of TSPAN8-mediated metastasis in pancreatic cancer. Oncogene 2021, (30), 4884–4893. [Google Scholar] [CrossRef]
- Aran, D.; Camarda, R.; Odegaard, J.; Paik, H.; Oskotsky, B.; Krings, G.; Goga, A.; Sirota, M.; Butte, A. J. Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat Commun 2017, (1), 1077. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, E. S.; Elhossiny, A. M.; Kadiyala, P.; Li, J.; McGue, J.; Griffith, B. D.; Zhang, Y.; Edwards, J.; Nelson, S.; Lima, F.; et al. Analysis of Donor Pancreata Defines the Transcriptomic Signature and Microenvironment of Early Neoplastic Lesions. Cancer Discov 2023, (6), 1324–1345. [Google Scholar] [CrossRef] [PubMed]
- Girish, V.; Lakhani, A. A.; Scaduto, C. M.; Thompson, S. L.; Brown, L. M.; Hagenson, R. A.; Sausville, E. L.; Mendelson, B. E.; Lukow, D. A.; Yuan, M. L.; et al. Oncogene-like addiction to aneuploidy in human cancers. bioRxiv 2023. [Google Scholar] [CrossRef] [PubMed]
- Replogle, J. M.; Zhou, W.; Amaro, A. E.; McFarland, J. M.; Villalobos-Ortiz, M.; Ryan, J.; Letai, A.; Yilmaz, O.; Sheltzer, J.; Lippard, S. J.; Ben-David, U.; Amon, A. Aneuploidy increases resistance to chemotherapeutics by antagonizing cell division. Proc Natl Acad Sci U S A 2020, (48), 30566–30576. [Google Scholar] [CrossRef] [PubMed]
- Bentkowski, P.; Radwan, J. Evolution of major histocompatibility complex gene copy number. PLoS Comput Biol 2019, (5), e1007015. [Google Scholar] [CrossRef] [PubMed]
- Alfieri, F.; Caravagna, G.; Schaefer, M. H. Cancer genomes tolerate deleterious coding mutations through somatic copy number amplifications of wild-type regions. Nat Commun 2023, (1), 3594. [Google Scholar] [CrossRef] [PubMed]
- Redon, R.; Ishikawa, S.; Fitch, K. R.; Feuk, L.; Perry, G. H.; Andrews, T. D.; Fiegler, H.; Shapero, M. H.; Carson, A. R.; Chen, W.; et al. Global variation in copy number in the human genome. Nature 2006, (7118), 444–54. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y. J.; Lu, X. F.; Chen, H.; Wang, X. Y.; Cheng, W.; Zhang, Q. W.; Chen, J. N.; Wang, X. Y.; Jin, J. Z.; Yan, F. R.; Chen, H.; Li, X. B. Single-cell Transcriptomics Reveals Early Molecular and Immune Alterations Underlying the Serrated Neoplasia Pathway Toward Colorectal Cancer. Cell Mol Gastroenterol Hepatol 2023, (2), 393–424. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Levitin, H. M.; Frattini, V.; Bush, E. C.; Boyett, D. M.; Samanamud, J.; Ceccarelli, M.; Dovas, A.; Zanazzi, G.; Canoll, P.; Bruce, J. N.; Lasorella, A.; Iavarone, A.; Sims, P. A. Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med 2018, (1), 57. [Google Scholar] [CrossRef] [PubMed]
- Tirosh, I.; Venteicher, A. S.; Hebert, C.; Escalante, L. E.; Patel, A. P.; Yizhak, K.; Fisher, J. M.; Rodman, C.; Mount, C.; Filbin, M. G.; Neftel, C.; Desai, N.; Nyman, J.; Izar, B.; Luo, C. C.; Francis, J. M.; Patel, A. A.; Onozato, M. L.; Riggi, N.; Livak, K. J.; Gennert, D.; Satija, R.; Nahed, B. V.; Curry, W. T.; Martuza, R. L.; Mylvaganam, R.; Iafrate, A. J.; Frosch, M. P.; Golub, T. R.; Rivera, M. N.; Getz, G.; Rozenblatt-Rosen, O.; Cahill, D. P.; Monje, M.; Bernstein, B. E.; Louis, D. N.; Regev, A.; Suva, M. L. Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 2016, (7628), 309–313. [Google Scholar] [CrossRef] [PubMed]
- Neftel, C.; Laffy, J.; Filbin, M. G.; Hara, T.; Shore, M. E.; Rahme, G. J.; Richman, A. R.; Silverbush, D.; Shaw, M. L.; Hebert, C. M.; Dewitt, J.; Gritsch, S.; Perez, E. M.; Gonzalez Castro, L. N.; Lan, X.; Druck, N.; Rodman, C.; Dionne, D.; Kaplan, A.; Bertalan, M. S.; Small, J.; Pelton, K.; Becker, S.; Bonal, D.; Nguyen, Q. D.; Servis, R. L.; Fung, J. M.; Mylvaganam, R.; Mayr, L.; Gojo, J.; Haberler, C.; Geyeregger, R.; Czech, T.; Slavc, I.; Nahed, B. V.; Curry, W. T.; Carter, B. S.; Wakimoto, H.; Brastianos, P. K.; Batchelor, T. T.; Stemmer-Rachamimov, A.; Martinez-Lage, M.; Frosch, M. P.; Stamenkovic, I.; Riggi, N.; Rheinbay, E.; Monje, M.; Rozenblatt-Rosen, O.; Cahill, D. P.; Patel, A. P.; Hunter, T.; Verma, I. M.; Ligon, K. L.; Louis, D. N.; Regev, A.; Bernstein, B. E.; Tirosh, I.; Suva, M. L., An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019, 178, (4), 835-849 e21.
