1. Introduction
Cassava, Manihot esculenta Crantz is tropical root crops and important source of carbohydrates for millions of people in the world [
1]. The crop can be grown in marginal soils with limited rainfall making it a food security crop for sub-Saharan African countries [
2]. Cassava production is seriously threatened by several viral diseases, with cassava brown streak disease (CBSD) being a major concern [
3]. CBSD has been a persistent issue in East Africa for over seven decades [
4] and its impact is steadily increasing in Central and southern Africa [
5,
6] and emerging as a threat to West Africa, the continent's largest cassava producer [
7].
The ipomoviruses Ugandan Cassava Brown Streak Virus (UCBSV) and Cassava Brown Streak Virus (CBSV) that cause the disease [
8] results in significant reduction in crop yield and economic loss of up to
$1 billion [
9]. The CBSD has a profound impact on the economic stability of the affected regions, disrupting important economic activities and undermining the livelihoods of local communities, posing a serious threat to food security by affecting the availability and accessibility of this essential staple food and thereby the nutritional well-being of the population in the affected areas [
10]. The disease has variable symptoms including yellow chlorosis in leaves, cork necrosis in tuberous roots, brown stripes in the stems and dieback [
11]. The variability of CBSD symptoms has led to different classifications depending on the specific screening method used, including resistance mechanisms such as insect vector avoidance, virus spread, virus replication, and immune response. The identification and quantification of resistance of selected cassava varieties to viral infections has been made possible by reliable techniques such as molecular virus detection [
12].
The use of CBSD-resistant varieties is considered a sustainable and effective control strategy. However, challenges remain due to whitefly vectors, variations in virus strain, and environmental conditions that can impact the consistent and durable expression of resistance across generations [
13]. The previous studies reported that the resistance to CBSD is controlled by additive gene action [
14,
15,
16] and non-additive gene action as reported in previous studies [
17,
18]. Other related diseases/pests of importance to cassava, Cassava Mosaic Disease (CMD) was reported to be influenced by additive gene action [
15] and non-additive [
19], and resistance to Cassava green mites reported to be influenced by additive gene action [
20] and the seventeen candidate genes associated with its resistance [
21]. The importance of both additive and non-additive gene effects in controlling the expression of agronomic traits was also reported [
17,
20]. Conflicting reports on gene effects can be attributed to factors such as population type, mating design, analytical methods, study locations, and genotype-environment interactions [
22,
23].
This study addressed a critical gap in understanding the disease resistance in cassava specifically focusing on Cassava Brown Streak Disease (CBSD). Despite ongoing research aimed at developing varieties that are resistant to CBSD, limited information is available on the use of mixed populations to determine inheritance patterns and gene actions controlling the resistance to CBSD. This study investigated the inheritance of CBSD resistance in cassava populations of biparental (F1) and selfed (S1) lines derived from known resistant parents and explored the variability in resistance to CBSD levels within the populations and identified superior sources of resistance.
2. Materials and Methods
2.1. Plant Materials
The breeding materials were obtained from the International Institute of Tropical Agriculture (IITA) in Uganda. Three resistant CBSD genotypes, namely MM06/0123, CBSD MM06/0130, and MM06/0128, along with one susceptible parent (TME14), were selected to develop populations of bi-parentals (F1), and selfed (S1) populations. The criteria for selecting these parents included their resistance to the diseases, high fresh root yield and high dry matter content (
Table 1). The total population developed was 703 and the checks NASE13 and NAROCASS1 were included in the experiments. Before planting the clonal evaluation trial in April 2019, the generated populations underwent screening for CBSD and CMD in a germplasm maintenance trial for two years at IITA.
2.2. Experimental Sites and Design
The experimental trials were conducted at National Semi-Arid Resources Research Institute (NaSARRI) and International Institute of Tropical Agriculture (IITA) research station. In eastern Uganda where NaSARRI is situated, the weather is characterized by tall savannah ecology with rainfall between 1000 mm and 1300 mm per annum and temperature between 18 °C to 31 °C with sandy loam soil of a PH between 5.8–6.0 [
24]. The place is located at latitude 01″320 N and longitude 32″250 E and is among the country's medium spot areas for CBSD screening. In central Uganda, IITA is located at Namulonge and characterized by tropical rainforest ecology with rainfall of about 1500mm per annum and a temperature average of 22.2 °C with sandy clay soil PH of 4.9–5.0 [
24]. The place is located at latitude 00″ 320 N and is the disease hot spot area for screening of CBSD.
The clonal evaluation trial was laid out in an augmented design comprised of test genotypes and checks planted together with parental genotypes in eight to ten blocks depending on the available genotypes in the specific year and location. The field experiments were planted for two cropping seasons from 2019 to 2022. The plot size of 5 m2 and the spacing of 1 m × 1 m were used. No fertilizer or irrigation were applied in the experiments and weeds were controlled using hand hoe.
2.3. Phenotypic Data Collection
2.3.1. Disease Assessment
Assessment of the disease was conducted by collecting data on Cassava brown streak disease and Cassava mosaic disease as presented below (
Table 2).
2.3.2. Assessment of Yield and Yield Components
At harvest twelve months after planting (12MAP) the following data on yield components for each genotype were collected; number of fresh storage roots per plot, weight of fresh storage root (kg plot
-1) which was converted to fresh root yield (t ha
-1), and weight of shoot (kg per plot). Other variables such as harvest index (HI) were calculated using the formula in equation 1 and dry matter content (DMC in %) was obtained using specific gravity calculated using the formula in equation 2 [
25].
Where, Wa = mass of roots in air (kg) and Ww = mass of roots in water (kg)
On virus detection and quantification, the selection of samples for detection was based on the observable foliar CBSD symptoms of score 1, 2 and 3 CBSD leaf severities and score of 1 only (Asymptomatic) for CBSD root necrosis. Root slices of 2 cm were cut and covered in aluminum foil, immediately stored on ice, and later refrigerated at 20 °C for short storage when in the field, then transferred to -80 °C freezer for long storage at TARI Kibaha laboratory. Before extraction, samples were removed from -80 °C and back to -200 °C. taken back to -20 °C. Total RNA was extracted using a cetyl trimetyl ammonium bromide (CTAB) protocol [
26]. The RNA quality and purity were determined using a NanoDrop2000 spectrophotometer (ThermoScientific). The RNA samples were used for testing the presence and absence of the CBSD virus using a Taqman assay. The tests for cassava brown streak virus (CBSV) and Ugandan cassava brown streak disease were done independently [
27]. Specific primers which anneal CBSV and UCBSV coat proteins were used (
Table 3) for virus detection.
