Submitted:
26 July 2024
Posted:
30 July 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Budding Yeast Septins and their Localisation
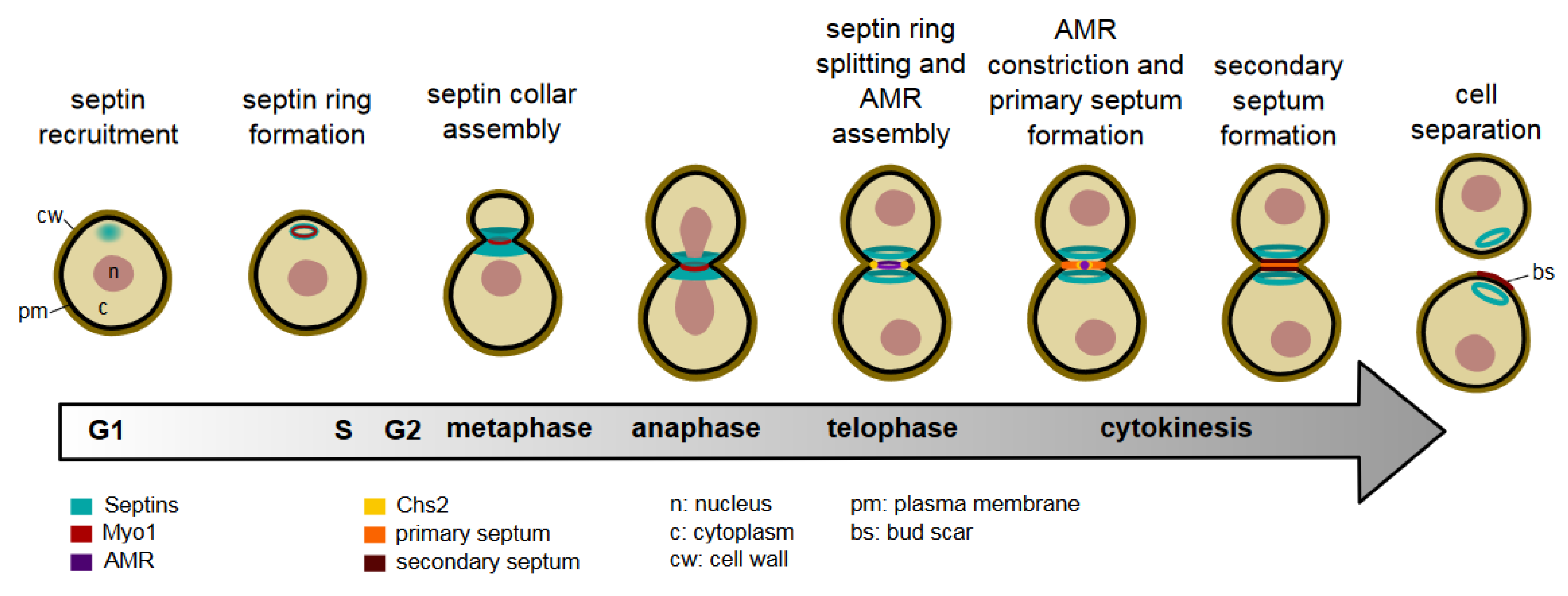
Septin Organisation and Cytokinesis in Budding Yeast
The control of Septin Dynamics During the Cell Cycle
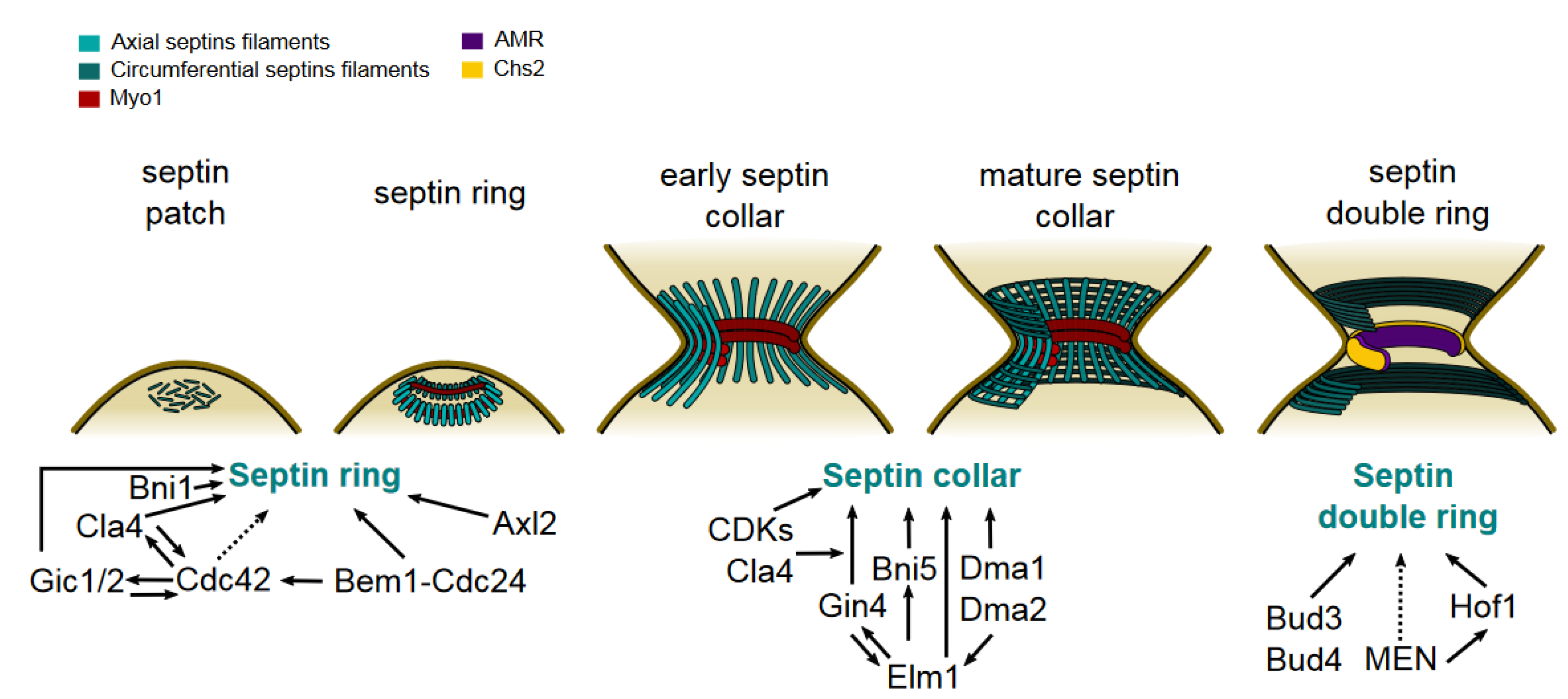
Septin Recruitment and Ring Assembly
Maturation Into the Septin Collar
Transition Into the Septin Double Ring
Conclusions and Perspectives
Acknowledgments
Conflicts of Interest
References
- Nishihama, R.; Onishi, M.; Pringle, J.R. New Insights into the Phylogenetic Distribution and Evolutionary Origins of the Septins. Biol Chem 2011, 392, 681–687. [Google Scholar] [CrossRef] [PubMed]
- Shuman, B.; Momany, M. Septins From Protists to People. Frontiers in Cell and Developmental Biology 2022, 9. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H.; Culotti, J.; Reid, B. Genetic Control of the Cell-Division Cycle in Yeast, I. Detection of Mutants. Proc Natl Acad Sci U S A 1970, 66, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H. Genetic Control of the Cell Division Cycle in Yeast. IV. Genes Controlling Bud Emergence and Cytokinesis. Exp Cell Res 1971, 69, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H.; Mortimer, R.K.; Culotti, J.; Culotti, M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of Cdc Mutants. Genetics 1973, 74, 267–286. [Google Scholar] [CrossRef] [PubMed]
- Byers, B.; Goetsch, L. A Highly Ordered Ring of Membrane-Associated Filaments in Budding Yeast. The Journal of cell biology 1976, 69, 717–721. [Google Scholar] [CrossRef] [PubMed]
- Ford, S.K.; Pringle, J.R. Cellular Morphogenesis in the Saccharomyces Cerevisiae Cell Cycle: Localization of the CDC11 Gene Product and the Timing of Events at the Budding Site. Dev Genet 1991, 12, 281–292. [Google Scholar] [CrossRef]
- Haarer, B.K.; Pringle, J.R. Immunofluorescence Localization of the Saccharomyces Cerevisiae CDC12 Gene Product to the Vicinity of the 10-Nm Filaments in the Mother-Bud Neck. Molecular and cellular biology 1987, 7, 3678–3687. [Google Scholar] [PubMed]
- De Virgilio, C.; DeMarini, D.J.; Pringle, J.R. SPR28, a Sixth Member of the Septin Gene Family in Saccharomyces Cerevisiae That Is Expressed Specifically in Sporulating Cells. Microbiology (Reading) 1996, 142 ( Pt 10) Pt 10, 2897–2905. [Google Scholar] [CrossRef]
- Mino, A.; Tanaka, K.; Kamei, T.; Umikawa, M.; Fujiwara, T.; Takai, Y. Shs1p: A Novel Member of Septin That Interacts with Spa2p, Involved in Polarized Growth inSaccharomyces Cerevisiae. Biochemical and Biophysical Research Communications 1998, 251, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Ozsarac, N.; Bhattacharyya, M.; Dawes, I.W.; Clancy, M.J. The SPR3 Gene Encodes a Sporulation-Specific Homologue of the Yeast CDC3/10/11/12 Family of Bud Neck Microfilaments and Is Regulated by ABFI. Gene 1995, 164, 157–162. [Google Scholar] [CrossRef]
- Barral, Y.; Mermall, V.; Mooseker, M.S.; Snyder, M. Compartmentalization of the Cell Cortex by Septins Is Required for Maintenance of Cell Polarity in Yeast. Molecular Cell 2000, 5, 841–851. [Google Scholar] [CrossRef] [PubMed]
- Castillon, G.A.; Adames, N.R.; Rosello, C.H.; Seidel, H.S.; Longtine, M.S.; Cooper, J.A.; Heil-Chapdelaine, R.A. Septins Have a Dual Role in Controlling Mitotic Exit in Budding Yeast. Curr Biol 2003, 13, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Longtine, M.S.; Theesfeld, C.L.; McMillan, J.N.; Weaver, E.; Pringle, J.R.; Lew, D.J. Septin-Dependent Assembly of a Cell Cycle-Regulatory Module in Saccharomyces Cerevisiae. Mol Cell Biol 2000, 20, 4049–4061. [Google Scholar] [CrossRef] [PubMed]
- Sakchaisri, K.; Asano, S.; Yu, L.-R.; Shulewitz, M.J.; Park, C.J.; Park, J.-E.; Cho, Y.-W.; Veenstra, T.D.; Thorner, J.; Lee, K.S. Coupling Morphogenesis to Mitotic Entry. Proceedings of the National Academy of Sciences 2004, 101, 4124–4129. [Google Scholar] [CrossRef]
- Kusch, J.; Meyer, A.; Snyder, M.P.; Barral, Y. Microtubule Capture by the Cleavage Apparatus Is Required for Proper Spindle Positioning in Yeast. Genes Dev 2002, 16, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Bi, E.; Maddox, P.; Lew, D.J.; Salmon, E.D.; McMillan, J.N.; Yeh, E.; Pringle, J.R. Involvement of an Actomyosin Contractile Ring in Saccharomyces Cerevisiae Cytokinesis. J Cell Biol 1998, 142, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- DeMarini, D.J.; Adams, A.E.M.; Fares, H.; Virgilio, C.D.; Valle, G.; Chuang, J.S.; Pringle, J.R. A Septin-Based Hierarchy of Proteins Required for Localized Deposition of Chitin in the Saccharomyces Cerevisiae Cell Wall. The Journal of Cell Biology 1997, 139, 75. [Google Scholar] [CrossRef] [PubMed]
- Wloka, C.; Nishihama, R.; Onishi, M.; Oh, Y.; Hanna, J.; Pringle, J.R.; Krauss, M.; Bi, E. Evidence That a Septin Diffusion Barrier Is Dispensable for Cytokinesis in Budding Yeast. Biol Chem 2011, 392, 813–829. [Google Scholar] [CrossRef] [PubMed]
- Estey, M.P.; Di Ciano-Oliveira, C.; Froese, C.D.; Bejide, M.T.; Trimble, W.S. Distinct Roles of Septins in Cytokinesis: SEPT9 Mediates Midbody Abscission. Journal of Cell Biology 2010, 191, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Joo, E.; Surka, M.C.; Trimble, W.S. Mammalian SEPT2 Is Required for Scaffolding Nonmuscle Myosin II and Its Kinases. Developmental Cell 2007, 13, 677–690. [Google Scholar] [CrossRef] [PubMed]
- Karasmanis, E.P.; Hwang, D.; Nakos, K.; Bowen, J.R.; Angelis, D.; Spiliotis, E.T. A Septin Double Ring Controls the Spatiotemporal Organization of the ESCRT Machinery in Cytokinetic Abscission. Current Biology 2019, 29, 2174–2182. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Froese, C.D.; Estey, M.P.; Trimble, W.S. SEPT9 Occupies the Terminal Positions in Septin Octamers and Mediates Polymerization-Dependent Functions in Abscission. The Journal of cell biology 2011, 195, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Renshaw, M.J.; Liu, J.; Lavoie, B.D.; Wilde, A. Anillin-Dependent Organization of Septin Filaments Promotes Intercellular Bridge Elongation and Chmp4B Targeting to the Abscission Site. Open Biol 2014, 4, 130190. [Google Scholar] [CrossRef] [PubMed]