- Caruso, F. P.; Garofano, L.; D’Angelo, F.; Yu, K.; Tang, F.; Yuan, J.; Zhang, J.; Cerulo, L.; Pagnotta, S. M.; Bedognetti, D.; Sims, P. A.; Suva, M.; Su, X. D.; Lasorella, A.; Iavarone, A.; Ceccarelli, M., A map of tumor-host interactions in glioma at single-cell resolution. Gigascience 2020, 9, (10).
- Nicoletti, A.; Vitale, F.; Quero, G.; Paratore, M.; Fiorillo, C.; Negri, M.; Carlino, A.; Inzani, F.; Gasbarrini, A.; Alfieri, S.; Zileri Dal Verme, L., Immunohistochemical Evaluation of the Expression of Specific Membrane Antigens in Patients with Pancreatic Ductal Adenocarcinoma. Cancers (Basel) 2023, 15, (18).
- Liu, H.; Shi, J.; Anandan, V.; Wang, H. L.; Diehl, D.; Blansfield, J.; Gerhard, G.; Lin, F. Reevaluation and identification of the best immunohistochemical panel (pVHL, Maspin, S100P, IMP-3) for ductal adenocarcinoma of the pancreas. Arch Pathol Lab Med 2012, (6), 601–9. [Google Scholar] [CrossRef] [PubMed]
- Luu, T. T. Review of Immunohistochemistry Biomarkers in Pancreatic Cancer Diagnosis. Front Oncol 2021, 11, 799025. [Google Scholar] [CrossRef] [PubMed]
| Patient ID | GEO ID | Age | Gender | Diagnosis | Primary or Metastatis | Stage | Grade |
|---|---|---|---|---|---|---|---|
| PDAC_1 | GSM4679538 | 56 | F | PDAC | Primary | IIA | 3 |
| PDAC_2 | GSM4679539 | 67 | F | PDAC | Primary | IIA | 2 |
| PDAC_3 | GSM4679541 | 74 | F | PDAC | Primary | IIA | 2 |
| PDAC_4 | GSM4679546 | 46 | M | PDAC | Liver Met | IV | ND |
| PDAC_5 | GSM4679547 | 67 | M | PDAC | Liver Met | IV | ND |
| AdjNorm_1 | GSM4710706 | - | - | Adjacent normal pancreas | - | - | |
| AdjNorm_2 | GSM4710707 | - | - | Adjacent normal pancreas | - | - | |
| Normal_N1 | GSM5032773 | 50 | M | Normal Pancreas Sample | - | - | |
| Met: metastasis; | ND: Not determined | ||||||
| Method | Model | Input | Output | Tool |
|---|---|---|---|---|
| sciCNV | -Smoothed averages of gene windows -RTAM normalization |
- UMI count - Cell annotation |
- matrix containing the inferred CNVs along with their score for each cell of the sample | R package https://rdrr.io/github/alimahdipour/sciCNV/f/vignettes/Introduction.Rmd |
| InferCNV | -Smoothed averages of gene windows | - UMI count - Cell annotation - a gene/chromosome positions file |
- matrix containing the inferred CNVs for each cell of the sample | R package https://github.com/broadinstitute/inferCNV/wiki |
| CopyKAT | Bayesian segmentation approach | - UMI count - Cell annotation |
- Matrix containing the inferred CNVs for each cell of the sample - cells inferred as aneuploid are considered tumoral - cluster of the predicted cancer cells |
R package https://github.com/navinlabcode/copykat |
| SCEVAN | -Multichannel segmentation approach | - UMI count - Cell annotation |