Complementary DNA (cDNA) was prepared from 1µg of template RNA using First strand cDNA synthesis (quick protocol). The complete reverse transcriptase reaction (2 µL) contained 50 µM Oligo dT18 (New England Biolabs), 10 x M-MuLV buffer, 200 U/µL of MuMLV reverse transcriptase (RT), 10 Mm dNTP Mix, 40 U/µL RNase inhibitor and Nuclease-free water. Samples detected with virus were analyzed by an absolute quantification qPCR reaction using a Taqman assay specific for CBSV and UCBSV. Quantification was done by measuring concentration of plasmid (6PK01-Fer2) using Qubit 3.0 florometer followed by ten-fold serial dilution started with original concentration of 21.5 ng/µL. A reaction for quantification was prepared using 25 µL contained Mg, 10 x PCR buffer, 25 Mm MgCl2, 10 Mm dNTP, 7.5 µM primers, 5 µM probe, 1 x reference dye, 5 U/µL Taq DNA polymerase and plasmid. The PCR program was set at 95 ᵒC for 10 minutes, 95 °C for 15 seconds and 60 °C for 1 minute for 40 PCR cycles. The reaction was performed using the Stratagene Mx3000P qPCR system (Agilent Technologies) and data was acquired using the Mx3000P qPCR software. The amplification of the RNA during the PCR process ranged from cycle 15 to 35 with amplification efficiency between 100% and 104.8% and standard curve of coefficient of correlation (R2) of 0.99 with slope of 3.21. Data obtained were assembled in Microsoft Excel and analyzed for detected genotypes with their specific virus and concentration.
2.4. Data Analysis
Data analysis was done at different phases. Phase one comprised analysis of variance using lmer package in R software R.4.1.2 [
27] (R Core Team. 2021) for the individual genotypes of all F1 and S1 were combined and the Best Linear Unbiased Prediction (BLUP) estimated a mixed model's random effects. On phase two, the genotypes of crosses F1’s and S1’s was analysed to determine variabilities using lmer package in R software R.4.1.2 and thirdly family bases where genotypes were averaged to assess the family performance. Analysis of variance was done using the model in Equation 3.
Whereas; Yijk is the observed phenotypic value, µ is the overall mean, Gi is genotype effect, Ej is environment effect, GEk is interaction effect of genotype and environment, ∈ is residual error term assumed normal distribution.
2.4.1. Analysis of Variance for Parents and Families
The combined data were analyzed using ASReml package of R software R.4.1.1. Genotypes were fitted as fixed factors, environments were treated as random factors and the mean squares for SCA, GCA1, GCA2 and the interactions with environments and seasons were fitted in the model in equation 4.
Whereas; Y is the observed phenotypic value of the progeny of the ith female crossed with jth male in the kth environment, µ is the overall mean, G is genotype effect, F is the GCA of the ith female, M is the GCA of the jth male, SCA is the cross between ith female and jth male, Ek is environment effect, GE is interaction effect of genotype and environment, ∈ is residual error term assumed normal distribution.
2.4.2. Estimation of Variance Components and Heritability
Phenotypic and genotypic variances were computed from the expected mean squares of the analysis of variances, the phenotypic coefficient of variation (PCV) and genotypic coefficient of variation (GCV) expressed in percent were estimated using the formula below; [
28].
Whereby
is the genotypic variance,
is the phenotypic variance, MSG is the genotypic mean square, MSE is the error mean square, and r is the replication number.
Whereas; PCV is the phenotypic coefficient of variation, GCV is the genotypic coefficient of variation, is the grand average of the traits, is the phenotypic variance and is the genotypic variance.
Heritability of the populations was estimated in two levels, level one was heritability across environments and level two was for specific years in the location and it was computed according to Falconer and Mackay [
29] using the formula;
Where is the genotypic variance and is the phenotypic variance
2.4.3. Selection of Resistant Genotypes and Level of Resistance in Populations
Absolute quantification of CBSV and UCBSV was determined by running the default settings of the MxPro qPCR software on the Stratagene Mx3000P qPCR system (Agilent Technologies). The data obtained were assembled in Microsoft Excel and correlation analysis was performed. The resistance level was categorized as the most resistant (MR), Resistant (R), most tolerant (MT) and tolerant (T) based on the foliar symptoms, root necrosis and virus detection test. The proportion of resistant and tolerant genotypes per family was computed by expressing the absolute number of resistant genotypes as a percentage of the total number of genotypes established in that family.
3. Results
3.1. Performance and Genetic Variation of Genotypes and Population Types
The results of descriptive statistics of the performance of genotypes across seasons and locations are presented in
Table 4. The CBSD incidences had a minimum of 0% and a maximum of 100% while that of CBSD severities had a minimum score of 1 and a maximum of 5 in CBSDL3S and CBSDRS respectively. The lowest mean CBSDLI (55.79%) was observed at 3MAP (CBSDL3I) and the highest (65.14%) at 9MAP (CBSDL91). The same trend was observed for CBSD leaf severities (CBSDLS), where the lowest mean (1.95) was observed at 3MAP and the highest (2.28) at 9MAP. For CBSD root necrosis incidence (CBSDRI) and severity (CBSDRS), the mean CBSDRI was 37.84% whereas the mean CBSDRS was 2.76. The trend for Cassava Mosaic Disease severity (CMDLS) followed a similar pattern, where the genotypes had the lowest severity of 1 and the highest of 5 at 3MAP and 9MAP respectively. The CMD incidences exhibit some fluctuations across different months after planting, with CMDL6I having the highest mean incidence of 19.31 ± 0.65 standard error. The CMD severities also vary, but generally remain relatively low compared to CBSD severities.
Other yield traits such as Dry Matter Content (DMC), Fresh Yield (FYLD), Harvest Index (HI) and Total Root Tuber Number (TRTN) show considerable variation. The DMC ranged from 16.3% to 68.16% with the average of 35.5% while the FYLD had an average of 15.46t/ha with few genotypes outperformed to the highest of 175 ta/ha. Harvest Index (HI) varies between 0 and I, indicating different proportions of biomass allocated to harvestable parts. The TRTN shows variability with a maximum value of 73.
Table 4.
Descriptive statistics for CBSD incidences and severities (3, 6, 9 MAP) and CMD incidences and severities (3, 6 MAP) and yield traits across two seasons and locations.
Table 4.