- Surka, M.C.; Tsang, C.W.; Trimble, W.S. The Mammalian Septin MSF Localizes with Microtubules and Is Required for Completion of Cytokinesis. Mol Biol Cell 2002, 13, 3532–3545. [Google Scholar] [CrossRef] [PubMed]
- Tokhtaeva, E.; Capri, J.; Marcus, E.A.; Whitelegge, J.P.; Khuzakhmetova, V.; Bukharaeva, E.; Deiss-Yehiely, N.; Dada, L.A.; Sachs, G.; Fernandez-Salas, E.; et al. Septin Dynamics Are Essential for Exocytosis. J Biol Chem 2015, 290, 5280–5297. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-W.; Yan, M.; Collins, R.F.; DiCiccio, J.E.; Grinstein, S.; Trimble, W.S. Mammalian Septins Are Required for Phagosome Formation. Mol Biol Cell 2008, 19, 1717–1726. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Bai, X.; Bowen, J.R.; Dolat, L.; Korobova, F.; Yu, W.; Baas, P.W.; Svitkina, T.; Gallo, G.; Spiliotis, E.T. Septin-Driven Coordination of Actin and Microtubule Remodeling Regulates the Collateral Branching of Axons. Curr Biol 2012, 22, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Li, C.-R.; Lee, R.T.-H.; Wang, Y.-M.; Zheng, X.-D.; Wang, Y. Candida Albicans Hyphal Morphogenesis Occurs in Sec3p-Independent and Sec3p-Dependent Phases Separated by Septin Ring Formation. J Cell Sci 2007, 120, 1898–1907. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, P.A.; Field, C.M.; Groen, A.C.; Mitchison, T.J.; Loose, M. Using Supported Bilayers to Study the Spatiotemporal Organization of Membrane Bound Proteins. Methods Cell Biol 2015, 128, 223–241. [Google Scholar] [CrossRef] [PubMed]
- Tada, T.; Simonetta, A.; Batterton, M.; Kinoshita, M.; Edbauer, D.; Sheng, M. Role of Septin Cytoskeleton in Spine Morphogenesis and Dendrite Development in Neurons. Curr Biol 2007, 17, 1752–1758. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Vessey, J.P.; Konecna, A.; Dahm, R.; Macchi, P.; Kiebler, M.A. The GTP-Binding Protein Septin 7 Is Critical for Dendrite Branching and Dendritic-Spine Morphology. Curr Biol 2007, 17, 1746–1751. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Milenkovic, L.; Jin, H.; Scott, M.P.; Nachury, M.V.; Spiliotis, E.T.; Nelson, W.J. A Septin Diffusion Barrier at the Base of the Primary Cilium Maintains Ciliary Membrane Protein Distribution. Science 2010, 329, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.K.; Shindo, A.; Park, T.J.; Oh, E.C.; Ghosh, S.; Gray, R.S.; Lewis, R.A.; Johnson, C.A.; Attie-Bittach, T.; Katsanis, N.; et al. Planar Cell Polarity Acts through Septins to Control Collective Cell Movement and Ciliogenesis. Science 2010, 329, 1337–1340. [Google Scholar] [CrossRef] [PubMed]
- Palander, O.; El-Zeiry, M.; Trimble, W.S. Uncovering the Roles of Septins in Cilia. Frontiers in Cell and Developmental Biology 2017, 5. [Google Scholar] [CrossRef] [PubMed]
- Tooley, A.J.; Gilden, J.; Jacobelli, J.; Beemiller, P.; Trimble, W.S.; Kinoshita, M.; Krummel, M.F. Amoeboid T Lymphocytes Require the Septin Cytoskeleton for Cortical Integrity and Persistent Motility. Nature cell biology 2009, 11, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Dolat, L.; Hunyara, J.L.; Bowen, J.R.; Karasmanis, E.P.; Elgawly, M.; Galkin, V.E.; Spiliotis, E.T. Septins Promote Stress Fiber–Mediated Maturation of Focal Adhesions and Renal Epithelial Motility. J Cell Biol 2014, 207, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Ihara, M.; Kinoshita, A.; Yamada, S.; Tanaka, H.; Tanigaki, A.; Kitano, A.; Goto, M.; Okubo, K.; Nishiyama, H.; Ogawa, O.; et al. Cortical Organization by the Septin Cytoskeleton Is Essential for Structural and Mechanical Integrity of Mammalian Spermatozoa. Dev Cell 2005, 8, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Yan, B.; Ren, J.; Lyu, R.; Wu, Y.; Guo, Y.; Li, D.; Zhang, H.; Hu, J. FIT2 Organizes Lipid Droplet Biogenesis with ER Tubule-Forming Proteins and Septins. J Cell Biol 2021, 220, e201907183. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Castellanos, N.; Rodríguez, A.; Rabanal-Ruiz, Y.; Fernández-Vega, A.; López-Miranda, J.; Vázquez-Martínez, R.; Frühbeck, G.; Malagón, M.M. The Cytoskeletal Protein Septin 11 Is Associated with Human Obesity and Is Involved in Adipocyte Lipid Storage and Metabolism. Diabetologia 2017, 60, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Nurullin, L.F.; Khuzakhmetova, V.F.; Khaziev, E.F.; Samigullin, D.V.; Tsentsevitsky, A.N.; Skorinkin, A.I.; Bukharaeva, E.A.; Vagin, O. Reorganization of Septins Modulates Synaptic Transmission at Neuromuscular Junctions. Neuroscience 2019, 404, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Tsang, C.W.; Estey, M.P.; DiCiccio, J.E.; Xie, H.; Patterson, D.; Trimble, W.S. Characterization of Presynaptic Septin Complexes in Mammalian Hippocampal Neurons. Biol Chem 2011, 392, 739–749. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-M.; Fedchyshyn, M.J.; Grande, G.; Aitoubah, J.; Tsang, C.W.; Xie, H.; Ackerley, C.A.; Trimble, W.S.; Wang, L.-Y. Septins Regulate Developmental Switching from Microdomain to Nanodomain Coupling of Ca2+ Influx to Neurotransmitter Release at a Central Synapse. Neuron 2010, 67, 100–115. [Google Scholar] [CrossRef] [PubMed]
- Mostowy, S.; Bonazzi, M.; Hamon, M.A.; Tham, T.N.; Mallet, A.; Lelek, M.; Gouin, E.; Demangel, C.; Brosch, R.; Zimmer, C.; et al. Entrapment of Intracytosolic Bacteria by Septin Cage-like Structures. Cell Host Microbe 2010, 8, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Krokowski, S.; Lobato-Márquez, D.; Chastanet, A.; Pereira, P.M.; Angelis, D.; Galea, D.; Larrouy-Maumus, G.; Henriques, R.; Spiliotis, E.T.; Carballido-López, R.; et al. Septins Recognize and Entrap Dividing Bacterial Cells for Delivery to Lysosomes. Cell Host Microbe 2018, 24, 866–874. [Google Scholar] [CrossRef] [PubMed]
- Dolat, L.; Hu, Q.; Spiliotis, E.T. Septin Functions in Organ System Physiology and Pathology. Biol Chem 2014, 395, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Versele, M.; Gullbrand, B.; Shulewitz, M.J.; Cid, V.J.; Bahmanyar, S.; Chen, R.E.; Barth, P.; Alber, T.; Thorner, J. Protein-Protein Interactions Governing Septin Heteropentamer Assembly and Septin Filament Organization in Saccharomyces Cerevisiae. Mol Biol Cell 2004, 15, 4568–4583. [Google Scholar] [CrossRef] [PubMed]
