Submitted:
30 July 2024
Posted:
31 July 2024
You are already at the latest version
Abstract
Keywords:
I. Introduction
II. Background
A. Anemia in Peru
B. ARIMA Models in Health Research
C. Parallel Computing in Data Analysis
III. Methodology
A. Data Collection and Preprocessing
- Insurance type
- Patient demographics
- Anemia diagnosis date
- Hemoglobin levels
- Follow-up dosage dates (1, 3, and 6 months)
- Recovery date
- Supplementation dates
- Treatment end date
- Healthcare facility information
- Geographic data (province, district)
B. ARIMA Modeling
- Stationarity testing using Augmented Dickey-Fuller test
- Model identification through ACF and PACF plots
- Parameter estimation
- Model diagnostics and validation
C. Parallel Computing Implementation
- Data partitioning based on geographic regions
- Distributed ARIMA model fitting across multiple cores
- Parallel processing of model diagnostics and forecasts
IV. Results
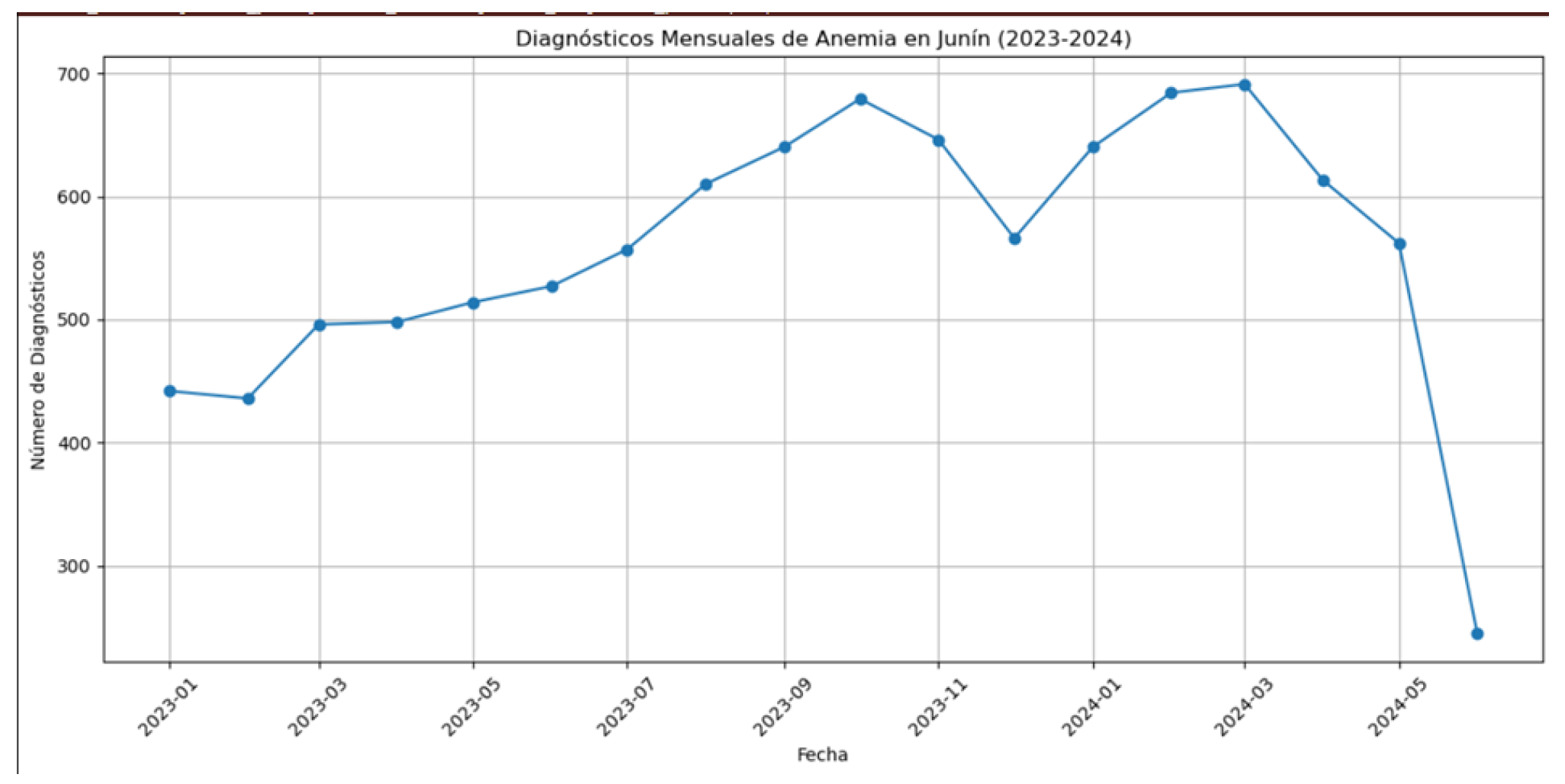
A. Anemia Prevalence Trends
B. ARIMA Model Performance
C. Forecasting Results
D. Parallel Computing Efficiency
V. Discussion
- Seasonal patterns in anemia diagnoses, with peaks typically occurring during winter months.
