1. Introduction
Infrastructure components for machine learning require mechanisms to manage and control data. This is due to the fact that artificial intelligence technologies provide reliable results, but require significant resources to operate. This applies to hardware, software, cloud systems and mechanisms of integration and configuration of the necessary software on cloud services. It is permissible to use ordinary personal computers to solve local problems of classification, segmentation, etc. Given the complexity of developing systems with elements of artificial intelligence, there was a need for MLOps - engineers. MLOps technology is the link between software developers and the engineering team.
The field of architecture development for automatic segmentation based on machine learning currently consists of numerous individual partial solutions. But there are practically no solutions for the full life cycle of the project. In addition, many solutions are customized and not adapted for running in cloud environments. The correct approach to infrastructure construction should provide for the possibility of “deployment” of the necessary environment on most cloud providers. The main components of the environment are the necessary libraries, models, and datasets for automatic segmentation.
Creating a dataset is a crucial step in developing any classification system and automatic data segmentation. Therefore, focusing attention on the development of a unified approach for forming the structure of image files is an urgent task, which will allow to increase in the number of images for processing. To standardize dataset formation processes, it was developed a dedicated software module for creating dataset rules. This module enables the modification of existing datasets to fit the required template.
To develop real projects, it is necessary to use cluster and cloud technologies. Therefore, the development of mechanisms for building an infrastructure for automatic segmentation of images based on U-net networks is an urgent task.
The main goal of this article is the formation of a mechanism for the development, training and supervision of neural network models for automatic segmentation of immunohistochemical images. The development of a unified approach to the formation of infrastructure will allow researchers to devote more time to the development of neural network architectures, and not to setting up the necessary environment.
The main contributions of the authors in this work can be summarized as follows:
The life cycle of the process for automatically segmenting biomedical images was developed using a combination of DevOps approaches and elements of machine learning. The developed life cycle will include the “Infrastructure as code” approach and a block for forming a dataset of original images and masks.
A software module was developed for forming requirements for the dataset for further automatic segmentation based on convolutional neural networks. The software module is implemented in the form of a web application that generates the result in JSON - a format for convenient use in any system.
The terraform file structure was developed for deploying the project on cloud platforms based on the IaC approach. Additionally, the project deployment process is described, taking into account the need to create a separate space for the dataset.
Continuous integration and delivery of code using CI/CD and creation of project code versions, which will allow automating the process of automatic segmentation of biomedical images, have been implemented.
Testing of the developed software module was carried out.
The article is organized as follows:
Section 2 presents the relevance of this research and describes the tasks addressed in this work;
Section 3 provides an analysis of existing solutions and a comparative analysis based on the most common criteria for solving the posed tasks;
Section 4 outlines the developed lifecycle of the automatic segmentation process;
Section 5 explains the principle of dataset formation for the automatic segmentation task;
Section 6 demonstrates the project infrastructure and examples of the proposed configuration files;
Section 7 presents a comparative analysis of the proposed approach; and the concluding section provides the conclusions of the article.
2. Statement of the Research Problem
The purpose of the article is to develop a mechanism for creating an environment for launching projects based on the U-net approach, which will include the following stages:
configuration of hardware parameters in the form of a script using the IaaC approach;
formation of rules for creating the necessary dataset structure for U-net networks;
creation of a pipeline for the implementation of automatic segmentation using machine learning libraries, models, and necessary components;
implementation of continuous integration and code delivery by means of CI/CD and creation of project code versions.
3. Analysis of Existing Solutions and Previous Studies
Most existing solutions for building neural network models are not free, as the learning process requires a large amount of hardware resources. A comparative analysis of systems for the use of artificial intelligence [
1] is given in the
Table 1.
Nyckel’s software system processes images and text and provides access to machine learning APIs. The Ximilar software system focuses on the development of systems with computer vision elements, has a set of existing models and allows you to develop your own. Roboflow specializes in computer vision. This system allows to customize the process of learning and training the model for your own needs. The Hasty software system offers a large number of libraries for searching objects in images and provides a convenient interface for configuring the learning process. The AWS Rekognition Custom Labels software system provides mechanisms for using elements of artificial intelligence in various tasks, in particular, finding objects in an image. This system has a large set of auxiliary resources in the form of video presentations. In [
2], the authors proposed a workflow for image classification using convolutional neural networks. The authors also offer a strategy and some 872 insights into choosing the optimal CNN hyperparameters. In the article [
3], the authors propose a workflow of acquisition of biomedical image data, analysis, storage, processing automatic diagnosis of biomedical images. In the article [
4], the authors present a workflow for the classification of 5 types of white blood cells based on a convolutional neural network for real-time image sorting. In work [
5], the authors presented a tool for processing biomedical images based on elements of machine learning. The elements of the developed tool include segmentation and object detection. The disadvantages of the work are that the authors do not detail the pipeline and elements of continuous integration of the code. The classification of histological images, divided into 4 classes, is given in [
6]. The authors provide a list of the main elements of the image processing process and tools for building classification architectures. Approaches to the classification of images of breast cancer are given in works [
7,
8,
9]. The works also consider the use of the Spark framework for working with Big Data and for speeding up image processing. In [
10], a hybrid Computer-Aided Diagnosis (CAD) model based on Digital X-ray Mammograms is proposed. Features of the development and use of convolutional neural network models for processing biomedical images are given in works [
11,
12,
13,
14,
15]. The analysis of the main functions of MLOps is considered in works [
16,
17]. End-to-end MLOps architecture and workflow are given in [
18]. The main tools used by the MLOps engineer are discussed in [
19,
20]. MLOps analysis of biomedical image processing approaches is given in the article [
21].
4. General Approaches
When developing a pipeline for Machine Learning - programs, you need to take into account the available resources (hardware and cloud). In the case of training models for automatic image segmentation, it is necessary to have sufficient computing resources. When creating a pipeline, it is necessary to create a list of processes that are used in the work. For example, in order to implement automatic segmentation, it is necessary to provide for the process of data preparation, formation of original images and corresponding masks, operation of Unet - network, selection of hyperparameters such as the number of epochs, batch size, etc. An equally important process is the visualization of the obtained results for further evaluation. The organization of teamwork requires a careful selection of tools for pipeline implementation. The main tools are containerization tools, approaches to implementing the infrastructure as code mechanism, CI/CD tools, such as Github CI/CD, etc. For automatic segmentation, need to consider frameworks for working with data and Unet networks.