Descriptive statistics for CBSD incidences and severities (3, 6, 9 MAP) and CMD incidences and severities (3, 6 MAP) and yield traits across two seasons and locations.
| Variable |
Min |
Max |
Mean ± SE |
STDEV |
CV (%) |
| CBSDL3I |
0 |
100 |
55.79 ± 0.8 |
42.75 |
76.64 |
| CBSDL6I |
0 |
100 |
63.35 ± 0.78 |
41.75 |
65.9 |
| CBSDL9I |
0 |
100 |
65.14 ± 0.78 |
41.45 |
63.63 |
| CBSDL3S |
1 |
5 |
1.95 ± 0.01 |
0.75 |
38.63 |
| CBSDL6S |
1 |
4 |
2.23 ± 0.02 |
0.89 |
39.88 |
| CBSDL9S |
1 |
4 |
2.28 ± 0.02 |
0.88 |
38.51 |
| CBSDR12S |
1 |
5 |
2.76 ± 0.02 |
1.32 |
47.8 |
| CBSDR12I |
0 |
100 |
37.84 ± 0.66 |
35.27 |
93.2 |
| CMDL3I |
0 |
100 |
18.33 ± 0.64 |
34.01 |
185.51 |
| CMDL6I |
0 |
100 |
19.31 ± 0.65 |
34.61 |
179.3 |
| CMDL9I |
0 |
100 |
13.11 ± 0.56 |
29.75 |
226.96 |
| CMDL3S |
1 |
5 |
1.5 ± 0.02 |
0.87 |
58.3 |
| CMDL6S |
1 |
4 |
1.52 ± 0.02 |
0.86 |
56.56 |
| CMDL9S |
1 |
5 |
1.36 ± 0.01 |
0.75 |
55.18 |
| DMC |
16.3 |
68.16 |
35.5 ± 0.11 |
5.03 |
77.63 |
| FYLD |
0 |
175 |
15.46 ± 0.34 |
17.98 |
116.27 |
| HI |
0 |
1 |
0.34 ± 0 |
0.17 |
48.51 |
| TRTN |
0 |
73 |
14.45 ± 0.21 |
11.22 |
14.16 |
The results of the analysis of variance detected highly significant differences (P < 0.001) between genotype, environment, and genotype by and environment interaction effect for CBSD incidences and severities and different stages of infections (3, 6 and 9), CBSD root severity (CBSDRS), CBSD root incidence (CBSDRI), cmd severities nad incidence at different stages of infections (
Table 5). Similarly, the significance levels for MSG, MSE, and MSG*E indicate significant genetic, environmental, and genotype-environment interaction effects on fresh yield (FRYLD), harvest index (HI) and dry matter content (DMC). Significant genotype and environment interaction (MSG*E) was observed in TRTN (P < 0.05) (
Table 5).
The results of analysis of variance for all the traits evaluated among the population types (PT), environments (E) and their interactions (PTxE) are presented in table 6. Highly significant differences (P < 0.001) were detected between the PT for all the traits except for CBSDRI, HI and DMC where no significant differences were detected. The differences between PT were significant (P < 0.005) for CBSDL3I and CMD3S. Highly significant differences (P < 0.001) were detected between environments for all the traits recorded. Furthermore, highly significant differences (P < 0.001) were detected for the interactions between PT and E for all the traits except for CMD3I, CMD6I, HI (P < 0.01) and CMD6S (P < 0.05).
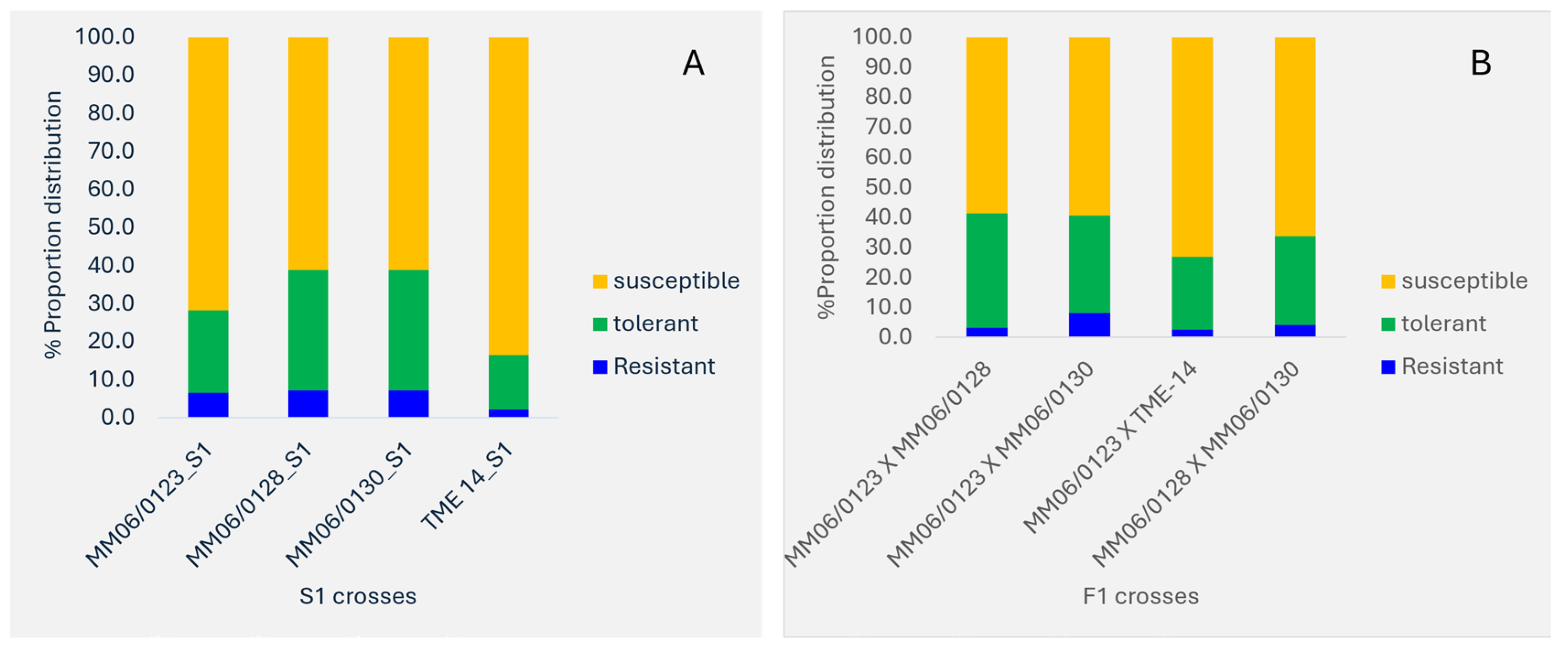
Frequency of CBSD root Necrosis Categories within Crosses
The proportion of CBSD root necrosis in fresh roots within crosses of the mapping populations were also compared (
Figure 1). The classification was categorized as resistant for score 1, tolerant for score 2 and susceptible for scores 3–5. Less than 10% resistant genotypes were observed among the F1 and S1 crosses. From a cross of susceptible by susceptible (TME14XTME14), a proportion of only 2% of resistant genotypes was observed. The S1 cross of resistant x resistant (MM060123) had a slightly higher number of susceptible genotypes of 71.7% compared to resistant x resistant parents of F1’s including MM060123XMM060128 (58.6%), MM060123XMM060130 (59.5), and MM060128XMM060130 (66.3%).
Table 6.
Mean squares for population types (PT) environment (E) and PTxE for disease resistance and yield traits evaluated across two seasons at Namulonge and Serere, Uganda.
Table 6.