- Cavini, I.A.; Leonardo, D.A.; Rosa, H.V.D.; Castro, D.K.S.V.; D’Muniz Pereira, H.; Valadares, N.F.; Araujo, A.P.U.; Garratt, R.C. The Structural Biology of Septins and Their Filaments: An Update. Frontiers in Cell and Developmental Biology 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Bertin, A.; McMurray, M.A.; Pierson, J.; Thai, L.; McDonald, K.L.; Zehr, E.A.; García, G.; Peters, P.; Thorner, J.; Nogales, E. Three-Dimensional Ultrastructure of the Septin Filament Network in Saccharomyces Cerevisiae. Mol. Biol. Cell 2012, 23, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Bridges, A.A.; Zhang, H.; Mehta, S.B.; Occhipinti, P.; Tani, T.; Gladfelter, A.S. Septin Assemblies Form by Diffusion-Driven Annealing on Membranes. Proc Natl Acad Sci U S A 2014, 111, 2146–2151. [Google Scholar] [CrossRef] [PubMed]
- John, C.M.; Hite, R.K.; Weirich, C.S.; Fitzgerald, D.J.; Jawhari, H.; Faty, M.; Schläpfer, D.; Kroschewski, R.; Winkler, F.K.; Walz, T.; et al. The Caenorhabditis Elegans Septin Complex Is Nonpolar. The EMBO Journal 2007, 26, 3296–3307. [Google Scholar] [CrossRef] [PubMed]
- Rodal, A.A.; Kozubowski, L.; Goode, B.L.; Drubin, D.G.; Hartwig, J.H. Actin and Septin Ultrastructures at the Budding Yeast Cell Cortex. MBoC 2005, 16, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Sirajuddin, M.; Farkasovsky, M.; Hauer, F.; Kühlmann, D.; Macara, I.G.; Weyand, M.; Stark, H.; Wittinghofer, A. Structural Insight into Filament Formation by Mammalian Septins. Nature 2007, 449, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Field, C.M.; al-Awar, O.; Rosenblatt, J.; Wong, M.L.; Alberts, B.; Mitchison, T.J. A Purified Drosophila Septin Complex Forms Filaments and Exhibits GTPase Activity. J Cell Biol 1996, 133, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Arbizzani, F.; Mavrakis, M.; Hoya, M.; Ribas, J.C.; Brasselet, S.; Paoletti, A.; Rincon, S.A. Septin Filament Compaction into Rings Requires the Anillin Mid2 and Contractile Ring Constriction. Cell Reports 2022, 39, 110722. [Google Scholar] [CrossRef] [PubMed]
- An, H.; Morrell, J.L.; Jennings, J.L.; Link, A.J.; Gould, K.L. Requirements of Fission Yeast Septins for Complex Formation, Localization, and Function. Mol Biol Cell 2004, 15, 5551–5564. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Q.; Kuhn, J.R.; Kovar, D.R.; Pollard, T.D. Spatial and Temporal Pathway for Assembly and Constriction of the Contractile Ring in Fission Yeast Cytokinesis. Dev Cell 2003, 5, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Bertin, A.; McMurray, M.A.; Grob, P.; Park, S.S.; Garcia, G., 3rd; Patanwala, I.; Ng, H.L.; Alber, T.; Thorner, J.; Nogales, E. Saccharomyces Cerevisiae Septins: Supramolecular Organization of Heterooligomers and the Mechanism of Filament Assembly. Proc Natl Acad Sci U S A 2008, 105, 8274–8279. [Google Scholar] [CrossRef] [PubMed]
- Garcia, G., 3rd; Bertin, A.; Li, Z.; Song, Y.; McMurray, M.A.; Thorner, J.; Nogales, E. Subunit-Dependent Modulation of Septin Assembly: Budding Yeast Septin Shs1 Promotes Ring and Gauze Formation. J Cell Biol 2011, 195, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Taveneau, C.; Blanc, R.; Péhau-Arnaudet, G.; Di Cicco, A.; Bertin, A. Synergistic Role of Nucleotides and Lipids for the Self-Assembly of Shs1 Septin Oligomers. Biochemical Journal 2020, 477, 2697–2714. [Google Scholar] [CrossRef] [PubMed]
- Beber, A.; Alqabandi, M.; Prévost, C.; Viars, F.; Lévy, D.; Bassereau, P.; Bertin, A.; Mangenot, S. Septin-Based Readout of PI(4,5)P2 Incorporation into Membranes of Giant Unilamellar Vesicles. Cytoskeleton 2019, 76, 92–103. [Google Scholar] [CrossRef] [PubMed]
- Szuba, A.; Bano, F.; Castro-Linares, G.; Iv, F.; Mavrakis, M.; Richter, R.P.; Bertin, A.; Koenderink, G.H. Membrane Binding Controls Ordered Self-Assembly of Animal Septins. eLife 2021, 10, e63349. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Kong, C.; Xie, H.; McPherson, P.S.; Grinstein, S.; Trimble, W.S. Phosphatidylinositol Polyphosphate Binding to the Mammalian Septin H5 Is Modulated by GTP. Current Biology 1999, 9, 1458–1467. [Google Scholar] [CrossRef] [PubMed]
- Bertin, A.; McMurray, M.A.; Thai, L.; Garcia, G.; Votin, V.; Grob, P.; Allyn, T.; Thorner, J.; Nogales, E. Phosphatidylinositol-4,5-Bisphosphate Promotes Budding Yeast Septin Filament Assembly and Organization. Journal of Molecular Biology 2010, 404, 711–731. [Google Scholar] [CrossRef] [PubMed]
- Tanaka-Takiguchi, Y.; Kinoshita, M.; Takiguchi, K. Septin-Mediated Uniform Bracing of Phospholipid Membranes. Current biology: CB 2009, 19, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Garrenton, L.S.; Stefan, C.J.; McMurray, M.A.; Emr, S.D.; Thorner, J. Pheromone-Induced Anisotropy in Yeast Plasma Membrane Phosphatidylinositol-4,5-Bisphosphate Distribution Is Required for MAPK Signaling. Proceedings of the National Academy of Sciences 2010, 107, 11805–11810. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Escudero, I.; Roelants, F.M.; Thorner, J.; Nombela, C.; Molina, M.; Cid, V.J. Reconstitution of the Mammalian PI3K/PTEN/Akt Pathway in Yeast. Biochem J 2005, 390, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Casamayor, A.; Snyder, M. Molecular Dissection of a Yeast Septin: Distinct Domains Are Required for Septin Interaction, Localization, and Function. Molecular and Cellular Biology 2003, 23, 2762–2777. [Google Scholar] [CrossRef] [PubMed]
- Omrane, M.; Camara, A.S.; Taveneau, C.; Benzoubir, N.; Tubiana, T.; Yu, J.; Guérois, R.; Samuel, D.; Goud, B.; Poüs, C.; et al. Septin 9 Has Two Polybasic Domains Critical to Septin Filament Assembly and Golgi Integrity. iScience 2019, 13, 138–153. [Google Scholar] [CrossRef] [PubMed]
- Alaoui, F.E.; Al-Akiki, I.; Ibanes, S.; Lyonnais, S.; Sanchez-Fuentes, D.; Desgarceaux, R.; Cazevieille, C.; Blanchard, M.-P.; Parmeggiani, A.; Carretero-Genevrier, A.; et al. Septin Filament Assembly Assist the Lateral Organization of Membranes 2024, 2024.03.19.585775.