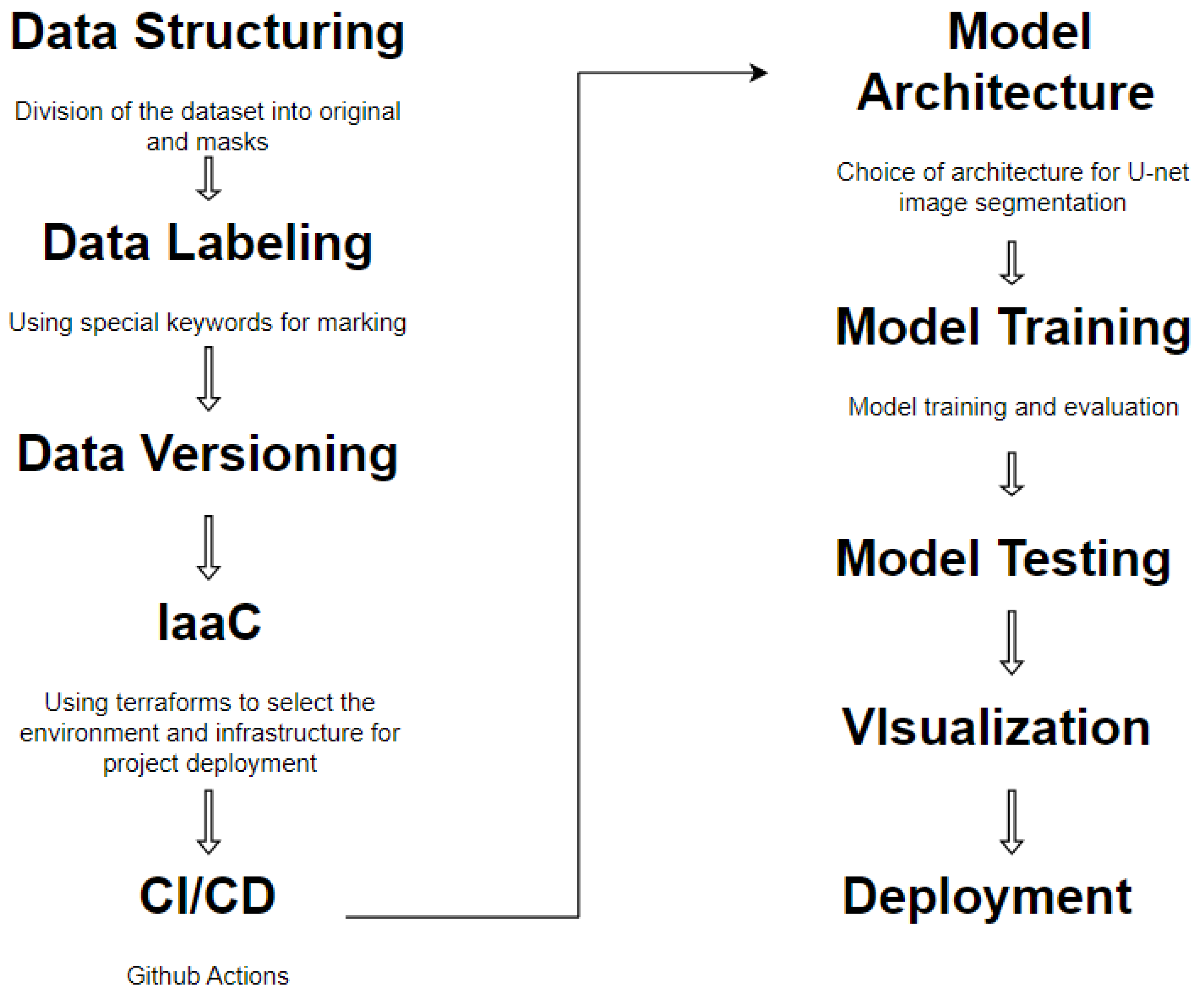
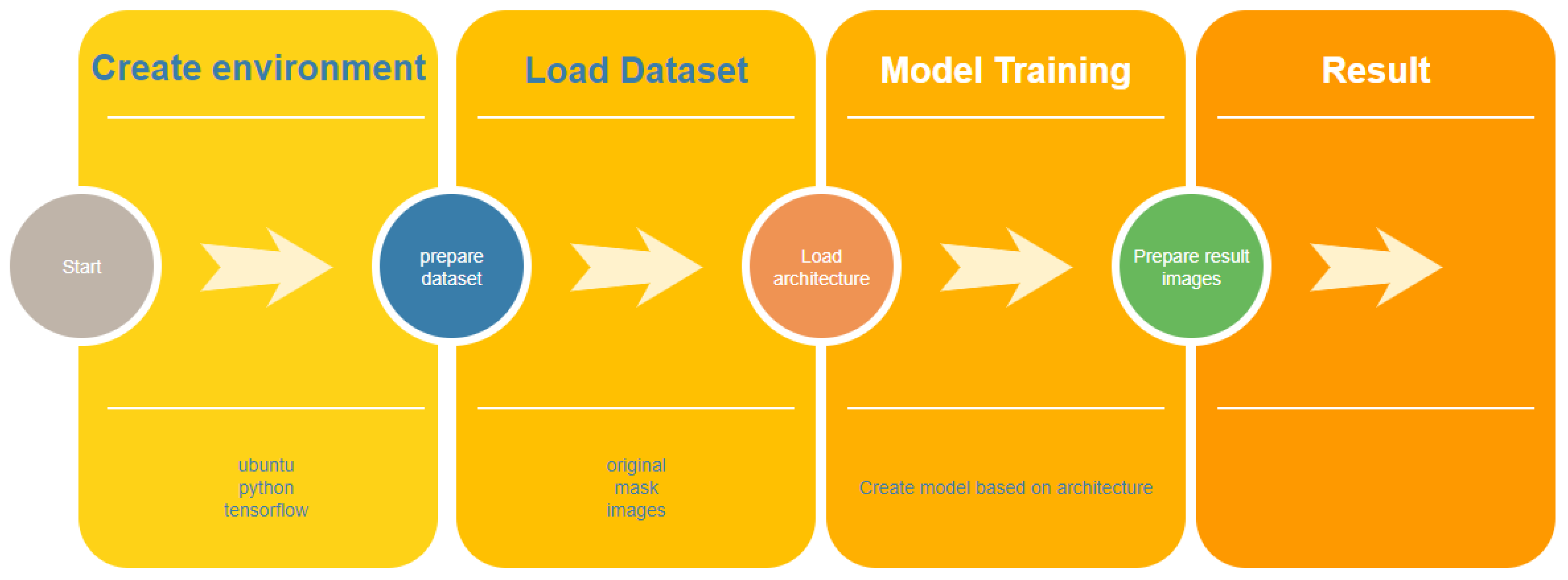
The life cycle of automatic image segmentation is shown in
Figure 1.