- Matrix containing the inferred CNVs for each cell of the sample - Tumor/normal cell classification likely based on aneuploidies - cluster of the predicted cancer cells |
R package https://github.com/AntonioDeFalco/SCEVAN |
| CELL TYPE | PDAC_1 | PDAC_2 | PDAC_3 | PDAC_4 (Met.) | PDAC_5 (Met.) | AdjNorm_1 | AdjNorm_2 | Normal_N1 |
|---|---|---|---|---|---|---|---|---|
| NON-IMMUNE CELLS | (34.80%) | (90%) | (93.20%) | (95.6%) | (93.0%) | (43.5%) | (43.8%) | (57.4%) |
| Epithelial cells | 196 | 844 | 1325 | 2774 | 2303 | 705 | 653 | 388 |
| Endothelial cells | 10 | 12 | 1 | 1 | 156 | 111 | 170 | |
| Fibroblasts | 95 | 83 | 84 | 1 | 31 | 8 | 84 | |
| Tissue stem cells | 23 | 27 | 9 | 529 | 62 | 127 | ||
| Others | 58 | 59 | 45 | 3 | 6 | 778 | 782 | 296 |
| IMMUNE CELLS |
716 (65.2%) |
114 (10.0%) |
107 (6.8%) |
127 (4.4%) |
173 (7.0%) |
2770 (55.7%) |
1974 (55.0%) |
790 (42.6%) |
| Total Number of Cells | 1098 | 1139 | 1570 | 2905 | 2484 | 4969 | 3590 | 1855 |
|
Sample |
Total no. of Cells in Sample |
Epithelial cells (SingleR) |
Cancer cells according to markers |
Intersection between the epithelial cells (SingleR) and cancer cells predicted by markers * |
InferCNV |
CopyKAT |
SCEVAN |
|---|---|---|---|---|---|---|---|
| PDAC_1 | 1098 | 196 (17.8%) |
146 (13.3%) |
145 (99.3%) |
979 (89.1%) |
899 (81.9%) |
176 (16.0%) |
| PDAC_2 | 1139 | 844 (74.1%) |
737 (64.8%) |
735 (99.7 %) |
1032 (90.6%) |
844 (74.1%) |
846 (74.3%) |
| PDAC_3 | 1570 | 1325 (84.4%) |
1184 (75.4%) |
1176 (99.3 %) |
1470 (93.6%) |
1066 (67.9%) | 605 (38.5%) |
| PDAC_4 (Met) | 2905 | 2774 (95.5%) |
2642 (91.0%) |
2639 (99.9 %) |
2799 (96.4%) |
1158 (39.9%) | 836 (28.8%) |
| PDAC_5 (Met) | 2484 | 2303 (92.7%) |
2191 (88.2%) |
2189 (99.9 %) |
1967 (79.2%) |
1019 (41.0%) | 867 (34.9%) |
| AdjNorm_1 | 4969 | 705 (14.2%) |
271 (5.5%) |
233 (86 %) |
1005 (25.9%) |
2282 (45.9%) | 2818 (56.7%) |
| AdjNorm_2 | 3590 | 653 (18.2%) |
50 (1.4%) |
39 (78 %) |
37 (1.0%) |
453 (12.6%) |
425 (11.8%) |
| Normal _N1 | 1855 | 388 (20.9%) |
0 (0%) |
0 (0 %) |
0 (0%) |
359 (19.4%) |
518 (27.9%) |
| Sample | InferCNV | CopyKAT | SCEVAN | |||
|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| PDAC_1 | 0.99 | 0.12 | 0.10 | 0.07 | 0.86 | 0.95 |
| PDAC_2 | 1.00 | 0.27 | 1.00 | 0.73 | 0.99 | 0.72 |
| PDAC_3 | 1.00 | 0.25 | 0.61 | 0.10 | 0.46 | 0.83 |
| PDAC_4 (Met) | 1.00 | 0.40 | 0.40 | 0.57 | 0.28 | 0.60 |
| PDAC_5 (Met) | 0.86 | 0.71 | 0.43 | 0.77 | 0.37 | 0.81 |
| AdjNorm_1 | 0.16 | 0.80 | 0.12 | 0.52 | 0.91 | 0.45 |
| AdjNorm_2 | 0.06 | 0.99 | 0.06 | 0.87 | 0.06 | 0.88 |
| Normal_N1 | 1.00 | 0.81 | 0.72 | |||
| MEAN | 0.72 | 0.57 | 0.39 | 0.55 | 0.56 | 0.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