- Beber, A.; Taveneau, C.; Nania, M.; Tsai, F.-C.; Di Cicco, A.; Bassereau, P.; Lévy, D.; Cabral, J.T.; Isambert, H.; Mangenot, S.; et al. Membrane Reshaping by Micrometric Curvature Sensitive Septin Filaments. Nat Commun 2019, 10, 420. [Google Scholar] [CrossRef] [PubMed]
- Bridges, A.A.; Jentzsch, M.S.; Oakes, P.W.; Occhipinti, P.; Gladfelter, A.S. Micron-Scale Plasma Membrane Curvature Is Recognized by the Septin Cytoskeleton. Journal of Cell Biology 2016, 213, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Cannon, K.S.; Woods, B.L.; Crutchley, J.M.; Gladfelter, A.S. An Amphipathic Helix Enables Septins to Sense Micrometer-Scale Membrane Curvature. Journal of Cell Biology 2019, 218, 1128–1137. [Google Scholar] [CrossRef] [PubMed]
- Ewers, H.; Tada, T.; Petersen, J.D.; Racz, B.; Sheng, M.; Choquet, D. A Septin-Dependent Diffusion Barrier at Dendritic Spine Necks. PLOS ONE 2014, 9, e113916. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.C.; Lin, Y.H.; Chen, H.I.; Wang, Y.Y.; Chiou, Y.W.; Lin, H.H.; Pan, H.A.; Wu, C.M.; Su, S.M.; Hsu, C.C.; et al. SEPT12 Mutations Cause Male Infertility with Defective Sperm Annulus. Hum Mutat 2012, 33, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Newby, J.; Gladfelter, A.S. Control of Septin Filament Flexibility and Bundling by Subunit Composition and Nucleotide Interactions. MBoC 2018, 29, 702–712. [Google Scholar] [CrossRef]
- Garcia, G., III; Bertin, A.; Li, Z.; Song, Y.; McMurray, M.A.; Thorner, J.; Nogales, E. Subunit-Dependent Modulation of Septin Assembly: Budding Yeast Septin Shs1 Promotes Ring and Gauze Formation. Journal of Cell Biology 2011, 195, 993–1004. [Google Scholar] [CrossRef] [PubMed]
- Frazier, J.A.; Wong, M.L.; Longtine, M.S.; Pringle, J.R.; Mann, M.; Mitchison, T.J.; Field, C. Polymerization of Purified Yeast Septins: Evidence That Organized Filament Arrays May Not Be Required for Septin Function. J Cell Biol 1998, 143, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Booth, E.A.; Vane, E.W.; Dovala, D.; Thorner, J. A Förster Resonance Energy Transfer (FRET)-Based System Provides Insight into the Ordered Assembly of Yeast Septin Hetero-Octamers *. Journal of Biological Chemistry 2015, 290, 28388–28401. [Google Scholar] [CrossRef] [PubMed]
- McMurray, M.A.; Bertin, A.; Garcia, G.; Lam, L.; Nogales, E.; Thorner, J. Septin Filament Formation Is Essential in Budding Yeast. Developmental Cell 2011, 20, 540–549. [Google Scholar] [CrossRef]
- Garcia, G.; Finnigan, G.C.; Heasley, L.R.; Sterling, S.M.; Aggarwal, A.; Pearson, C.G.; Nogales, E.; McMurray, M.A.; Thorner, J. Assembly, Molecular Organization, and Membrane-Binding Properties of Development-Specific Septins. J Cell Biol 2016, 212, 515–529. [Google Scholar] [CrossRef] [PubMed]
- Heasley, L.R.; McMurray, M.A. Roles of Septins in Prospore Membrane Morphogenesis and Spore Wall Assembly in Saccharomyces Cerevisiae. MBoC 2016, 27, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Cid, V.J.; Adamikova, L.; Sanchez, M.; Molina, M.; Nombela, C. Cell Cycle Control of Septin Ring Dynamics in the Budding Yeast. Microbiology 2001, 147, 1437–1450. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.B.; Haarer, B.K.; Pringle, J.R. Cellular Morphogenesis in the Saccharomycescerevisiae Cell Cycle: Localization of the CDC3 Gene Product and the Timing of Events at the Budding Site.
- Caviston, J.P.; Longtine, M.; Pringle, J.R.; Bi, E. The Role of Cdc42p GTPase-Activating Proteins in Assembly of the Septin Ring in Yeast. Mol Biol Cell 2003, 14, 4051–4066. [Google Scholar] [CrossRef]
- Dobbelaere, J.; Gentry, M.S.; Hallberg, R.L.; Barral, Y. Phosphorylation-Dependent Regulation of Septin Dynamics during the Cell Cycle. Dev Cell 2003, 4, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Longtine, M.S.; Fares, H.; Pringle, J.R. Role of the Yeast Gin4p Protein Kinase in Septin Assembly and the Relationship between Septin Assembly and Septin Function. J Cell Biol 1998, 143, 719–736. [Google Scholar] [CrossRef] [PubMed]
- Tartakoff, A.M.; Aylyarov, I.; Jaiswal, P. Septin-Containing Barriers Control the Differential Inheritance of Cytoplasmic Elements. Cell Reports 2013, 3, 223–236. [Google Scholar] [CrossRef]
- Fares, H.; Goetsch, L.; Pringle, J.R. Identification of a Developmentally Regulated Septin and Involvement of the Septins in Spore Formation in Saccharomyces Cerevisiae. J Cell Biol 1996, 132, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Spiliotis, E.T.; Nakos, K. Cellular Functions of Actin- and Microtubule-Associated Septins. Current Biology 2021, 31, R651–R666. [Google Scholar] [CrossRef]
- Ong, K.; Wloka, C.; Okada, S.; Svitkina, T.; Bi, E. Architecture and Dynamic Remodelling of the Septin Cytoskeleton during the Cell Cycle. Nat Commun 2014, 5, 5698. [Google Scholar] [CrossRef] [PubMed]
- Vrabioiu, A.M.; Mitchison, T.J. Structural Insights into Yeast Septin Organization from Polarized Fluorescence Microscopy. Nature 2006, 443, 466–469. [Google Scholar] [CrossRef] [PubMed]
- McQuilken, M.; Jentzsch, M.S.; Verma, A.; Mehta, S.B.; Oldenbourg, R.; Gladfelter, A.S. Analysis of Septin Reorganization at Cytokinesis Using Polarized Fluorescence Microscopy. Front Cell Dev Biol 2017, 5, 42. [Google Scholar] [CrossRef] [PubMed]
- Demay, B.S.; Bai, X.; Howard, L.; Occhipinti, P.; Meseroll, R.A.; Spiliotis, E.T.; Oldenbourg, R.; Gladfelter, A.S. Septin Filaments Exhibit a Dynamic, Paired Organization That Is Conserved from Yeast to Mammals. J Cell Biol 2011, 193, 1065–1081. [Google Scholar] [CrossRef] [PubMed]
- Tamborrini, D.; Juanes, M.A.; Ibanes, S.; Rancati, G.; Piatti, S. Recruitment of the Mitotic Exit Network to Yeast Centrosomes Couples Septin Displacement to Actomyosin Constriction. Nat Commun 2018, 9, 4308. [Google Scholar] [CrossRef] [PubMed]
- Marquardt, J.; Chen, X.; Bi, E. Architecture, Remodeling, and Functions of the Septin Cytoskeleton. Cytoskeleton 2019, 76, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Bi, E.; Park, H.O. Cell Polarization and Cytokinesis in Budding Yeast. Genetics 2012, 191, 347–387. [Google Scholar] [CrossRef] [PubMed]
- Cvrckova, F.; De Virgilio, C.; Manser, E.; Pringle, J.R.; Nasmyth, K. Ste20-like Protein Kinases Are Required for Normal Localization of Cell Growth and for Cytokinesis in Budding Yeast. Genes Dev 1995, 9, 1817–1830. [Google Scholar] [CrossRef] [PubMed]
- Kang, P.J.; Mullner, R.; Lian, K.; Park, H.-O. Cdc42 Couples Septin Recruitment to the Axial Landmark Assembly via Axl2 in Budding Yeast. Journal of Cell Science 2023, 137, jcs261080. [Google Scholar] [CrossRef] [PubMed]
- Chant, J.; Mischke, M.; Mitchell, E.; Herskowitz, I.; Pringle, J.R. Role of Bud3p in Producing the Axial Budding Pattern of Yeast. J Cell Biol 1995, 129, 767–778. [Google Scholar] [CrossRef] [PubMed]
- Sanders, S.L.; Herskowitz, I. The BUD4 Protein of Yeast, Required for Axial Budding, Is Localized to the Mother/BUD Neck in a Cell Cycle-Dependent Manner. J Cell Biol 1996, 134, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Kang, P.J.; Angerman, E.; Jung, C.H.; Park, H.O. Bud4 Mediates the Cell-Type-Specific Assembly of the Axial Landmark in Budding Yeast. Journal of cell science 2012, 125, 3840–3849. [Google Scholar] [CrossRef] [PubMed]
- Okada, S.; Leda, M.; Hanna, J.; Savage, N.S.; Bi, E.; Goryachev, A.B. Daughter Cell Identity Emerges from the Interplay of Cdc42, Septins, and Exocytosis. Developmental Cell 2013, 26, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Lippincott, J.; Li, R. Sequential Assembly of Myosin II, an IQGAP-like Protein, and Filamentous Actin to a Ring Structure Involved in Budding Yeast Cytokinesis. J Cell Biol 1998, 140, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Vallen, E.A.; Dravis, C.; Tcheperegine, S.E.; Drees, B.; Bi, E. Identification and Functional Analysis of the Essential and Regulatory Light Chains of the Only Type II Myosin Myo1p in Saccharomyces Cerevisiae. The Journal of cell biology 2004, 165, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Watts, F.Z.; Shiels, G.; Orr, E. The Yeast MYO1 Gene Encoding a Myosin-like Protein Required for Cell Division. The EMBO journal 1987, 6, 3499–3505. [Google Scholar] [CrossRef] [PubMed]