Formation of a dataset is one of the most important stages when working with big data. The main task in the formation of the dataset is the distribution of the sample into test and training in accordance with the rules for the formation of directories.
The “Data labeling” stage involves the process of labeling the file names with certain keywords, for example “_mask”. Taking into account the features of automatic segmentation, it is necessary to divide the sample into original images and mask images.
In most cases, the tasks of classification and segmentation of images in large volumes require special hardware: processor, RAM, GPU, etc. To run the functionality of the program, it is also necessary to create additional frameworks, libraries, etc. Therefore, using a regular PC for this type of task is impractical. Manual configuration of a hardware environment or cloud system takes a lot of time and requires the necessary knowledge.
The mechanism of continuous integration and continuous code delivery is a convenient mechanism for updating software code versions on cloud systems, which allows for maximum automation of the process of code delivery and deployment on the server.
The stage of development of the convolutional network architecture of the U-net type is important, which forms the rules of the neural network. The process of selecting parameters for U-net training is one of the longest.
The neural network training process is the most time-consuming and requires a lot of resources, including CPU time, RAM, etc.
After all the analyzed stages, there is the stage of introducing the final neural network in production to work with the test sample and new images.
5. Materials & Methods
5.1. Formation of the Dataset
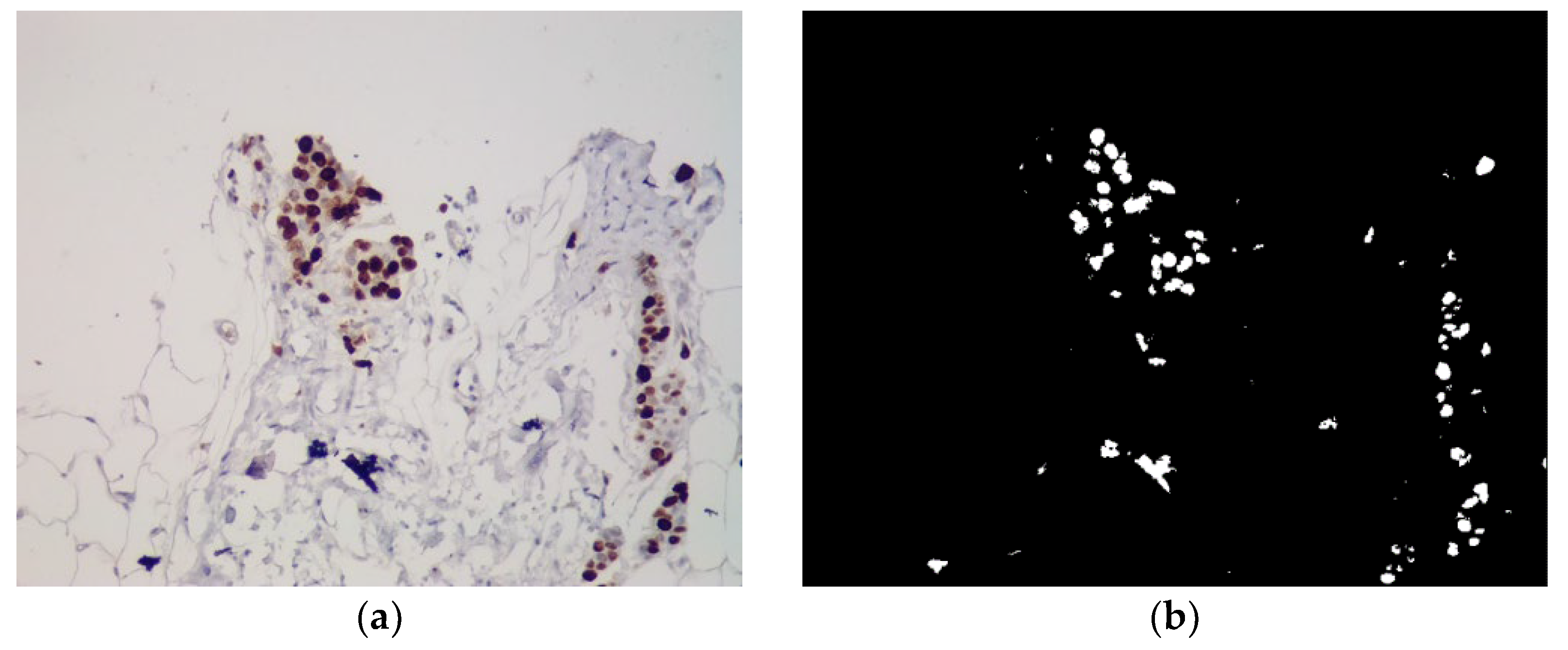
Machine learning requires the availability of both software tools for the implementation of a specific task and the availability of a prepared dataset. Dataset preparation depends on the task at hand. For example, for the task of classification, the sample is usually divided into training and test in the form of separate directories in the file system. For the task of implementing automatic segmentation of images based on U-net networks, it is necessary to take into account such an additional factor as “images - masks”. Currently, there are a large number of datasets in open access, but their files are formed in a different manner, which implies the need for their additional processing. An example of immunohistochemical images for automatic segmentation is shown in
Figure 2.