- Geographic variations in anemia prevalence, with certain districts showing persistently higher rates.
- A correlation between supplementation adherence and recovery rates, highlighting the importance of consistent treatment [13].
VI. Conclusions and Future Work
- ARIMA models are effective for short-term forecasting of anemia prevalence, aiding in timely decision-making and resource allocation.
- Parallel computing reduces processing time, enabling faster analysis and decision-making.
- The integration of these methods can inform public health strategies and optimize resource allocation for anemia prevention and treatment programs.
References
- World Health Organization, “Nutritional Anaemias: Tools for Effective Prevention and Control,” 2017.
- L. A. Celi et al., “Big Data in Healthcare: Prospects and Challenges,” BMJ Innovations, vol. 1, no. 1, pp. 9-16, 2015.
- G. E. P. Box, G. M. Jenkins, G. C. Reinsel, and G. M. Ljung, Time Series Analysis: Forecasting and Control, 5th ed. Wiley, 2015.
- Ministerio de Salud del Perú, “Informe de Anemia Infantil en Perú,” 2017.
- J. P. Aparco, “Determinants of Anemia in Peruvian Children,” Revista Peruana de Medicina Experimental y Salud Pública, vol. 33, no. 2, pp. 273-280, 2016.
- R. J. Hyndman and G. Athanasopoulos, Forecasting: Principles and Practice, 2nd ed. OTexts, 2018.
- Z. Ceylan, “Forecasting the COVID-19 Spread in Turkey Using ARIMA Models,” International Journal of Environmental Research and Public Health, vol. 17, no. 19, 2020.
- J. Dean and S. Ghemawat, “MapReduce: Simplified Data Processing on Large Clusters,” Communications of the ACM, vol. 51, no. 1, pp. 107-113, 2008.
- M. Zaharia et al., “Apache Spark: A Unified Engine for Big Data Processing,” Communications of the ACM, vol. 59, no. 11, pp. 56-65, 2016.
- W. McKinney, “Data Structures for Statistical Computing in Python,” in Proceedings of the 9th Python in Science Conference, 2010, pp. 51-56.
- R. Adhikari and R. K. Agrawal, “An Introductive Survey on Time Series Modeling and Forecasting,” arXiv preprint arXiv:1302.6613, 2013.
- I. Foster, Designing and Building Parallel Programs: Concepts and Tools for Parallel Software Engineering. Addison-Wesley, 1995.
- L. L. Iannotti et al., “Iron Supplementation in Early Childhood: Health Benefits and Risks,” American Journal of Clinical Nutrition, vol. 84, no. 6, pp. 1261-1270, 2006.
- Makridakis, E. Spiliotis, and V. Assimakopoulos, “Statistical and Machine Learning Forecasting Methods: Concerns and Ways Forward,” PLoS One, vol. 13, no. 3, 2018.
| Geographic Level | RMSE | MAE | MAPE |
|---|---|---|---|
| Regional | 0.15 | 0.12 | 5.2% |
| Provincial | 0.18 | 0.14 | 6.7% |
| District | 0.22 | 0.17 | 8.1% |
| Process | Serial (s) | Parallel (s) | Speedup |
|---|---|---|---|
| Data Preprocessing | 120 | 35 | 3.43x |
| Model Fitting | 450 | 85 | 5.29x |
| Forecasting | 180 | 40 | 4.50x |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