- Boyne, J.R.; Yosuf, H.M.; Bieganowski, P.; Brenner, C.; Price, C. Yeast Myosin Light Chain, Mlc1p, Interacts with Both IQGAP and Class II Myosin to Effect Cytokinesis. Journal of cell science 2000, 113 Pt 24, 4533–4543. [Google Scholar] [CrossRef] [PubMed]
- Epp, J.A.; Chant, J. An IQGAP-Related Protein Controls Actin-Ring Formation and Cytokinesis in Yeast. Current Biology 1997, 7, 921–929. [Google Scholar] [CrossRef]
- Fang, X.; Luo, J.; Nishihama, R.; Wloka, C.; Dravis, C.; Travaglia, M.; Iwase, M.; Vallen, E.A.; Bi, E. Biphasic Targeting and Cleavage Furrow Ingression Directed by the Tail of a Myosin II. J Cell Biol 2010, 191, 1333–1350. [Google Scholar] [CrossRef] [PubMed]
- Shannon, K.B.; Li, R. A Myosin Light Chain Mediates the Localization of the Budding Yeast IQGAP-like Protein during Contractile Ring Formation. Curr Biol 2000, 10, 727–730. [Google Scholar] [CrossRef] [PubMed]
- Tolliday, N.; VerPlank, L.; Li, R. Rho1 Directs Formin-Mediated Actin Ring Assembly during Budding Yeast Cytokinesis. Curr Biol 2002, 12, 1864–1870. [Google Scholar] [CrossRef] [PubMed]
- Vallen, E.A.; Caviston, J.; Bi, E. Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in Cytokinesis in Saccharomyces Cerevisiae. Mol Biol Cell 2000, 11, 593–611. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Okada, S.; Cai, G.; Zhou, B.; Bi, E. MyosinII Heavy Chain and Formin Mediate the Targeting of Myosin Essential Light Chain to the Division Site before and during Cytokinesis. Molecular biology of the cell 2015, 26, 1211–1224. [Google Scholar] [CrossRef] [PubMed]
- Chuang, J.S.; Schekman, R.W. Differential Trafficking and Timed Localization of Two Chitin Synthase Proteins, Chs2p and Chs3p. The Journal of cell biology 1996, 135, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Meitinger, F.; Petrova, B.; Lombardi, I.M.; Bertazzi, D.T.; Hub, B.; Zentgraf, H.; Pereira, G. Targeted Localization of Inn1, Cyk3 and Chs2 by the Mitotic-Exit Network Regulates Cytokinesis in Budding Yeast. J Cell Sci 2010, 123, 1851–1861. [Google Scholar] [CrossRef] [PubMed]
- Foltman, M.; Filali-Mouncef, Y.; Crespo, D.; Sanchez-Diaz, A. Cell Polarity Protein Spa2 Coordinates Chs2 Incorporation at the Division Site in Budding Yeast. PLOS Genetics 2018, 14, e1007299. [Google Scholar] [CrossRef]
- VerPlank, L.; Li, R. Cell Cycle-Regulated Trafficking of Chs2 Controls Actomyosin Ring Stability during Cytokinesis. Mol Biol Cell 2005, 16, 2529–2543. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.F.; Bennett, A.M.; Ma, W.K.; Hall, M.C.; Yeong, F.M. Dependence of Chs2 ER Export on Dephosphorylation by Cytoplasmic Cdc14 Ensures That Septum Formation Follows Mitosis. Molecular biology of the cell 2012, 23, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Nishihama, R.; Schreiter, J.H.; Onishi, M.; Vallen, E.A.; Hanna, J.; Moravcevic, K.; Lippincott, M.F.; Han, H.; Lemmon, M.A.; Pringle, J.R.; et al. Role of Inn1 and Its Interactions with Hof1 and Cyk3 in Promoting Cleavage Furrow and Septum Formation in S. Cerevisiae. J Cell Biol 2009, 185, 995–1012. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Schreiter, J.H.; Okada, H.; Wloka, C.; Okada, S.; Yan, D.; Duan, X.; Bi, E. Hof1 and Chs4 Interact via F-BAR Domain and Sel1-like Repeats to Control Extracellular Matrix Deposition during Cytokinesis. Current Biology 2017, 27, 2878–2886. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Nishihama, R.; Onishi, M.; Pringle, J.R. Role of the Hof1–Cyk3 Interaction in Cleavage-Furrow Ingression and Primary-Septum Formation during Yeast Cytokinesis. Molecular Biology of the Cell 2018, 29, 597–609. [Google Scholar] [CrossRef]
- Devrekanli, A.; Foltman, M.; Roncero, C.; Sanchez-Diaz, A.; Labib, K. Inn1 and Cyk3 Regulate Chitin Synthase during Cytokinesis in Budding Yeasts. Journal of cell science 2012, 125, 5453–5466. [Google Scholar] [CrossRef] [PubMed]
- Jendretzki, A.; Ciklic, I.; Rodicio, R.; Schmitz, H.P.; Heinisch, J.J. Cyk3 Acts in Actomyosin Ring Independent Cytokinesis by Recruiting Inn1 to the Yeast Bud Neck. Molecular genetics and genomics: MGG 2009, 282, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Onishi, M.; Ko, N.; Nishihama, R.; Pringle, J.R. Distinct Roles of Rho1, Cdc42, and Cyk3 in Septum Formation and Abscission during Yeast Cytokinesis. J Cell Biol 2013, 202, 311–329. [Google Scholar] [CrossRef]
- Sanchez-Diaz, A.; Marchesi, V.; Murray, S.; Jones, R.; Pereira, G.; Edmondson, R.; Allen, T.; Labib, K. Inn1 Couples Contraction of the Actomyosin Ring to Membrane Ingression during Cytokinesis in Budding Yeast. Nature cell biology 2008, 10, 395–406. [Google Scholar] [CrossRef]
- Foltman, M.; Molist, I.; Arcones, I.; Sacristan, C.; Filali-Mouncef, Y.; Roncero, C.; Sanchez-Diaz, A. Ingression Progression Complexes Control Extracellular Matrix Remodelling during Cytokinesis in Budding Yeast. PLOS Genetics 2016, 12, e1005864. [Google Scholar] [CrossRef] [PubMed]
- Lippincott, J.; Li, R. Dual Function of Cyk2, a Cdc15/PSTPIP Family Protein, in Regulating Actomyosin Ring Dynamics and Septin Distribution. J Cell Biol 1998, 143, 1947–1960. [Google Scholar] [CrossRef] [PubMed]
- Bi, E.; Maddox, P.; Lew, D.J.; Salmon, E.D.; McMillan, J.N.; Yeh, E.; Pringle, J.R. Involvement of an Actomyosin Contractile Ring in Saccharomyces Cerevisiae Cytokinesis. J Cell Biol 1998, 142, 1301–1312. [Google Scholar] [CrossRef] [PubMed]
- Lippincott, J.; Li, R. Sequential Assembly of Myosin II, an IQGAP-like Protein, and Filamentous Actin to a Ring Structure Involved in Budding Yeast Cytokinesis. Journal of Cell Biology 1998, 140, 355–366. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Vallen, E.A.; Dravis, C.; Tcheperegine, S.E.; Drees, B.; Bi, E. Identification and Functional Analysis of the Essential and Regulatory Light Chains of the Only Type II Myosin Myo1p in Saccharomyces Cerevisiae. Journal of Cell Biology 2004, 165, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Liu, W.; Bretscher, A. The Yeast Formin Bnr1p Has Two Localization Regions That Show Spatially and Temporally Distinct Association with Septin Structures. Mol Biol Cell 2010, 21, 1253–1262. [Google Scholar] [CrossRef] [PubMed]
- Pruyne, D.; Gao, L.; Bi, E.; Bretscher, A. Stable and Dynamic Axes of Polarity Use Distinct Formin Isoforms in Budding Yeast. Molecular biology of the cell 2004, 15, 4971–4989. [Google Scholar] [CrossRef] [PubMed]
- Lippincott, J.; Li, R. Dual Function of Cyk2, a Cdc15/PSTPIP Family Protein, in Regulating Actomyosin Ring Dynamics and Septin Distribution. Journal of Cell Biology 1998, 143, 1947–1960. [Google Scholar] [CrossRef] [PubMed]
- Huxley, H.E. The Mechanism of Muscular Contraction. Science 1969, 164, 1356–1365. [Google Scholar] [CrossRef] [PubMed]
- Huxley, H.; Hanson, J. Changes in the Cross-Striations of Muscle during Contraction and Stretch and Their Structural Interpretation. Nature 1954, 173, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Mendes Pinto, I.; Rubinstein, B.; Kucharavy, A.; Unruh, J.R.; Li, R. Actin Depolymerization Drives Actomyosin Ring Contraction during Budding Yeast Cytokinesis. Developmental Cell 2012, 22, 1247–1260. [Google Scholar] [CrossRef] [PubMed]
- Ko, N.; Nishihama, R.; Tully, G.H.; Ostapenko, D.; Solomon, M.J.; Morgan, D.O.; Pringle, J.R. Identification of Yeast IQGAP (Iqg1p) as an Anaphase-Promoting-Complex Substrate and Its Role in Actomyosin-Ring-Independent Cytokinesis. MBoC 2007, 18, 5139–5153. [Google Scholar] [CrossRef] [PubMed]
- Tully, G.H.; Nishihama, R.; Pringle, J.R.; Morgan, D.O. The Anaphase-Promoting Complex Promotes Actomyosin-Ring Disassembly during Cytokinesis in Yeast. MBoC 2009, 20, 1201–1212. [Google Scholar] [CrossRef] [PubMed]