Mean squares for population types (PT) environment (E) and PTxE for disease resistance and yield traits evaluated across two seasons at Namulonge and Serere, Uganda.
| Trait |
Sources of Variation |
| MSPT (3) |
MSE (3) |
MSPT*E (9) |
MSR (2138) |
| CBSDL3S |
1.76 * |
46.99 *** |
2.06 *** |
0.49 |
| CBSDL6S |
3.55 *** |
67.71 *** |
3.41 *** |
0.71 |
| CBSDL9S |
3.87 *** |
31.07 *** |
2.32 *** |
0.75 |
| CBSDRS |
10.02 *** |
25.53 *** |
4.62 *** |
1.65 |
| CBSDL3I |
4332 * |
19.933 *** |
5629 *** |
1551 |
| CBSDL6I |
9521 *** |
11161 *** |
7460 *** |
1603 |
| CBSDL9I |
11298 *** |
135177 *** |
6052 *** |
1573 |
| CBSDRI |
2157.3 |
18811.6 *** |
2763 *** |
1115.3 |
| CMD3S |
1.76 * |
46.99 *** |
2.06 *** |
0.49 |
| CMD6S |
12.44 *** |
30.71 *** |
1.47 * |
0.711 |
| CMD3I |
46803 *** |
92391 *** |
2795 ** |
1080 |
| CMD6I |
20097 *** |
94448 *** |
2934 ** |
1098 |
| FRYLD (ta/ha) |
1033 *** |
530.25 *** |
145.6 *** |
33.15 |
| HI |
0.03 |
3.77 *** |
0.045 ** |
0.02 |
| DMC |
35.73 |
768.78 *** |
116.32 *** |
23.83 |
| TRTN |
2421.1 *** |
7863.7 *** |
407.3 *** |
112.8 |
Frequency of CBSD within populations of F1 and S1
Figure 1.
Proportion of CBSD root necrosis in biparental F1 and Selfing crosses (S1), Classification of scores: score 1 = resistant, score 2 = tolerant and score 3-5 = susceptible. A = S1 and B = F1.
Figure 1.
Proportion of CBSD root necrosis in biparental F1 and Selfing crosses (S1), Classification of scores: score 1 = resistant, score 2 = tolerant and score 3-5 = susceptible. A = S1 and B = F1.
Combining Abilities and Mode of Gene Action for Resistance to Cassava Brown Streak Disease
The analysis of variance for combining abilities showed that environment was significant (p < 0.05) for all the traits measured (
Table 7). The mean squares for GCA1, GCA2, and SCA represent the genetic variability within the parents and families for each trait. The GCA1 (female) effects were significant (p < 0.05) for all CBSD incidence and severities on foliar, roots, and the root yield traits. The GCA2 (male) effects showed significant difference only on CBSDRI and CBSDRS, on the other traits non-significant effects were observed. SCA indicates the specific combining ability, representing the non-additive genetic effects resulting from the interaction between parental genotypes The SCA effects were not significant for all traits measured. The GCA/SCA ratio provides insights into the relative importance of additive (GCA) and non-additive (SCA) genetic effects in trait inheritance The GCA/SCA ratio was high and positive. The percentage contribution to Sum of Square showed that GCA1 had a high contribution than its corresponding GCA2 and SCA for all the traits under investigation.
Genetic components: The genetic parameters related to disease and yield traits in cassava populations are summarized in
Table 8. The results showed that the phenotypic variance (PV) for traits associated with Cassava Brown Streak Disease (CBSD), Cassava Mosaic Disease (CMD), fresh yield (FYLD), and dry matter content (DMC) were higher than the corresponding genotypic variances (GV) and environmental variances (EV). The high PCV (>20%) was recorded for all trait except for DMC whose PVC was moderate (12.39%). The highest GCV (67.05%) was observed for CMDI6 and the lowest for DMC (5.49%).
The genetic advance (GA) ranged from 0.02 for HI to 17.54 for CBSDL6I and the genetic advance as percent of mean (GAM) ranged from 3.21% for CBSDRI to 62.23% for CMD6I. The GAM was classified as low (below 10%), medium (10–20%) and high (above 20%). Low GAM (<10%) was observed for DMC, HI and CBSDRI, it was medium for CBSDL6S, CBSDS9, FRYLD and CBSDRS, whereas it was high for CBSDL6I, CMD6I and CMD6S (
Table 8). Low heritability estimates (≤30) were observed across seasons and locations for all the traits presented and ranged from 0.01 (CBSDRI) to 0.27 (CMDS6).
The heritability estimates for CBSD traits was further expanded to locations in specific growing season and the results are presented in
Table 9. Almost all broad sense Heritability for CBSD severities and incidence at IITA were higher than NaSARRI. The highest of 69.2% was observed for CBSDL61 and the lowest of 27.3% was on CBSDL3I in 2019/2022 (across locations). The estimates of heritability at IITA location was higher compared to those at NaSARRI. The broad sense heritability estimates ranged from 14.6 (CBSDRS at NaSARRI) to 69.2 (CBSDI6 at IITA).
Table 7.
Mean square of parents and families for CBSD, CMD and root yield traits.
Table 7.
Mean square of parents and families for CBSD, CMD and root yield traits.
| SOV |
CBSDLS |
CBSDLI |
CBSDRS |
CBSDRI |
CMDS |
CMDI |
FYLD |
HI |
DMC |
TRTN |
| Environment |
81.333 *** |
141870 *** |
25.9728 *** |
23519 *** |
50.116 *** |
130085 *** |
23491.5 *** |
5.9935 *** |
749.31 *** |
11551.2 *** |
| GCA1 |
7.061 *** |
23094 *** |
14.7296 *** |
13209.4 *** |
5.205 *** |
7535 *** |
5999.9 *** |
0.2998 *** |
178.08 *** |
1988.7 *** |
| GCA2 |
8.159 *** |
31911 *** |
8.398 *** |
8952.5 *** |
1.161 * |
1903 * |
2520 *** |
0.1514 *** |
34.77 |
1866.6 *** |
| SCA |
1.385 * |
3715 ** |
3.7492 * |
1919.2 |
0.403 |
349 |
1292.5 *** |
0.029 |
9.62 |
784.1 *** |
| Environment:GCA1 |
2.06 *** |
4237 *** |
2.4087 * |
2670.9 *** |
0.693 * |
1441 ** |
592.9 *** |
0.0369 ** |
86.64 *** |
277.4 *** |
| Environment:GCA2 |
0.422 |
2185 |
3.0499 * |
2535.4 ** |
0.616 |
981 |
308.5 |
0.0095 |
5.8 |
89.3 |
| Environment: SCA |
0.481 |
998 |
1.0776 |
549.7 |
0.358 |
645 |
387.7 |
0.0045 |
8.47 |
135 |
| Residuals |
0.659 |
1438 |
1.6394 |
1145.1 |
0.471 |
754 |
268.3 |
0.0199 |
22.94 |
100.7 |
| GCA/SCA ratio |
9.3 |
12.1 |
6.03 |
10.9 |
17.6 |
29.7 |
6.8 |
15.5 |
24.9 |
4.3 |
| % SS GCA1 |
61.76 |
59.47 |
69.84 |
72.08 |
86.87 |
87.87 |
74.61 |
78.17 |
89.18 |
59.14 |
| % SS GCA2 |
28.55 |
32.87 |
15.93 |
19.54 |
7.74 |
8.88 |
12.53 |
15.78 |
6.96 |
22.2 |
| % SS SCA |
9.69 |
7.65 |
14.22 |
8.38 |
5.39 |
3.26 |
12.86 |
6.05 |
3.86 |
18.66 |
Table 8.