The criteria for forming a dataset for the task of automatic segmentation are as follows:
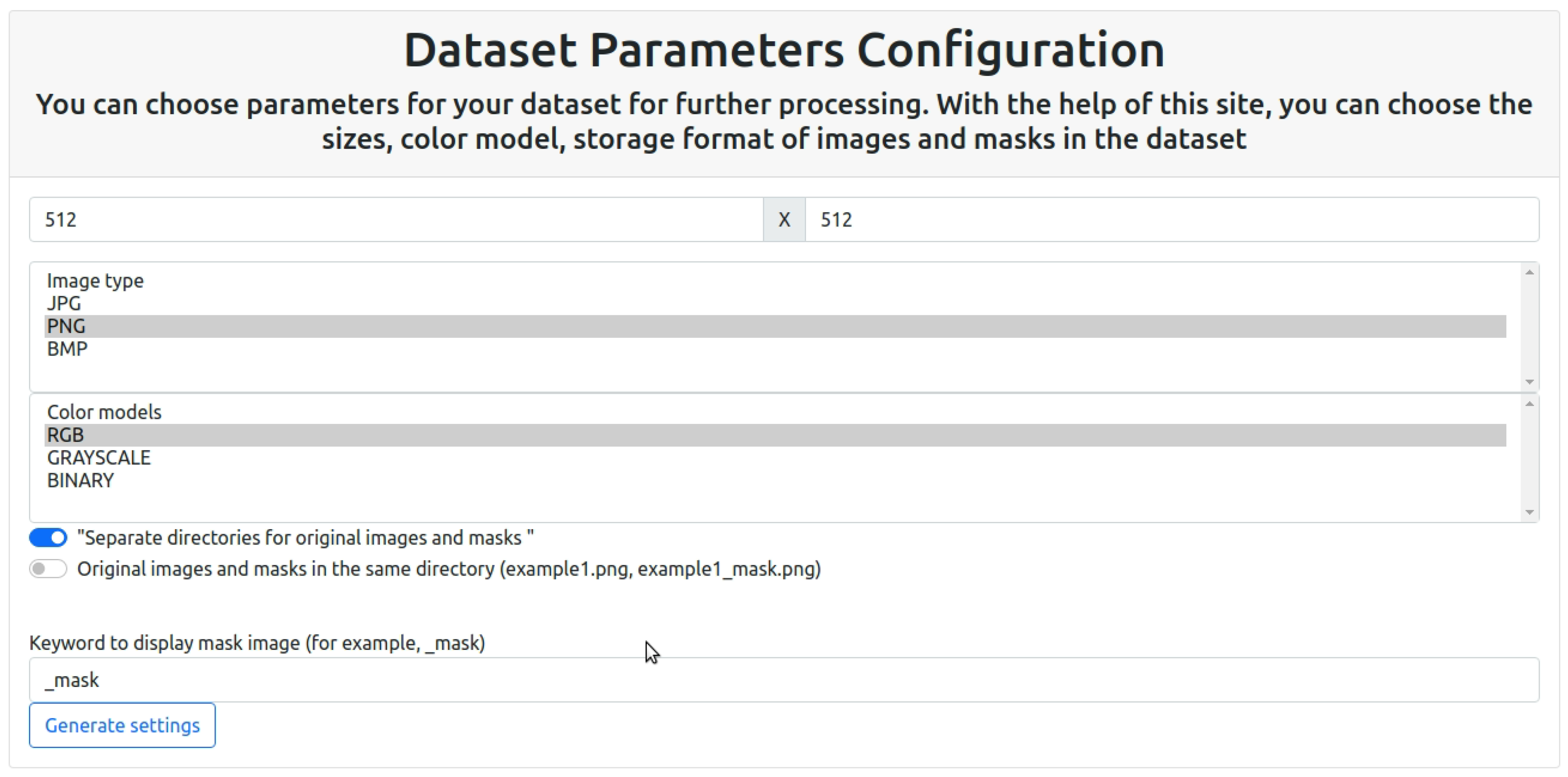
In order to unify the dataset, in this work, a separate software module was developed for the formation of dataset rules, which will allow reworking existing datasets according to the required template. The formation of a unified template will speed up the learning process and expand the training sample, which will improve the accuracy and quality of the automatic segmentation software module. The software module’s graphic interface for creating dataset requirements is displayed in
Figure 3.
A feature of this software module is the ability to set the color model of images of training and test samples. The color model allows you to store information about the image, as well as highlight the objects under study and the background. Detailed color information helps machine learning algorithms more accurately recognize and distinguish different micro-objects in images.
The aspect ratio of the input image is an important aspect that affects the quality of data processing, classification, and segmentation. Uniform size improves quality and facilitates the learning process. Images that are too small can lose detail, while images that are too large can be computationally expensive. Another important factor is that different neural network architectures work with predefined image sizes. Therefore, the ability to select parameters for the dataset greatly simplifies the learning process. Image size can affect the distribution of classes and proportions of objects in the image. For example, when reducing the size, the proportions of objects can change, this can affect the quality of segmentation.
The result the dataset parameters formation is a JSON file (
Figure 4), which can be further used in the program module, which will allow the formation of the necessary directory structure.
This is only a set of parameters for the dataset, and image processing is directly assigned to a separate software module.
The purpose of data unification is to improve the quality of data, which allows to reduce the number of errors. A unified structure simplifies data processing processes such as cleaning, normalization and transformation. A critical advantage of unified data is the reproducibility of research results. This is especially important at the stage of development of any system. Standardized data is easier to scale and adapt to new tasks and requirements. The unification of the structure of datasets is a key factor for increasing the efficiency and reliability of machine learning processes.
5.2. Project Infrastructure
Artificial intelligence allows you to solve many problems and significantly improves the quality of computational experiments. But the price of this is the need for a large number of resources and their diversity. The most important elements to pay attention to when conducting computational experiments are:
Manually creating a set of instructions for deploying the required computing environment is a deprecated mechanism.