- Cabib, E. The Septation Apparatus, a Chitin-Requiring Machine in Budding Yeast. Archives of biochemistry and biophysics 2004, 426, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Kashimshetty, R.; Ng, K.E.; Tan, H.B.; Yeong, F.M. Exit from Mitosis Triggers Chs2p Transport from the Endoplasmic Reticulum to Mother-Daughter Neck via the Secretory Pathway in Budding Yeast. The Journal of cell biology 2006, 174, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Bi, E. Cytokinesis in Budding Yeast: The Relationship between Actomyosin Ring Function and Septum Formation. Cell Struct Funct 2001, 26, 529–537. [Google Scholar] [CrossRef]
- Schmidt, M.; Bowers, B.; Varma, A.; Roh, D.H.; Cabib, E. In Budding Yeast, Contraction of the Actomyosin Ring and Formation of the Primary Septum at Cytokinesis Depend on Each Other. J Cell Sci 2002, 115, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Roh, D.-H.; Bowers, B.; Schmidt, M.; Cabib, E. The Septation Apparatus, an Autonomous System in Budding Yeast. Mol Biol Cell 2002, 13, 2747–2759. [Google Scholar] [CrossRef] [PubMed]
- Lippincott, J.; Shannon, K.B.; Shou, W.; Deshaies, R.J.; Li, R. The Tem1 Small GTPase Controls Actomyosin and Septin Dynamics during Cytokinesis. J Cell Sci 2001, 114, 1379–86. [Google Scholar] [CrossRef] [PubMed]
- Tamborrini, D.; Piatti, S. Septin Clearance from the Division Site Triggers Cytokinesis in Budding Yeast. Microbial Cell 2019, 6, 296–298. [Google Scholar] [CrossRef] [PubMed]
- Dobbelaere, J.; Barral, Y. Spatial Coordination of Cytokinetic Events by Compartmentalization of the Cell Cortex. Science 2004, 305, 393–396. [Google Scholar] [CrossRef] [PubMed]
- Onishi, M.; Ko, N.; Nishihama, R.; Pringle, J.R. Distinct Roles of Rho1, Cdc42, and Cyk3 in Septum Formation and Abscission during Yeast Cytokinesis. Journal of Cell Biology 2013, 202, 311–329. [Google Scholar] [CrossRef] [PubMed]
- Okada, H.; MacTaggart, B.; Ohya, Y.; Bi, E. The Kinetic Landscape and Interplay of Protein Networks in Cytokinesis. iScience 2021, 24, 101917. [Google Scholar] [CrossRef] [PubMed]
- Weiss, E.L. Mitotic Exit and Separation of Mother and Daughter Cells. Genetics 2012, 192, 1165–1202. [Google Scholar] [CrossRef] [PubMed]
- Martin-Cuadrado, A.B.; Morrell, J.L.; Konomi, M.; An, H.; Petit, C.; Osumi, M.; Balasubramanian, M.; Gould, K.L.; Del Rey, F.; de Aldana, C.R. Role of Septins and the Exocyst Complex in the Function of Hydrolytic Enzymes Responsible for Fission Yeast Cell Separation. Mol Biol Cell 2005, 16, 4867–4881. [Google Scholar] [CrossRef] [PubMed]
- Gladfelter, A.S.; Moskow, J.J.; Zyla, T.R.; Lew, D.J. Isolation and Characterization of Effector-Loop Mutants of CDC42 in Yeast. Mol Biol Cell 2001, 12, 1239–1255. [Google Scholar] [CrossRef]
- Iwase, M.; Luo, J.; Nagaraj, S.; Longtine, M.; Kim, H.B.; Haarer, B.K.; Caruso, C.; Tong, Z.; Pringle, J.R.; Bi, E. Role of a Cdc42p Effector Pathway in Recruitment of the Yeast Septins to the Presumptive Bud Site. Mol Biol Cell 2006, 17, 1110–1125. [Google Scholar] [CrossRef] [PubMed]
- Gladfelter, A.S.; Bose, I.; Zyla, T.R.; Bardes, E.S.; Lew, D.J. Septin Ring Assembly Involves Cycles of GTP Loading and Hydrolysis by Cdc42p. The Journal of cell biology 2002, 156, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Benton, B.K.; Tinkelenberg, A.; Gonzalez, I.; Cross, F.R. Cla4p, a Saccharomyces Cerevisiae Cdc42p-Activated Kinase Involved in Cytokinesis, Is Activated at Mitosis. Mol Cell Biol 1997, 17, 5067–76. [Google Scholar] [CrossRef] [PubMed]
- Kadota, J.; Yamamoto, T.; Yoshiuchi, S.; Bi, E.; Tanaka, K. Septin Ring Assembly Requires Concerted Action of Polarisome Components, a PAK Kinase Cla4p, and the Actin Cytoskeleton in Saccharomyces Cerevisiae. Molecular biology of the cell 2004, 15, 5329–5345. [Google Scholar] [CrossRef] [PubMed]
- Versele, M.; Thorner, J. Septin Collar Formation in Budding Yeast Requires GTP Binding and Direct Phosphorylation by the PAK, Cla4. J Cell Biol 2004, 164, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Bose, I.; Irazoqui, J.E.; Moskow, J.J.; Bardes, E.S.; Zyla, T.R.; Lew, D.J. Assembly of Scaffold-Mediated Complexes Containing Cdc42p, the Exchange Factor Cdc24p, and the Effector Cla4p Required for Cell Cycle-Regulated Phosphorylation of Cdc24p. The Journal of biological chemistry 2001, 276, 7176–7186. [Google Scholar] [CrossRef] [PubMed]
- Gulli, M.-P.; Jaquenoud, M.; Shimada, Y.; Niederhäuser, G.; Wiget, P.; Peter, M. Phosphorylation of the Cdc42 Exchange Factor Cdc24 by the PAK-like Kinase Cla4 May Regulate Polarized Growth in Yeast. Molecular Cell 2000, 6, 1155–1167. [Google Scholar] [CrossRef] [PubMed]
- Kozubowski, L.; Saito, K.; Johnson, J.M.; Howell, A.S.; Zyla, T.R.; Lew, D.J. Symmetry-Breaking Polarization Driven by a Cdc42p GEF-PAK Complex. Current biology: CB 2008, 18, 1719–1726. [Google Scholar] [CrossRef] [PubMed]
- Chollet, J.; Dünkler, A.; Bäuerle, A.; Vivero-Pol, L.; Mulaw, M.A.; Gronemeyer, T.; Johnsson, N. Cdc24 Interacts with the Septins to Create a Positive Feedback during Bud Site Assembly in Yeast. Journal of Cell Science, 2402. [Google Scholar] [CrossRef]
- Brown, J.L.; Jaquenoud, M.; Gulli, M.P.; Chant, J.; Peter, M. Novel Cdc42-Binding Proteins Gic1 and Gic2 Control Cell Polarity in Yeast. Genes Dev 1997, 11, 2972–2982. [Google Scholar] [CrossRef] [PubMed]
- Orlando, K.; Sun, X.; Zhang, J.; Lu, T.; Yokomizo, L.; Wang, P.; Guo, W. Exo-Endocytic Trafficking and the Septin-Based Diffusion Barrier Are Required for the Maintenance of Cdc42p Polarization during Budding Yeast Asymmetric Growth. Mol Biol Cell 2011, 22, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Sadian, Y.; Gatsogiannis, C.; Patasi, C.; Hofnagel, O.; Goody, R.S.; Farkasovsky, M.; Raunser, S. The Role of Cdc42 and Gic1 in the Regulation of Septin Filament Formation and Dissociation. Elife 2013, 2, e01085. [Google Scholar] [CrossRef] [PubMed]
- Kang, P.J.; Miller, K.E.; Guegueniat, J.; Beven, L.; Park, H.-O. The Shared Role of the Rsr1 GTPase and Gic1/Gic2 in Cdc42 Polarization. MBoC 2018, 29, 2359–2369. [Google Scholar] [CrossRef] [PubMed]
- Daniels, C.N.; Zyla, T.R.; Lew, D.J. A Role for Gic1 and Gic2 in Cdc42 Polarization at Elevated Temperature. PLoS ONE 2018, 13, e0200863. [Google Scholar] [CrossRef] [PubMed]
- Lemmon, M.A.; Ferguson, K.M.; O’Brien, R.; Sigler, P.B.; Schlessinger, J. Specific and High-Affinity Binding of Inositol Phosphates to an Isolated Pleckstrin Homology Domain. Proceedings of the National Academy of Sciences of the United States of America 1995, 92, 10472–10476. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.-D.; Sperber, L.M.; Kane, S.A.; Tong, Z.; Tong, A.H.Y.; Boone, C.; Bi, E. Sequential and Distinct Roles of the Cadherin Domain-Containing Protein Axl2p in Cell Polarization in Yeast Cell Cycle. MBoC 2007, 18, 2542–2560. [Google Scholar] [CrossRef] [PubMed]
- Lai, H.; Chiou, J.-G.; Zhurikhina, A.; Zyla, T.R.; Tsygankov, D.; Lew, D.J. Temporal Regulation of Morphogenetic Events in Saccharomyces Cerevisiae. Mol Biol Cell 2018, 29, 2069–2083. [Google Scholar] [CrossRef] [PubMed]
- Kang, P.J.; Mullner, R.; Lian, K.; Park, H.-O. Cdc42 Couples Septin Recruitment to the Axial Landmark Assembly via Axl2 in Budding Yeast. Journal of Cell Science 2024, 137, jcs261080. [Google Scholar] [CrossRef]
- Merlini, L.; Bolognesi, A.; Juanes, M.A.; Vandermoere, F.; Courtellemont, T.; Pascolutti, R.; Seveno, M.; Barral, Y.; Piatti, S. Rho1- and Pkc1-Dependent Phosphorylation of the F-BAR Protein Syp1 Contributes to Septin Ring Assembly. Molecular biology of the cell 2015, 26, 3245–3262. [Google Scholar] [CrossRef] [PubMed]
- Qiu, W.; Neo, S.P.; Yu, X.; Cai, M. A Novel Septin-Associated Protein, Syp1p, Is Required for Normal Cell Cycle-Dependent Septin Cytoskeleton Dynamics in Yeast. Genetics 2008, 180, 1445–1457. [Google Scholar] [CrossRef] [PubMed]
- Ibanes, S.; El-Alaoui, F.; Lai-Kee-Him, J.; Cazevieille, C.; Hoh, F.; Lyonnais, S.; Bron, P.; Cipelletti, L.; Picas, L.; Piatti, S. The Syp1/FCHo2 Protein Induces Septin Filament Bundling through Its Intrinsically Disordered Domain. Cell Rep 2022, 41, 111765. [Google Scholar] [CrossRef] [PubMed]