Genetic parameters for diseases and yield-related traits in cassava populations.
Table 8.
Genetic parameters for diseases and yield-related traits in cassava populations.
| Traits |
Mean |
SE |
EV |
GV |
PV |
ECV |
GCV |
PCV |
H |
GA |
GAM |
| CBSDL6I |
61.49 |
11.28 |
1018.6 |
310.47 |
1329.07 |
51.91 |
28.66 |
59.29 |
0.23 |
17.54 |
28.53 |
| CBSDL6S |
2.2 |
0.25 |
0.5 |
0.13 |
0.62 |
32.09 |
16.17 |
35.94 |
0.2 |
0.33 |
14.99 |
| CMD6I |
20.88 |
9.81 |
769.72 |
196.06 |
965.78 |
132.85 |
67.05 |
148.81 |
0.2 |
13 |
62.23 |
| CMD6S |
1.56 |
0.23 |
0.41 |
0.15 |
0.55 |
40.69 |
24.51 |
47.5 |
0.27 |
0.41 |
26.04 |
| FYLD |
19.24 |
5.92 |
280.26 |
30.96 |
311.22 |
87.01 |
28.92 |
91.69 |
0.1 |
3.61 |
18.79 |
| HI |
0.38 |
0.05 |
0.02 |
0 |
0.02 |
35.39 |
10.2 |
36.87 |
0.08 |
0.02 |
5.82 |
| DMC |
35.5 |
1.39 |
15.56 |
3.8 |
19.36 |
11.11 |
5.49 |
12.39 |
0.2 |
1.78 |
5.01 |
| CBSDRI |
20.72 |
10.48 |
878.08 |
9.62 |
887.7 |
143.04 |
14.97 |
143.82 |
0.01 |
0.67 |
3.21 |
| CBSDRS |
1.48 |
0.19 |
0.29 |
0.05 |
0.33 |
36.07 |
14.65 |
38.93 |
0.14 |
0.17 |
11.37 |
Table 9.
Broad-sense Heritability of CBSD across years and locations.
Table 9.
Broad-sense Heritability of CBSD across years and locations.
| Trait |
Year |
Location |
H (%) |
| CBSDI3 |
2019 to 2022 |
IITA |
65.0 |
| CBSDI3 |
2020 to 2022 |
NaSARRI |
32.7 |
| CBSDI3 |
2019 to 2022 |
Multilocation |
27.3 |
| CBSDS3 |
2019 to 2022 |
IITA |
54.0 |
| CBSDS3 |
2020 to 2022 |
NaSARRI |
32.5 |
| CBSDS3 |
2019 to 2022 |
Multilocation |
29.2 |
| CBSDI6 |
2019 to 2022 |
IITA |
69.2 |
| CBSDI6 |
2020 to 2022 |
NaSARRI |
49.4 |
| CBSDI6 |
2019 to 2022 |
Multilocation |
39.3 |
| CBSDS6 |
2019 to 2022 |
IITA |
63.2 |
| CBSDS6 |
2020 to 2022 |
NaSARRI |
43.4 |
| CBSDS6 |
2019 to 2022 |
Multilocation |
36.4 |
| CBSDI9 |
2019 to 2022 |
IITA |
53.3 |
| CBSDI9 |
2020 to 2022 |
NaSARRI |
44.6 |
| CBSDI9 |
2019 to 2022 |
Multilocation |
39.4 |
| CBSDS9 |
2019 to 2022 |
IITA |
46.8 |
| CBSDS9 |
2020 to 2022 |
NaSARRI |
58.4 |
| CBSDS9 |
2019 to 2022 |
Multilocation |
42.4 |
| CBSDRI |
2019 to 2022 |
IITA |
62.1 |
| CBSDRI |
2020 to 2022 |
NaSARRI |
31.9 |
| CBSDRI |
2019 to 2022 |
Multilocation |
48.2 |
| CBSDRS |
2019 to 2022 |
IITA |
57.9 |
| CBSDRS |
2020 to 2022 |
NaSARRI |
14.6 |
| CBSDRS |
2019 to 2022 |
Multilocation |
36.1 |
Virus Detection and Quantification
The results of virus detection and quantification revealed prevalence of CBSV and less occurrence of UCBSV in both sites (
Table 10). The sample of roots chosen for virus detection was selected during the root assessment, and the one with a score of 1 was selected.
The UCBSV was observed on leaves at NaSARRI and on roots at IITA. At NaSARRI site, one sample was observed to have co-infection with both CBSV and UCBSV on leaves. Root samples collected at NaSARRI for season one had 66.3% clean genotypes while during season two 82.54% of leaves and 39.3% of roots were not detected with the viruses. At IITA, 51.35% of leaves and only 9.41% of root samples collected were found to be clean from virus.
Based on the observed leaf symptoms, root necrosis, and virus detection, 9 genotypes were either most resistant (MR), resistant (R), most tolerant (MT) or were tolerant (T) to CBSD. Their phenotypes (apart from the class score of 1-2 root necrosis) ranged from negative virus detection, low leaf incidence (less than 20%) and maximum leaf severity score of class 2 (
Table 11). Genotypes MM160145, MM160582B and MM160089A tested negative (no cq) for leaf and roots virus detection, however they showed root symptoms (necrosis) implying that they are infected by a new strain that cannot be picked by the current primers.
Sixteen asymptomatic genotypes including checks, were identified with MM160031 exhibited asymptomatic positivity for both foliar and root necrosis (
Table 12). These were observations were made at IITA. The rest of genotypes displayed asymptomatic positivity for either foliar symptoms or root necrosis at either of the sites (Table 14). The check varieties NAROCASS1 (IITA) and NAROCASS 2 (NaSARRI) were also found to be asymptomatic positive for root necrosis
Table 11.
Performance of nine most promising genotypes with ≤20% CBSD incidence and ≤2 CBSD severity, CBSDRS ≤2, CMD severity and negative virus detected at IITA and NaSARRI.
Table 11.