A modern approach is to use the Infrastructure as Code mechanism, that is, to describe the necessary parameters and dependencies in the form of code for further deployment on the cloud. This approach and the Terraform tool are also used in our development. An example of the code part of the main file main.tf for deploying the digitalocean cloud environment is shown on
Figure 5.
In this case, the dataset set of biomedical images, in particular immunohistochemical, is placed on the Zenodo service [
29]. However, the use of this particular resource is not a mandatory requirement, and it is possible to use any other resources.
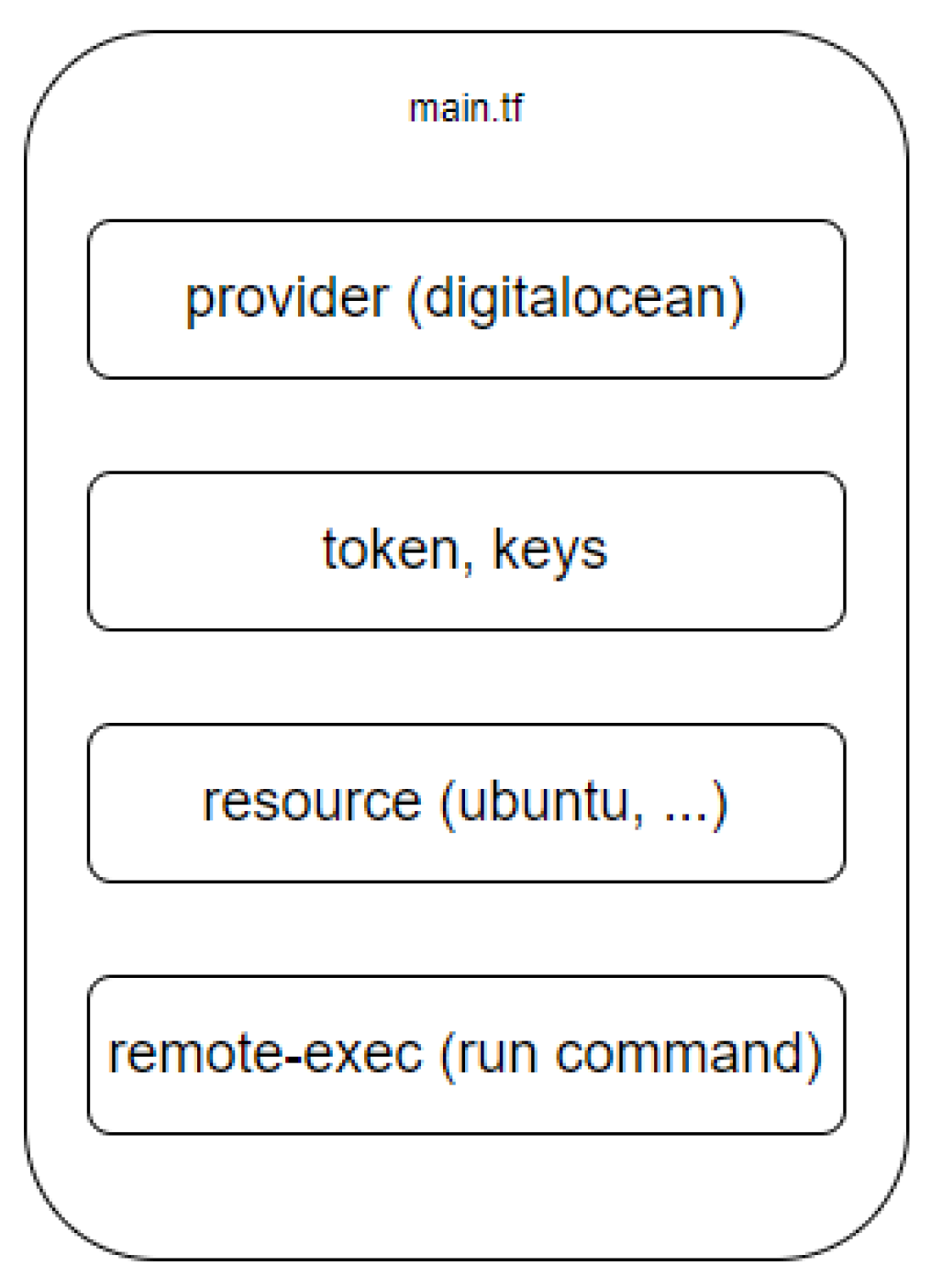
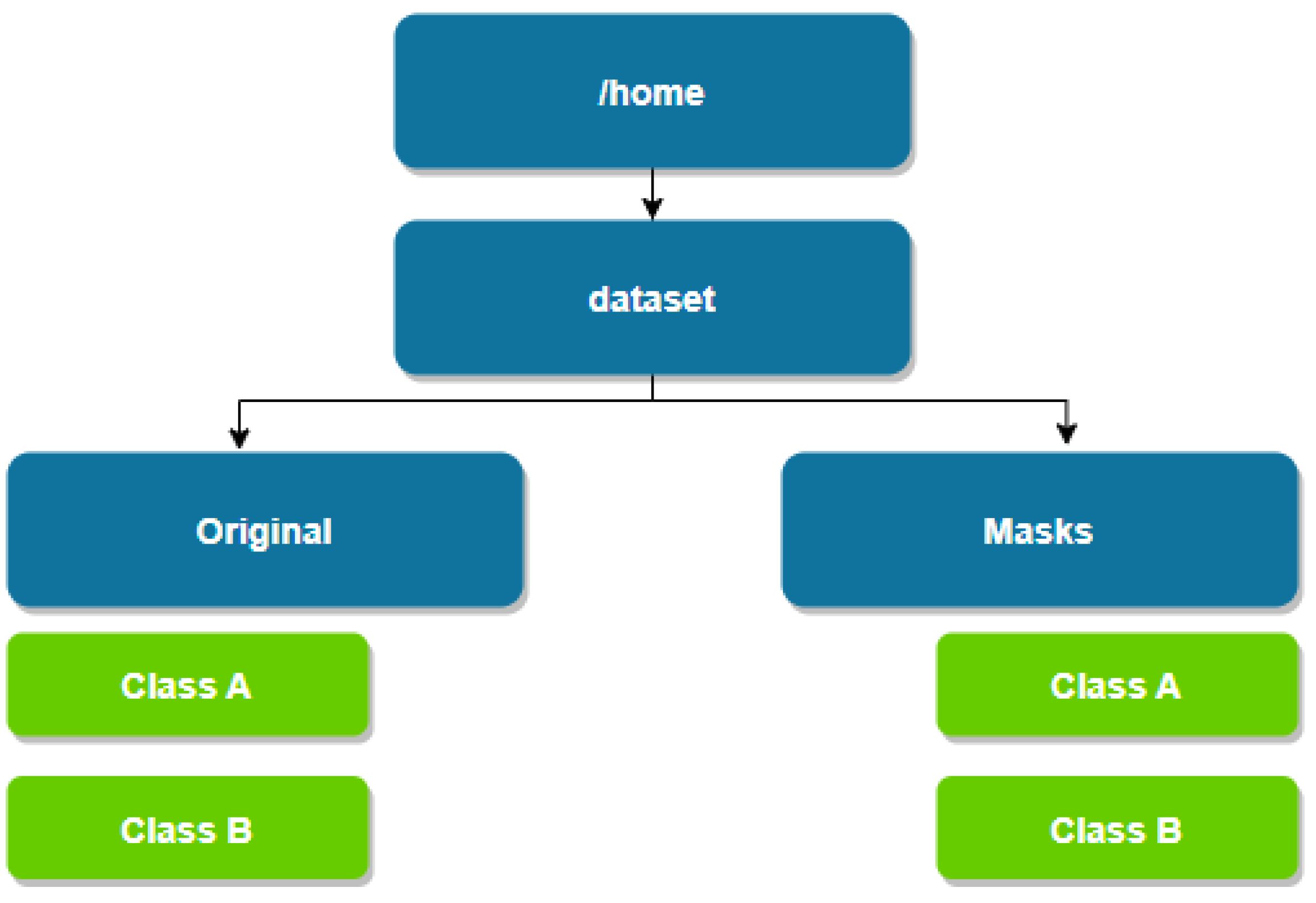
The distribution of the main.tf components file is shown in the
Figure 6.
In the “provider” section, the provider is indicated, in our case it is digitalocean. Providers in Terraform act as abstractions for various cloud platforms (e.g., AWS, Google Cloud, Azure) as well as for other services (e.g., GitHub, Kubernetes). It is also possible to work with several providers or with different accounts of the same provider.
The “token, keys” section is responsible for storing tokens and ssh keys for connecting to the cloud. Typically, this section is used to provide the credentials and authentication required to access various cloud services and providers.