- Mortensen, E.M.; McDonald, H.; Yates, J., 3rd; Kellogg, D.R. Cell Cycle-Dependent Assembly of a Gin4-Septin Complex. Mol Biol Cell 2002, 13, 2091–2105. [Google Scholar] [CrossRef] [PubMed]
- Asano, S.; Park, J.E.; Yu, L.R.; Zhou, M.; Sakchaisri, K.; Park, C.J.; Kang, Y.H.; Thorner, J.; Veenstra, T.D.; Lee, K.S. Direct Phosphorylation and Activation of a Nim1-Related Kinase Gin4 by Elm1 in Budding Yeast. J Biol Chem 2006, 281, 27090–27098. [Google Scholar] [CrossRef] [PubMed]
- Egelhofer, T.A.; Villen, J.; McCusker, D.; Gygi, S.P.; Kellogg, D.R. The Septins Function in G1 Pathways That Influence the Pattern of Cell Growth in Budding Yeast. PloS one 2008, 3, e2022. [Google Scholar] [CrossRef] [PubMed]
- Shulewitz, M.J.; Inouye, C.J.; Thorner, J. Hsl7 Localizes to a Septin Ring and Serves as an Adapter in a Regulatory Pathway That Relieves Tyrosine Phosphorylation of Cdc28 Protein Kinase in Saccharomyces Cerevisiae. Mol Cell Biol 1999, 19, 7123–7137. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.L.; Blacketer, M.J.; Edgington, N.P.; Myers, A.M. Assembly Interdependence among the S. Cerevisiae Bud Neck Ring Proteins Elm1p, Hsl1p and Cdc12p. Yeast 2003, 20, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Finnigan, G.C.; Duvalyan, A.; Liao, E.N.; Sargsyan, A.; Thorner, J. Detection of Protein-Protein Interactions at the Septin Collar in Saccharomyces Cerevisiae Using a Tripartite Split-GFP System. Molecular biology of the cell 2016, 27, 2708–2725. [Google Scholar] [CrossRef] [PubMed]
- Barral, Y.; Parra, M.; Bidlingmaier, S.; Snyder, M. Nim1-Related Kinases Coordinate Cell Cycle Progression with the Organization of the Peripheral Cytoskeleton in Yeast. Genes Dev 1999, 13, 176–187. [Google Scholar] [CrossRef] [PubMed]
- Breitkreutz, A.; Choi, H.; Sharom, J.R.; Boucher, L.; Neduva, V.; Larsen, B.; Lin, Z.Y.; Breitkreutz, B.J.; Stark, C.; Liu, G.; et al. A Global Protein Kinase and Phosphatase Interaction Network in Yeast. Science 2010, 328, 1043–1046. [Google Scholar] [CrossRef] [PubMed]
- Finnigan, G.C.; Sterling, S.M.; Duvalyan, A.; Liao, E.N.; Sargsyan, A.; Garcia, G.; Nogales, E.; Thorner, J. Coordinate Action of Distinct Sequence Elements Localizes Checkpoint Kinase Hsl1 to the Septin Collar at the Bud Neck in Saccharomyces Cerevisiae. Mol Biol Cell 2016, 27, 2213–2233. [Google Scholar] [CrossRef]
- Okuzaki, D.; Nojima, H. Kcc4 Associates with Septin Proteins of Saccharomyces Cerevisiae. FEBS Letters 2001, 489, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Bouquin, N.; Barral, Y.; Courbeyrette, R.; Blondel, M.; Snyder, M.; Mann, C. Regulation of Cytokinesis by the Elm1 Protein Kinase in Saccharomyces Cerevisiae. J Cell Sci 2000, 113 ( Pt 8) Pt 8, 1435–1445. [Google Scholar] [CrossRef]
- Marquardt, J.; Yao, L.-L.; Okada, H.; Svitkina, T.; Bi, E. The LKB1-like Kinase Elm1 Controls Septin Hourglass Assembly and Stability by Regulating Filament Pairing. Current Biology 2020, 30, 2386–2394. [Google Scholar] [CrossRef] [PubMed]
- Sreenivasan, A.; Kellogg, D. The Elm1 Kinase Functions in a Mitotic Signaling Network in Budding Yeast. Mol Cell Biol 1999, 19, 7983–7994. [Google Scholar] [CrossRef]
- Marquardt, J.; Chen, X.; Bi, E. Reciprocal Regulation by Elm1 and Gin4 Controls Septin Hourglass Assembly and Remodeling. Journal of Cell Biology 2024, 223, e202308143. [Google Scholar] [CrossRef] [PubMed]
- Szkotnicki, L.; Crutchley, J.M.; Zyla, T.R.; Bardes, E.S.; Lew, D.J. The Checkpoint Kinase Hsl1p Is Activated by Elm1p-Dependent Phosphorylation. Mol Biol Cell 2008, 19, 4675–4686. [Google Scholar] [CrossRef] [PubMed]
- Fraschini, R.; Bilotta, D.; Lucchini, G.; Piatti, S. Functional Characterization of Dma1 and Dma2, the Budding Yeast Homologues of Schizosaccharomyces Pombe Dma1 and Human Chfr. Mol Biol Cell 2004, 15, 3796–3810. [Google Scholar] [CrossRef] [PubMed]
- Merlini, L.; Fraschini, R.; Boettcher, B.; Barral, Y.; Lucchini, G.; Piatti, S. Budding Yeast Dma Proteins Control Septin Dynamics and the Spindle Position Checkpoint by Promoting the Recruitment of the Elm1 Kinase to the Bud Neck. PLoS Genet 2012, 8, e1002670. [Google Scholar] [CrossRef] [PubMed]
- Patasi, C.; Godocikova, J.; Michlikova, S.; Nie, Y.; Kacerikova, R.; Kvalova, K.; Raunser, S.; Farkasovsky, M. The Role of Bni5 in the Regulation of Septin Higher-Order Structure Formation. Biological chemistry 2015, 396, 1325–1337. [Google Scholar] [CrossRef] [PubMed]
- Lee, P.R.; Song, S.; Ro, H.S.; Park, C.J.; Lippincott, J.; Li, R.; Pringle, J.R.; De Virgilio, C.; Longtine, M.S.; Lee, K.S. Bni5p, a Septin-Interacting Protein, Is Required for Normal Septin Function and Cytokinesis in Saccharomyces Cerevisiae. Mol Cell Biol 2002, 22, 6906–6920. [Google Scholar] [CrossRef]
- Booth, E.A.; Sterling, S.M.; Dovala, D.; Nogales, E.; Thorner, J. Effects of Bni5 Binding on Septin Filament Organization. Journal of Molecular Biology 2016, 428, 4962–4980. [Google Scholar] [CrossRef] [PubMed]
- Finnigan, G.C.; Booth, E.A.; Duvalyan, A.; Liao, E.N.; Thorner, J. The Carboxy-Terminal Tails of Septins Cdc11 and Shs1 Recruit Myosin-II Binding Factor Bni5 to the Bud Neck in Saccharomyces Cerevisiae. Genetics 2015, 200, 843–862. [Google Scholar] [CrossRef] [PubMed]
- Touati, S.A.; Kataria, M.; Jones, A.W.; Snijders, A.P.; Uhlmann, F. Phosphoproteome Dynamics during Mitotic Exit in Budding Yeast. EMBO J 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Renz, C.; Oeljeklaus, S.; Grinhagens, S.; Warscheid, B.; Johnsson, N.; Gronemeyer, T. Identification of Cell Cycle Dependent Interaction Partners of the Septins by Quantitative Mass Spectrometry. PLOS ONE 2016, 21. [Google Scholar] [CrossRef] [PubMed]
- Swaney, D.L.; Beltrao, P.; Starita, L.; Guo, A.; Rush, J.; Fields, S.; Krogan, N.J.; Villen, J. Global Analysis of Phosphorylation and Ubiquitylation Cross-Talk in Protein Degradation. Nature methods 2013, 10, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, C.P.; Smolka, M.B.; Payne, S.H.; Bafna, V.; Eng, J.; Zhou, H. A Multidimensional Chromatography Technology for In-Depth Phosphoproteome Analysis. Molecular & cellular proteomics: MCP 2008, 7, 1389–1396. [Google Scholar] [CrossRef]
- Holt, L.J.; Tuch, B.B.; Villen, J.; Johnson, A.D.; Gygi, S.P.; Morgan, D.O. Global Analysis of Cdk1 Substrate Phosphorylation Sites Provides Insights into Evolution. Science 2009, 325, 1682–1686. [Google Scholar] [CrossRef] [PubMed]
- Smolka, M.B.; Albuquerque, C.P.; Chen, S. -h.; Zhou, H. Proteome-Wide Identification of in Vivo Targets of DNA Damage Checkpoint Kinases. Proceedings of the National Academy of Sciences 2007, 104, 10364–10369. [Google Scholar] [CrossRef] [PubMed]
- Bodenmiller, B.; Wanka, S.; Kraft, C.; Urban, J.; Campbell, D.; Pedrioli, P.G.; Gerrits, B.; Picotti, P.; Lam, H.; Vitek, O.; et al. Phosphoproteomic Analysis Reveals Interconnected System-Wide Responses to Perturbations of Kinases and Phosphatases in Yeast. Science signaling 2010, 3, rs4. [Google Scholar] [CrossRef] [PubMed]
- Soufi, B.; Kelstrup, C.D.; Stoehr, G.; Frohlich, F.; Walther, T.C.; Olsen, J.V. Global Analysis of the Yeast Osmotic Stress Response by Quantitative Proteomics. Molecular bioSystems 2009, 5, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Paulo, J.A.; Nusinow, D.P.; Huttlin, E.L.; Gygi, S.P. Investigation of Proteomic and Phosphoproteomic Responses to Signaling Network Perturbations Reveals Functional Pathway Organizations in Yeast. Cell Reports 2019, 29, 2092–2104. [Google Scholar] [CrossRef] [PubMed]