Performance of nine most promising genotypes with ≤20% CBSD incidence and ≤2 CBSD severity, CBSDRS ≤2, CMD severity and negative virus detected at IITA and NaSARRI.
| Accession |
Pedigrees |
CBSDS3 |
CMDS3 |
CBSDS6 |
CMDS6 |
CBSDS9 |
CMDS9 |
CBSDRS |
Status |
Location |
| MM160145 |
S1_MM060130 |
1 |
1 |
1 |
1 |
2 |
1 |
2 |
MT |
both |
| MM161487 |
S1_MM060123 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
MR |
both |
| MM160227 |
S1_MM060130 |
2 |
3 |
1 |
1 |
1 |
1 |
1 |
R |
both |
| MM160582B |
S1_ MM060128 |
1 |
1 |
1 |
1 |
1 |
1 |
2 |
MT |
both |
| MM161113 |
F1_MM060123XMM060130 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
MR |
NAS |
| MM161247 |
F1_MM060123XMM060130 |
2 |
1 |
1 |
1 |
1 |
1 |
1 |
MR |
NAS |
| MM160089A |
S1_MM060130 |
1 |
1 |
1 |
1 |
1 |
1 |
2 |
MT |
IITA |
| MM160406 |
F1_MM060128XMM060130 |
1 |
3 |
1 |
3 |
1 |
1 |
1 |
T |
IITA |
| MM160530 |
S1_MM060128 |
1 |
1 |
1 |
1 |
2 |
1 |
1 |
MT |
IITA |
Table 12.
Sixteen asymptomatic cassava genotypes (including two checks) at IITA and NaSSARI.
Table 12.
Sixteen asymptomatic cassava genotypes (including two checks) at IITA and NaSSARI.
| Genotype |
Pedigrees |
CBSDL3S |
CBSDL6S |
CBSDL6S |
CBSDRS |
CBSV_L |
CBSV_R |
Location |
| MM160031 |
MM060130 X MM060130 |
1 |
1 |
1 |
1 |
24.03 |
26.25 |
IITA |
| MM160371 |
MM060128 X MM060130 |
1 |
1 |
1 |
1 |
No Cq |
36.82 |
NaSARRI |
| MM160234 |
MM060130 X MM060130 |
1 |
1 |
1 |
1 |
No Cq |
31.92 |
IITA |
| MM161627 |
MM060128 HS |
1 |
1 |
1 |
1 |
No Cq |
29.52 |
IITA |
| MM160760 |
MM060123 X MM060128 |
1 |
1 |
1 |
1 |
No Cq |
34.27 |
NaSARRI |
| MM161145 |
MM060123 X MM060130 |
1 |
1 |
1 |
1 |
No Cq |
37.61 |
NaSARRI |
| MM160969 |
MM060123 X MM060128 |
1 |
1 |
1 |
1 |
|
28.45 |
IITA |
| MM161030 |
MM060123 X MM060128 |
1 |
1 |
1 |
1 |
|
29.26 |
IITA |
| MM160909 |
MM060123 X MM060128 |
1 |
1 |
1 |
1 |
|
30.8 |
IITA |
| LTG 5 |
UNKNOWN |
1 |
1 |
1 |
1 |
|
32.11 |
IITA |
| MM160668A |
MM060128 X MM060128 |
1 |
1 |
1 |
1 |
|
28.68 |
IITA |
| NAROCASS1 |
NDL9036 |
1 |
1 |
1 |
1 |
|
36.66 |
IITA |
| MM160069 |
MM060130 X MM060130 |
1 |
1 |
1 |
1 |
|
34.96 |
NaSARRI |
| MM160877 |
MM060123 X MM060128 |
1 |
1 |
1 |
1 |
|
35.76 |
NaSARRI |
| NAROCASS2 |
Kitumbua |
1 |
1 |
1 |
1 |
|
36.68 |
NaSARRI |
| MM160602 |
MM060128 X MM060128 |
1 |
1 |
1 |
1 |
|
25.37 |
NaSARRI |
4. Discussion
The observed genetic variation within and between populations emphasized the role of the selfed populations in enhancing selection breeding. The results indicated that resistance to CBSD was primarily controlled by an additive mode of gene action, consistent with previous studies by Chipeta (2018) and Nduwumuremyi (2018). Furthermore, the identification of the most resistant genotypes to CBSD across various locations highlighted the importance of utilizing different locations in the selection process [
29].
The performance of cassava genotypes under varying environmental conditions revealed an essential dimension of their adaptability [
30] and enabled the assessment of the performance of the genotypes for a range of traits. The significant differences (p < 0.05) detected among the genotypes in response to both Cassava Brown Streak Disease (CBSD) and Cassava Mosaic Disease (CMD) indicated a diverse array of resistance levels among the genotypes. These variations hold significant implications for breeding programs efforts to make genetic progress in breeding for resistance against the two economically important diseases [
31]. Consistent results were also observed for traits such as CBSDLS, CBSDRS, and fresh yield (FRYLD), further strengthening the validity and robustness of the results. The repetition of similar patterns across multiple traits underscored the stability of the findings and enhanced the validity in the observed trend. The identification of the quantitataive loci associated with the the resistance to CBSD will further elucidate the genetic architecture of the resistance, paving the way for further investigation.
The interaction between genotypes and the environment, particularly in the context of CBSD, revealed the influence of quantitative traits [
32] which underscored the multifaceted nature of genotype-environment relationships and emphasized for the need for a nuanced approach in assessing and selecting genotypes for desired traits and also underscoring the complex interplay between genetics and the environment which is crucial for informed decision-making in cassava genotypes selection for superior resistance to disease and overall performance.
The performance of the resistant parent at IITA in 2019, which showed similar mean scores to the susceptible parent for CBSDLS suggested that prolonged drought might directly contribute to weakening plant defenses in some resistant varieties within population [
33]. This highlights the complexity of the interaction between drought stress and plant susceptibility to CBSD. Further investigation into the deeper insights into how environmental factors such as drought and soil conditions influence the expression of resistance traits is needed. This will allow the development of cassava varieties that are resilient to both biotic and biotic stresses. Additionally, developing environment-specific breeding strategies that take into account local environmental conditions to optimize resistance and yield is essential.
The results from S1 and F1 indicated that higher accumulation of resistant genes might be attainable from crosses between lines with existing resistance rather than through biparental crosses of resistant and susceptible lines but resistant lines were also generated from selfing susceptible line (TME14XTME14) suggesting that the resistance to CBSD was likely recessive as reported by Sheat and Winter (2023). In addition, the presence of multiple genes conferring resistance to CBSD was indicated by segregation patterns in F1s and S1s populations through the distribution of scores which exhibited similarities between crosses and S1s indicating the complex genetic basis of CBSD resistance. The identification of a higher number of resistant progenies from S1 generation of MM160123 suggested a strong expression of the recessive alleles for the resistance trait transferred from parent MM160123.
The analysis of phenotypic variance (PV) demonstrated the allocation of total variation into genotypic (GV) and environmental variance (EV) which provided valuable insights into the underlying sources of variability that contributed to the observed phenotypic variation. The high proportion of PV indicated the strong environmental influence on the traits under study and the relatively lower Proportion of Genotypic Variance Components (GVC) observed in some traits, particularly DMC, suggested a comparatively weaker genetic influence on their expression [
34]. This observation highlights the potential for genetic improvement through hybridization, followed by rigorous selection, rather than relying solely on the phenotypic performance of individual genotypes. Similar conclusions were reported by Nduwumuremyi et al. (2018a). The substantial differences observed between Phenotypic Coefficient of Variation (PCV) and its corresponding Genotypic Coefficient of Variation (GCV) highlighted the significant impact of environmental factors on the expression of these traits indicating the sensitivity of genotypes to variations in the environments suggesting that, selecting genotypes based solely on phenotypic performance may yield limited genetic improvement of the traits due to the confounding influence of environmental factors [
35].