The “resource” section is responsible for storing information about the operating system on which the project will be hosted. Resources can be diverse, including virtual machines, databases, network components, user accounts. Description of infrastructure as code (IaC) makes it easy to manage configurations. Terraform automatically calculates the dependency graph and ensures that resources are created and deleted in the correct order.
The “remote-exec” section stores a set of commands that should be executed automatically when a droplet is created: downloading a dataset, software code, individual programs, etc. The remote-exec section of Terraform configurations is used to execute commands on remote machines after the resource has been created. It is part of the provisioner block, which allows you to perform actions on resources during their creation, update or deletion.
The project deployment process for automatic segmentation is shown in
Figure 7.
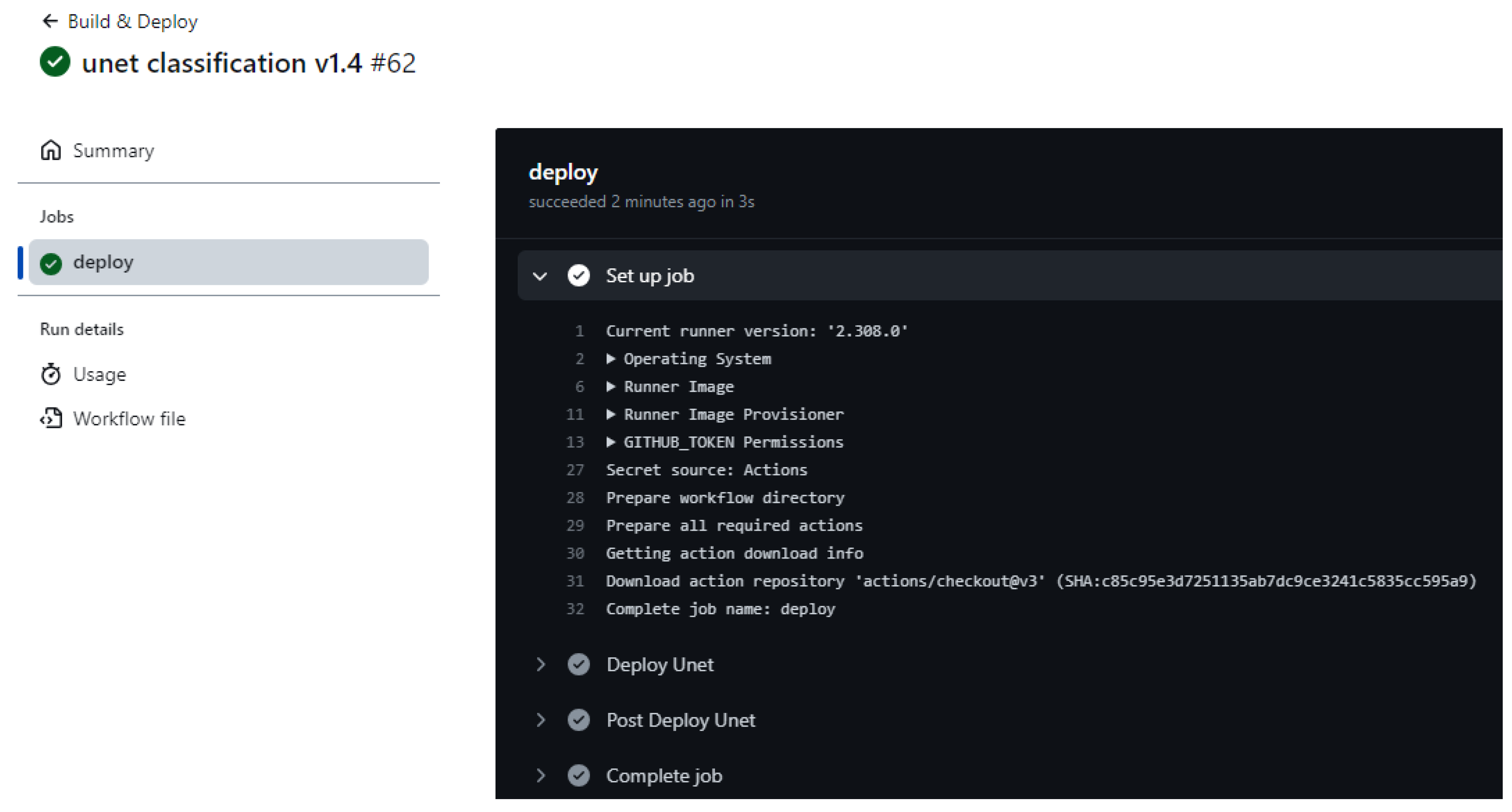
The CI/CD mechanism ensures continuous code delivery and continuous code integration on the server. An example of a GitHub action is shown in
Figure 8. Mechanisms for implementing CI are the code repository, and CI is the server. The main concepts of CI are frequent integrations, automated tests, project assembly. The primary goal of CI is to ensure continuous integration of developers’ code into a shared repository with automated testing at every step. The purpose of CD is to prepare code for deployment to a production environment after passing automated tests, often with manual approval.