- Lanz, M.C.; Yugandhar, K.; Gupta, S.; Sanford, E.J.; Faça, V.M.; Vega, S.; Joiner, A.M.N.; Fromme, J.C.; Yu, H.; Smolka, M.B. In-Depth and 3-Dimensional Exploration of the Budding Yeast Phosphoproteome. EMBO reports 2021, 22, e51121. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.S.; Blobel, G. Cell Cycle-Regulated Attachment of the Ubiquitin-Related Protein SUMO to the Yeast Septins. J Cell Biol 1999, 147, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, L.; Lau, A.; Lambert, J.P.; Zhou, H.; Fong, Y.; Couture, J.F.; Figeys, D.; Baetz, K. Regulation of Septin Dynamics by the Saccharomyces Cerevisiae Lysine Acetyltransferase NuA4. PloS one 2011, 6, e25336. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, K.; Svitkina, T.; Bi, E. Critical Roles of a RhoGEF-Anillin Module in Septin Architectural Remodeling during Cytokinesis. Current Biology 2020, 30, 1477–1490. [Google Scholar] [CrossRef] [PubMed]
- Kang, P.J.; Hood-DeGrenier, J.K.; Park, H.O. Coupling of Septins to the Axial Landmark by Bud4 in Budding Yeast. Journal of cell science 2013, 126, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Guo, J.; Zhou, Y.T.; Gao, X.D. The Anillin-Related Region of Bud4 Is the Major Functional Determinant for Bud4’s Function in Septin Organization during Bud Growth and Axial Bud Site Selection in Budding Yeast. Eukaryotic cell 2015, 14, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Chant, J.; Herskowitz, I. Genetic Control of Bud Site Selection in Yeast by a Set of Gene Products That Constitute a Morphogenetic Pathway. Cell 1991, 65, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Gong, T.; Gao, X.-D. Identification of an Amphipathic Helix Important for the Formation of Ectopic Septin Spirals and Axial Budding in Yeast Axial Landmark Protein Bud3p. PLoS ONE 2011, 6, e16744. [Google Scholar] [CrossRef] [PubMed]
- Schrock, M.N.; Yan, Y.; Goeckel, M.E.; Basgall, E.M.; Lewis, I.C.; Leonard, K.G.; Halloran, M.; Finnigan, G.C. Characterization of Bud3 Domains Sufficient for Bud Neck Targeting in S. Cerevisiae. Access Microbiol 2022, 4, 000341. [Google Scholar] [CrossRef] [PubMed]
- Stegmeier, F.; Amon, A. Closing Mitosis: The Functions of the Cdc14 Phosphatase and Its Regulation. Annu Rev Genet 2004, 38, 203–232. [Google Scholar] [CrossRef] [PubMed]
- Jaspersen, S.L.; Morgan, D.O. Cdc14 Activates Cdc15 to Promote Mitotic Exit in Budding Yeast. Curr Biol 2000, 10, 615–618. [Google Scholar] [CrossRef] [PubMed]
- König, C.; Maekawa, H.; Schiebel, E. Mutual Regulation of Cyclin-Dependent Kinase and the Mitotic Exit Network. J Cell Biol 2010, 188, 351–368. [Google Scholar] [CrossRef] [PubMed]
- Visintin, R.; Craig, K.; Hwang, E.S.; Prinz, S.; Tyers, M.; Amon, A. The Phosphatase Cdc14 Triggers Mitotic Exit by Reversal of Cdk- Dependent Phosphorylation. Mol Cell 1998, 2, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Gray, C.H.; Good, V.M.; Tonks, N.K.; Barford, D. The Structure of the Cell Cycle Protein Cdc14 Reveals a Proline-Directed Protein Phosphatase. Embo J 2003, 22, 3524–3535. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.P.; Hall, H.; Chaparian, R.; Mara, M.; Mueller, A.; Hall, M.C.; Shannon, K.B. Dephosphorylation of Iqg1 by Cdc14 Regulates Cytokinesis in Budding Yeast. Molecular biology of the cell 2015, 26, 2913–2926. [Google Scholar] [CrossRef] [PubMed]
- Bloom, J.; Cristea, I.M.; Procko, A.L.; Lubkov, V.; Chait, B.T.; Snyder, M.; Cross, F.R. Global Analysis of Cdc14 Phosphatase Reveals Diverse Roles in Mitotic Processes *. Journal of Biological Chemistry 2011, 286, 5434–5445. [Google Scholar] [CrossRef]
- Palani, S.; Meitinger, F.; Boehm, M.E.; Lehmann, W.D.; Pereira, G. Cdc14-Dependent Dephosphorylation of Inn1 Contributes to Inn1–Cyk3 Complex Formation. Journal of Cell Science 2012, 125, 3091–3096. [Google Scholar] [CrossRef] [PubMed]
- Shou, W.; Seol, J.H.; Shevchenko, A.; Baskerville, C.; Moazed, D.; Chen, Z.W.; Jang, J.; Charbonneau, H.; Deshaies, R.J. Exit from Mitosis Is Triggered by Tem1-Dependent Release of the Protein Phosphatase Cdc14 from Nucleolar RENT Complex. Cell 1999, 97, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Kuilman, T.; Maiolica, A.; Godfrey, M.; Scheidel, N.; Aebersold, R.; Uhlmann, F. Identification of Cdk Targets That Control Cytokinesis. EMBO J 2015, 34, 81–96. [Google Scholar] [CrossRef] [PubMed]
- Varela Salgado, M.; Adriaans, I.E.; Touati, S.A.; Ibanes, S.; Lai-Kee-Him, J.; Ancelin, A.; Cipelletti, L.; Picas, L.; Piatti, S. Phosphorylation of the F-BAR Protein Hof1 Drives Septin Ring Splitting in Budding Yeast. Nat Commun 2024, 15, 1–17. [Google Scholar] [CrossRef]
- Meitinger, F.; Boehm, M.E.; Hofmann, A.; Hub, B.; Zentgraf, H.; Lehmann, W.D.; Pereira, G. Phosphorylation-Dependent Regulation of the F-BAR Protein Hof1 during Cytokinesis. Genes Dev 2011, 25, 875–888. [Google Scholar] [CrossRef] [PubMed]
- Meitinger, F.; Palani, S.; Hub, B.; Pereira, G. Dual Function of the NDR-Kinase Dbf2 in the Regulation of the F-BAR Protein Hof1 during Cytokinesis. Mol Biol Cell 2013, 24, 1290–1304. [Google Scholar] [CrossRef] [PubMed]
- Fankhauser, C.; Reymond, A.; Cerutti, L.; Utzig, S.; Hofmann, K.; Simanis, V. The S. Pombe Cdc15 Gene Is a Key Element in the Reorganization of F-Actin at Mitosis. Cell 1995, 82, 435–444. [Google Scholar] [CrossRef] [PubMed]
- Willet, A.H.; McDonald, N.A.; Bohnert, K.A.; Baird, M.A.; Allen, J.R.; Davidson, M.W.; Gould, K.L. The F-BAR Cdc15 Promotes Contractile Ring Formation through the Direct Recruitment of the Formin Cdc12. The Journal of cell biology 2015, 208, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Carnahan, R.H.; Gould, K.L. The PCH Family Protein, Cdc15p, Recruits Two F-Actin Nucleation Pathways to Coordinate Cytokinetic Actin Ring Formation in Schizosaccharomyces Pombe. The Journal of cell biology 2003, 162, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Graziano, B.R.; Yu, H.Y.; Alioto, S.L.; Eskin, J.A.; Ydenberg, C.A.; Waterman, D.P.; Garabedian, M.; Goode, B.L. The F-BAR Protein Hof1 Tunes Formin Activity to Sculpt Actin Cables during Polarized Growth. Molecular biology of the cell 2014, 25, 1730–1743. [Google Scholar] [CrossRef] [PubMed]
- Garabedian, M.V.; Wirshing, A.; Vakhrusheva, A.; Turegun, B.; Sokolova, O.S.; Goode, B.L. A Septin-Hof1 Scaffold at the Yeast Bud Neck Binds and Organizes Actin Cables. Molecular Biology of the Cell 2020, 31, 1988–2001. [Google Scholar] [CrossRef] [PubMed]
- Garabedian, M.V.; Stanishneva-Konovalova, T.; Lou, C.; Rands, T.J.; Pollard, L.W.; Sokolova, O.S.; Goode, B.L. Integrated Control of Formin-Mediated Actin Assembly by a Stationary Inhibitor and a Mobile Activator. Journal of Cell Biology 2018, 217, 3512–3530. [Google Scholar] [CrossRef] [PubMed]
- Bohnert, K.A.; Gould, K.L. Cytokinesis-Based Constraints on Polarized Cell Growth in Fission Yeast. PLOS Genetics 2012, 8, e1003004. [Google Scholar] [CrossRef] [PubMed]
- Vallen, E.A.; Caviston, J.; Bi, E. Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in Cytokinesis in Saccharomyces Cerevisiae. Mol Biol Cell 2000, 11, 593–611. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Schreiter, J.; Nishihama, R.; Wloka, C.; Bi, E. Targeting and Functional Mechanisms of the Cytokinesis-related F-BAR Protein Hof1 during the Cell Cycle. Molecular Biology of the Cell 2013, 24, 1305–1320. [Google Scholar] [CrossRef] [PubMed]
- Frost, A.; Unger, V.M.; De Camilli, P. The BAR Domain Superfamily: Membrane-Molding Macromolecules. Cell 2009, 137, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, S.; Toyooka, K.; Senju, Y. Subcellular Membrane Curvature Mediated by the BAR Domain Superfamily Proteins. Seminars in cell & developmental biology 2010, 21, 340–349. [Google Scholar] [CrossRef]
- Zhao, H.; Michelot, A.; Koskela, E.V.; Tkach, V.; Stamou, D.; Drubin, D.G.; Lappalainen, P. Membrane-Sculpting BAR Domains Generate Stable Lipid Microdomains. Cell reports 2013, 4, 1213–1223. [Google Scholar] [CrossRef] [PubMed]
- Picas, L.; Viaud, J.; Schauer, K.; Vanni, S.; Hnia, K.; Fraisier, V.; Roux, A.; Bassereau, P.; Gaits-Iacovoni, F.; Payrastre, B.; et al. BIN1/M-Amphiphysin2 Induces Clustering of Phosphoinositides to Recruit Its Downstream Partner Dynamin. Nat Commun 2014, 5, 5647. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