The analysis of general combining ability (GCA) and specific combining ability (SCA) results provided valuable insights into the underlying gene effects and their interactions in the expression of the studied traits. The significance of GCA results indicated the presence of additive gene effects and additive x additive interactions. The substantial significance of specific combining ability (SCA) results, on the other hand, suggested the presence of dominance and epistatic effects. The observed significance of GCA across all the traits underscored the predominance of additive genes in the expression of the traits for CBSDLS, CBSDRS, and fresh yield (FRYLD) [
17], CBSDLI (Chipeta et al., 2018) and HI [
23]. The non-significance of SCA for CBSDRI, CMDS, CMDI, HI and DMC indicated that parental interactions do not significantly influence the performance of their hybrids, suggesting a relatively stronger influence of additive gene effects compared to non-additive effects. The high positive GCA/SCA ratio indicated a greater contribution of additive gene effects in shaping the expression of the studied traits which underscores the substantial role of additive genetic components in the variability of these traits.
The predominance of additive gene effects suggested the potential for selection based on additive genetic contributions and contributed to our understanding of the genetic architecture underlying the traits which provided a foundation for informed breeding strategies.
Across different seasons and locations, the majority of heritability estimates were found to be low suggesting a pronounced influence of environmental factors [
36]. At the specific location, however, moderate to high broad sense heritability estimates were recorded for the CBSD trait which indicated a stronger genetic influence on the expression of this trait in that specific environment, implying that improvement through simple selection could be a viable approach for enhancing resistance to CBSD as suggested by Kayondo et al. (2018) [
37].
The identification of resistant genotypes to CBSD including MM160145, MM161487, MM160227, and MM160582B, exhibiting minimal foliar symptoms and root necrosis (≤2), underscored their high level of resistance to CBSD. These genotypes came from the full-sib population among CBSD resistant, parents MM060123, MM060128, and MM060130. The resistance observed in these genotypes was derived from S1 populations, surpassing their S0 parents in performance. Similar results were reported by Pariyo (2015) [
38] and Kawesi (2016) [
39], indicating the effectiveness of utilizing S1 populations for breeding for resistance to CBSD. The stability of the four genotypes across contrasting environments indicated the potential for making genetic progress in resistance to CBSD through inbreeding.
The observed root necrosis in genotypes testing negative for CBSD virus on both roots and leaves indicated that the roots necrosis might be caused by a distinct biotype of virus which could not be detected by the primers used. The study was limited by the inability to develop primers that matched the observed results. Additionally, the coexistence of genotypes testing positive for CBSD on leaves but negative on roots indicated a concentration of virus in the leaves, possibly because the virus had not yet reached the roots or that the roots had developed resistance mechanisms. Conversely, genotypes testing positive for CBSD on roots but negative on leaves suggested differential resistance mechanisms between the two plant parts. Similar observations were reported by Shirima (2017). Understanding the molecular and physiological basis of these differential resistance mechanisms is important, as this knowledge can lead to more targeted breeding efforts and the development of cassava varieties with comprehensive resistance profiles.
5. Recommendations
The study recommends leveraging selfed populations to enhance resistance breeding as they have shown strong expressions of recessive alleles and greater resistance compared to their parental lines. Additionally, the use of cassava genotypes MM161487, MM160227, MM170098, MM160582B, and MM160145 should be prioritized in breeding programs aimed at developing varieties with high yield potential and resistance to CBSD and CMDS.
Author Contributions
The authors made the following contributions to the study: Conceptualization K.L.S.; methodology K.L.S. and E.K.; Software. I.K.; validation and formal analysis K.L.S. and I.K.; resources, E.K., R.E. and H.K.; data curation K.L.S.; writing original draft preparation, K.L.S.; writing, reviewing and editing, K.L.S., P.R., E.K. and M.O.; visualization D.M.; supervision, E.K., P.R. and I.K.; funding acquisation H.K. and E.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Cornell University through a sub-award agreement (N0.84941-11056) between TARI and Cornell University through Next Generation Cassava Breeding Project
Data Availability Statement
The data presented in this study are available on request from the corresponding author due to the legal and institution regulations. Th data is apart of ongoing research projects, sharing it can compromise the integrity of these studies and violate data usage agreements.
Acknowledgments
The research grant was provided by NextGen cassava Project to Makerere Regional Centre for Crop improvement (MaRCCI) for the research and training of the first author. The International Institute for Tropical Agriculture (IITA) Uganda hosted field experiments at their experimental field stations and field assistance. and IITA Dar es salaam provided necessary equipment for virus activity analysis in their Biotechnology Laboratory.
Conflicts of Interest
The authors declare no conflict of interest with any one and the funders had no role in the design of the study, data collection, analysis or interpretation of the results or in the writing of the manuscript.
References
- Morgan, N.K.; Choct, M. Cassava: Nutrient composition and nutritive value in poultry diets. Anim. Nutr. 2016, 2, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, G.; et al. Roots, Tubers and Bananas Contributions to Food Security Article. 2019.
- Daya, P. Tackling Cassava Brown Streak Disease in Uganda with Nuclear Techniques.pdf. 2022.