GitHub Actions is an automation tool built into GitHub that allows you to configure, create, and execute various workflows directly from your repository. Workflows are YAML files located in the .github/workflows directory.
This mechanism is necessary in order not to transfer the new version of the project code manually, but to do it automatically. After creating a new commit in the main branch, the project code is delivered to the cloud.
6. Results
Roboflow is characterized by a wider functionality for the development and formation of a dataset in comparison with analogues. Hasty.ai has a wide range of functionality, including segmentation and labeling. AWS Rekognition Custom Labels uses AWS’s own Lambda tool for machine learning.
Comparative analysis of the developed system with advantages and disadvantages among known analogues (
Table 1) is given in
Table 2.
As a result of running the software module, the dataset’s directory structure is initially generated (see
Figure 9).
The results of image segmentation are shown in
Figure 10.
Masks serve as labels for model training. The network learns to compare its predictions with the masks and adjust the weights to reduce the difference between the predictions and the actual labels. Image - masks can be formed in the following ways: manual annotation (experts manually annotate the image, highlighting the contours of objects), semi-automatic methods, automatic methods (using segmentation algorithms to automatically create masks).
U-Net is able to accurately segment objects of various shapes and sizes thanks to the use of masks, which allows taking into account complex contours and boundaries of objects. The presence of masks allows you to use methods of automatic selection of model parameters, such as cross-validation, to achieve better segmentation accuracy.
To obtain information about the nuclei of the studied cells, threshold segmentation is additionally used to create a binary image. After that, the quantitative and qualitative characteristics of micro-objects can be determined for further analysis.
Automating the processes of deploying models into the production environment ensures fast and seamless integration of new or updated models. MLOps lets you build and deploy machine learning models faster, more efficiently, and more reliably. This is achieved by automating routine processes, ensuring continuous integration and deployment, monitoring model performance, supporting scalability and improving collaboration between teams.
7. Conclusions
At present, the number of software systems using artificial intelligence has sharply increased. The field of biomedical research actively employs neural networks for tasks such as classification and segmentation. However, the process of setting up the development environment is complex and labor-intensive, requiring the technical expertise of DevOps engineers.
The approach developed in the article helps to quickly organize the necessary environment for implementing programs for automatic segmentation of immunohistochemical images using modern libraries. This work presents the development of a lifecycle process for the automatic segmentation of biomedical images. The developed lifecycle is based on the “Infrastructure as Code” approach. A distinctive feature of this approach is the block for creating a dataset of original images and masks. The lifecycle for implementing automatic segmentation combines DevOps approaches with elements of machine learning.
A web application module has been developed to define dataset requirements, generating results in JSON format for convenient use in any systems. A “lightweight” configuration file has also been created to launch the infrastructure based on Terraform technology. The advantage of the developed system is that it is oriented towards the process of automatic segmentation using U-net. At the same time, the image processing and dataset formation stages for segmentation tasks clearly distinguish this system from others.
Thus, the developed mechanisms allow for the acceleration, standardization, and optimization of the software setup process for implementing automatic image segmentation using machine learning elements. The use of such a unified approach improves the quality of the development process and enables the reproducibility of experiments on various cloud platforms.
Author Contributions
Conceptualization, O.P.; methodology, O.P. and O.B.; software, O.P.; validation, Y.B.; formal analysis, O.P.; investigation, O.P.; resources, G.M.; data curation, G.M.; writing—original draft preparation, M.B.; writing—review and editing, O.B.; visualization, Y.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The datasets used in this study are available here: Ref. [
29].
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Computer Vision SaaS Landscape: Comparison of the Top 9 Players. Nyckel. Available online: https://www.nyckel.com/blog/computer-vision-platforms/ (accessed on 25 July 2024).
- Lunga, D.; Yang, H.L.; Reith, A.; Weaver, J.; Yuan, J.; Bhaduri, B. Domain-Adapted Convolutional Networks for Satellite Image Classification: A Large-Scale Interactive Learning Workflow. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 962–977. [Google Scholar] [CrossRef]
- Kouanou, A.T.; Tchiotsop, D.; Kengne, R.; Zephirin, D.T.; Armele, N.M.A.; Tchinda, R. An Optimal Big Data Workflow for Biomedical Image Analysis. Inform. Med. Unlocked 2018, 11, 68–74. [Google Scholar] [CrossRef]
- Zordan, M.; Chiang, S.-H.; Tang, H.; Huang, A.; Liu, M.-C. Cellular Image Classification Workflow for Real-Time Image Based Sort Decisions. In Imaging, Manipulation, and Analysis of Biomolecules, Cells, and Tissues XX.; SPIE, 2022; Vol. 11964, p 119640F. [CrossRef]
- Berg, S.; Kutra, D.; Kroeger, T.; Straehle, C.N.; Kausler, B.X.; Haubold, C.; Schiegg, M.; Ales, J.; Beier, T.; Rudy, M.; others. Ilastik: Interactive Machine Learning for (Bio) Image Analysis. Nat. Methods 2019, 16, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Vesal, S.; Ravikumar, N.; Davari, A.; Ellmann, S.; Maier, A. Classification of Breast Cancer Histology Images Using Transfer Learning. Proceedings of the Image Analysis and Recognition: 15th International Conference, ICIAR 2018, PoVoa de Varzim, Portugal, June 27-29; Springer International Publishing, 2018; pp 812–819. [CrossRef]
- Tchapga, C.T.; Mih, T.A.; Kouanou, A.T.; Fonzin, T.F.; Fogang, P.K.; Mezatio, B.A.; Tchiotsop, D. Biomedical Image Classification in a Big Data Architecture Using Machine Learning Algorithms. J. Healthc. Eng. 2021, 2021, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Wu, M.; Gopukumar, D.; Zhao, Y. Big Data Application in Biomedical Research and Health Care: A Literature Review. Biomed. Inform. Insights 2016, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Oussous, A.; Benjelloun, F.-Z.; Ait Lahcen, A.; Belfkih, S. Big Data Technologies: A Survey. J. King Saud Univ. 2018, 30, 431–448. [Google Scholar] [CrossRef]
- Al-Tam, R.M.; Al-Hejri, A.M.; Narangale, S.M.; Samee, N.A.; Mahmoud, N.F.; Al-masni, M.A.; Al-antari, M.A. A Hybrid Workflow of Residual Convolutional Transformer Encoder for Breast Cancer Classification Using Digital X-Ray Mammograms. Biomedicines 2022, 10, 2971. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Ren, F.; Wang, Z.; Wang, L.; Zhang, T.; Liu, Y.; Rao, X.; Zheng, C.; Zhang, F. Breast Cancer Histopathological Image Classification Using a Hybrid Deep Neural Network. Methods 2020, 173, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Berezsky, O.; Pitsun, O.; Melnyk, G.; Datsko, T.; Izonin, I.; Derysh, B. An Approach toward Automatic Specifics Diagnosis of Breast Cancer Based on an Immunohistochemical Image. J. Imaging 2023, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Ghahremani, P.; Li, Y.; Kaufman, A.; Vanguri, R.; Greenwald, N.; Angelo, M.; Hollmann, T.J.; Nadeem, S. Deep Learning-Inferred Multiplex ImmunoFluorescence for Immunohistochemical Image Quantification. Nat. Mach. Intell. 2022, 4, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Izonin, I.; Tkachenko, R.; Yemets, K.; Havryliuk, M. An Interpretable Ensemble Structure with a Non-Iterative Training Algorithm to Improve the Predictive Accuracy of Healthcare Data Analysis. Sci Rep 2024, 14, 12947. [Google Scholar] [CrossRef] [PubMed]
- Berezsky, O.; Pitsun, O.; Liashchynskyi, P.; Derysh, B.; Batryn, N. Computational Intelligence in Medicine. In Lecture Notes in Data Engineering, Computational Intelligence, and Decision Making; Babichev, S., Lytvynenko, V., Eds.; Springer International Publishing: Cham, 2023; pp 488–510. [CrossRef]
- Alla, S.; Adari, S.K. What Is MLOps? In Beginning MLOps with MLFlow; Apress, Berkeley, CA: Berkeley, CA, 2021; pp 79–124. [CrossRef]
- Mäkinen, S.; Skogström, H.; Laaksonen, E.; Mikkonen, T. Who Needs MLOps: What Data Scientists Seek to Accomplish and How Can MLOps Help? Proceedings of the 2021 IEEE/ACM 1st Workshop on AI Engineering-Software Engineering for AI (WAIN), 22-30 May 2021; IEEE, 2021; pp 109–112. [CrossRef]
- Kreuzberger, D.; Kühl, N.; Hirschl, S. Machine Learning Operations (MLOps): Overview, Definition, and Architecture. IEEE Access 2023, 11, 31866–31879. [Google Scholar] [CrossRef]
- Symeonidis, G.; Nerantzis, E.; Kazakis, A.; Papakostas, G.A. MLOps - Definitions, Tools and Challenges. Proceedings of the 2022 IEEE 12th Annual Computing and Communication Workshop and Conference (CCWC), 26th-29th January 2022; IEEE, 2022; pp 0453–0460. [CrossRef]
- Treveil, M.; Omont, N.; Stenac, C.; Lefevre, K.; Phan, D.; Zentici, J.; Lavoillotte, A.; Miyazaki, M.; Heidmann, L. Introducing MLOps; O’Reilly Media, 2020.
- Pitsun, O.; Berezsky, O.; Melnyk, G. ; others. Application Of MLOps Practices For Biomedical Image Classification. In CEUR Workshop Proceedings; 2022; Vol. 3302, pp 69–77.
- Classify anything using Nyckel’s classification API. Available online: https://www.nyckel.com/ (accessed on 25 July 2024).
- Image Recognition & Visual Search API For Business – Ximiliar. Available online: https://www.ximilar.com/ (accessed on 25 July 2024).
- Roboflow: Computer vision tools for developers and enterprises. Available online: https://roboflow.com/ (accessed on 25 July 2024).
- Hasty. A single application for all your vision AI needs. Available online: https://hasty.cloudfactory.com/ (accessed on 25 July 2024).
- Levity. Available online: https://levity.ai/ (accessed on 25 July 2024).
- Google Cloud Vertex, AI. Available online: https://cloud.google.com/vertex-ai#section-8 (accessed on 25 July 2024).
- Amazon Rekognition Custom Labels. Available online: https://aws.amazon.com/rekognition/custom-labels-features/ (accessed on 25 July 2024).
- Berezsky, O.; Datsko, T.; Melnyk, G. Cytological and Histological Images of Breast Cancer, 2023. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).