- Alicai, T.; et al. Expansion of the cassava brown streak pandemic in Uganda revealed by annual field survey data for 2004 to 2017. Sci Data 2019, 1–8. [Google Scholar] [CrossRef]
- Mulenga, R.M.; Boykin, L.M.; Chikoti, P.C.; Sichilima, S.; Ng’Uni, D.; Alabi, O.J. Cassava brown streak disease and Ugandan cassava brown streak virus reported for the first time in Zambia. Plant Dis 2018, 102, 1410–1418. [Google Scholar] [CrossRef] [PubMed]
- Legg, J.P.; et al. Cassava virus diseases: Biology, epidemiology, and management, 1st ed.; Elsevier Inc., 2015; volume 91, no. 1. [Google Scholar] [CrossRef]
- Ano, C.U.; et al. Cassava Brown Streak Disease Response and Association With Agronomic Traits in Elite Nigerian Cassava Cultivars. Front Plant Sci 2021, 12, 1–15. [Google Scholar] [CrossRef]
- Liu, Q.; Liu, J.; Zhang, P.; He, S. Root and Tuber Crops. Encyclopedia of Agriculture and Food Systems 2014, 5, 46–61. [Google Scholar] [CrossRef]
- Tomlinson, K.R.; Bailey, A.M.; Alicai, T.; Seal, S.U.E.; Foster, G.D. Review Cassava brown streak disease: historical timeline, current knowledge and future prospects. Mol. Plant Pathol. 2017, 9, 1282–1294. [Google Scholar] [CrossRef]
- Kanju, E.E.; Masumba, E.; Tumwegamire, S. Performance of cassava brown streak disease-tolerant varieties in Zanzibar, Tanzania. Indian Journal of Horticulture 2017, 74, 557–561. [Google Scholar] [CrossRef]
- Mohammed, I.U.; Abarshi, M.M.; Muli, B.; Hillocks, R.J.; Maruthi, M.N. The Symptom and Genetic Diversity of Cassava Brown Streak Viruses Infecting Cassava in East Africa. Advances in Virology 2012, 2012, 795697. [Google Scholar] [CrossRef] [PubMed]
- Samar, S.; Fuerholzner, B.; Stein, B.; Winter, S. Resistance against cassava brown streak viruses from africa in cassava germplasm from South America. Front Plant Sci 2019, 10, 567. [Google Scholar] [CrossRef]
- Ceballos, H.; et al. Excellence in Cassava Breeding: Perspectives for the Future. Crop Breed Genet Genom 2020, 2, e200008. [Google Scholar] [CrossRef]
- Chipeta, M.M.; Melis, R.; Shanahan, P. Gene action controlling cassava brown streak disease resistance and storage root yield in cassava. Euphytica 2018, 1–15. [Google Scholar] [CrossRef]
- Ceballos, H.; Kawuki, R.S.; Gracen, V.E.; Yencho, G.C.; Hershey, C.H. Conventional breeding, marker-assisted selection, genomic selection and inbreeding in clonally propagated crops: a case study for cassava. Theoretical and Applied Genetics 2015, 128, 1647–1667. [Google Scholar] [CrossRef] [PubMed]
- Kulembeka, H.P.; et al. Diallel analysis of field resistance to brown streak disease in cassava (Manihot esculenta Crantz) landraces from Tanzania. Euphytica 2012, 277–288. [Google Scholar] [CrossRef]
- Nduwumuremyi, A.; Melis, R.; Shanahan, P.; Theodore, A. Genetic inheritance of pulp colour and selected traits of cassava (Manihot esculenta Crantz) at early generation selection. J Sci Food Agric 2018, 98, 3190–3197. [Google Scholar] [CrossRef] [PubMed]
- Zacarias, A.M.; Labuschagne, M.T. Diallel analysis of cassava brown streak disease, yield and yield related characteristics in Mozambique. Euphytica 2010, 176, 309–320. [Google Scholar] [CrossRef]
- Kamau, J.; Melis, R.; Laing, M.; Derera, J.; Shanahan, P.; Ngugi, E. Combining the yield ability and secondary traits of selected cassava genotypes in the semi-arid areas of Eastern Kenya. J Plant Breed Crop Sci 2010, 2, 181–191, http://www.academicjournals.org/jpbcs. [Google Scholar]
- Chalwe, A.; Melis, R.; Shanahan, P.; Chiona, M. Inheritance of resistance to cassava green mite and other useful agronomic traits in cassava grown in Zambia. Euphytica 2015. [Google Scholar] [CrossRef]
- Ezenwaka, L.; et al. Genome-wide association study of resistance to cassava green mite pest and related traits in cassava. Crop Sci 2018, 58. [Google Scholar] [CrossRef]
- Chikoti, P.C.; Mulenga, R.M.; Tembo, M.; Sseruwagi, P. Cassava mosaic disease: a review of a threat to cassava production in Zambia. Journal of Plant Pathology 2019, 101, 467–477. [Google Scholar] [CrossRef]
- Tumuhimbise, R. Breeding and evaluation of cassava for high storage root yield and early bulking in Uganda. Doctoral dissertation, University of KwaZulu-Natal, Pietermaritzburg, 2013; pp. 44–58. [Google Scholar]
- Gerald, S.; Robert, M.; Michael, U.; Mark, W. Using improved varieties and fertility enhancements for increasing yield of common beans (Phaseolus vulgaris L.) grown by small-landholder farmers in Uganda. Afr J Agric Res 2015, 10, 4795–4805. [Google Scholar] [CrossRef]
- Okogbenin, E.; Ekanayake, I.J.; Porto, M.C.M. Genotypic Variability in Adaptation Responses of Selected Clones of Cassava to Drought Stress in the Sudan Savanna Zone of Nigeria. J Agron Crop Sci 2003, 189, 376–389. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Report 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Shirima, R.; Maeda, D.; Kanju, E.; Ceasar, G.; Tibazarwa, F.; Legg, J. Absolute quantification of cassava brown streak virus mRNA by real-time qPCR. J Virol Methods 2017, 245, 5–13. [Google Scholar] [CrossRef]
- Sighhh 1985.pdf.
- Das, A.; et al. Deciphering genotype-by- Environment interaction for targeting test environments and rust resistant genotypes in field pea (Pisum sativum l.). Front Plant Sci 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Matova, P.M.; Kamutando, C.N.; Mutari, B.; Magorokosho, C.; Labuschagne, M. Adaptability and Stability Analysis of Commercial Cultivars, Experimental Hybrids and Lines under Natural Fall Armyworm Infestation in Zimbabwe Using Different Stability Models. Agronomy 2022, 12, 1724. [Google Scholar] [CrossRef]
- Musopole, H.; Mtonga, A. Resistance levels of cassava landraces to CMD, CBSD and vector whiteflies in Malawi. 2023.
- Pervin 2007.pdf.
- Seleiman, M.F.; et al. plants Drought Stress Impacts on Plants and Different Approaches to Alleviate Its Adverse Effects. Plants 2020, 10, 259. [Google Scholar] [CrossRef]
- Mohammed, J.; Bulti, T.; Girma, M. Genetic variability, heritability, and genetic advance for quantitative traits of sorghum [Sorghum Bicolor (L.) Moench] genotypes at Fedis, Eastern Ethiopia. International Journal of Agricultural Science and Food Technology 2023, 9, 064–075. [Google Scholar] [CrossRef]
- Temesgen, B. Speed breeding to accelerate crop improvement. International Journal of Agricultural Science and Food Technology 2022, 8, 178–186. [Google Scholar] [CrossRef]
- Lamara, A.; Fellahi, Z.E.A.; Hannachi, A.; Benniou, R. Assessing the phenotypic variation, heritability and genetic advance in bread wheat (Triticum aestivum L.) candidate lines grown under rainfed semi-arid region of Algeria. Rev Fac Nac Agron Medellin 2022, 75, 10107–10118. [Google Scholar] [CrossRef]
- Kayondo, S.I.; et al. Genome-wide association mapping and genomic prediction for CBSD resistance in Manihot esculenta. Sci. Rep. 2018, 1–11. [Google Scholar] [CrossRef]
- Pariyo, A.; et al. Stability of resistance to cassava brown streak disease in major agro-ecologies of Uganda. J. Plant Breed. Crop. Sci. 2015, 7, 67–78. [Google Scholar] [CrossRef]
- Kaweesi, T.; Kyaligonza, V.; Baguma, Y.; Kawuki, R.; Morag, F. Inbreeding enhances field resistance to cassava brown streak viruses. J Plant Breed Crop Sci 2016, 8, 138–149. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).