1. Introduction
Genetic diversity is the presence of different forms of genes found in all ecosystems [
1,
2]. This means that regardless of the type of environment in which specific species live, their population can exhibit variable genetic traits [
3]. The essence of this diversity is that most of the variable gene forms are inherited from generation to generation [
4]. The impact of genetic diversity directly affects the entire population [
5,
6,
7]. It is crucial for the process of selection – a process in which some genotype variants have an advantage over others. Selection is an important mechanism acting on animal and plant populations [
8,
9,
10,
11]. This effect can be observed both at the population level and at the individual level. The main factor driving selection is the availability of resources in a given environment [
12,
13]. Naturally, if only limited environmental resources are available, the only way to survive and thrive is to gain an advantage through a genotype is greater suitability to the environment [
14,
15,
16].
In the case of plants and animals, genetic diversity has a crucial impact on selection [
17]. In the process of natural selection, the force of choice operates, where unfavorable traits are eliminated, and an individual with the most advantageous traits for a given environment has a greater chance of survival [
18,
19]. This natural mechanism favors variants that are best adapted to living in a particular environment [
20,
21].
As a result of genetic diversity, the selection process begins with the recurrent genetic components present in the population [
22,
23]. Over time, these become important factors driving the diverse selection process [
24]. Existing adaptive capabilities, including genetic traits, determine a population's ability to survive in challenging conditions and provide an advantage based on genetic components [
25,
26]. Consequently, a diverse genotype has a greater chance of survival and the ability to cope with environmental conditions through reproduction, leading to the emergence of new generations from this genotype [
25]. Another factor influencing genotype selection is polymorphism, the phenomenon of genetic discontinuity manifested by the presence of different forms within a single species that differ functionally or structurally [
27,
28,
29]. Polymorphism involves the formation of hierarchies and the division of functions within a population. It can occur at the phenotypic level or at the genetic or molecular level [
30,
31]. When the frequency of two (or more) variations in a given population is too high to be caused by mutation, the population is considered polymorphic.
In general, greater genetic diversity in a population may result in a higher chance of gaining an advantage in genotype selection [
30,
32]. In reality, genetic diversity arising from the combination of environmental conditions and the influence of biological factors is crucial for the adaptation of a population to new conditions [
33,
34]. A high level of genetic diversity will help minimize the effects of changing environmental conditions, consequently increasing the chances of survival for an individual or population [
2,
35].
The results of genetic similarity (genetic variability) have great significance in making selection decisions, which is related to the choice of the method for calculating this characteristic [
2]. The method of genetic similarity is used in genetics and zoology to determine the degree of relatedness between individuals [
36,
37,
38]. It involves comparing the levels of concordance in the genome of a given species, or gene or DNA sequences, to determine the phylogenetic distance between individuals. This method is commonly used in studies of human genealogy, species phylogeny, and research on evolutionary mechanisms and the tracing of species origins [
39,
40]. The genetic similarity method contributes to evolutionary studies on the origins of species, their lineage, and migrations [
41,
42]. Along with other methods, it is used to assess the impact of environmental changes and biological processes on biodiversity. Due to the potentially multi-level analysis of all DNA sequences, this method is particularly useful in zoology and molecular biology.
Different measures of calculating genetic similarity represent a specific type of statistic that compares genetic data between individuals [
43,
44,
45]. Each measure has its advantages and disadvantages and reflects a particular methodology. The commonly encountered measures of genetic similarity can be divided into four main categories: classical, contemporary, dendrographic, and index-based. Classical measures include standard units, mean components, and the inbreeding index [
46]. The first is a measure of identity, while the other two reflect the genetic composition relative to a given subset or population. Contemporary measures of genetic similarity, such as jump length and interdependence level, reflect the relationship between individual data and general data [
47]. These measures identify tendencies to occur in groups and calculate the likelihood of specific objects appearing within a population. These measures are often used, particularly in studies of genetic authenticity and microorganism identity. Dendrographic measures are very similar to classical measures but reflect similarities and differences at the genetic level [
48,
49]. Dendrographic measures help in understanding genetic relationships between populations and other groups or individuals [
50]. Index-based measures are multidisciplinary measures that examine data structure at the population level [
51]. They utilize elements of previous measures, estimating levels of similarities and differences within a population. Among other things, they estimate the ratio of individuals sharing a common ancestor or provide a detailed view of kinship and mismatch levels within a population.
The aim of the study was to compare six statistical measures (Euclidean, Jaccard, Kulczyński, Sokal and Michener, Nei, and Rogers) to assess the genetic similarity of 21 rapeseed objects (ten interspecific Brassicaceae hybrids of F2 generation, and 11 of their parental forms) based on observations of 146 alleles obtained using 21 ISSR molecular markers.
3. Results
The analyzed material was examined using 21 primers. The number of amplification products for individual primers ranged from 4 (for UBC889, ISSR856, ISSR857, and ISSR890) to 11 (for mstg13, UBC835, and UBC891). In total, 146 markers were obtained.
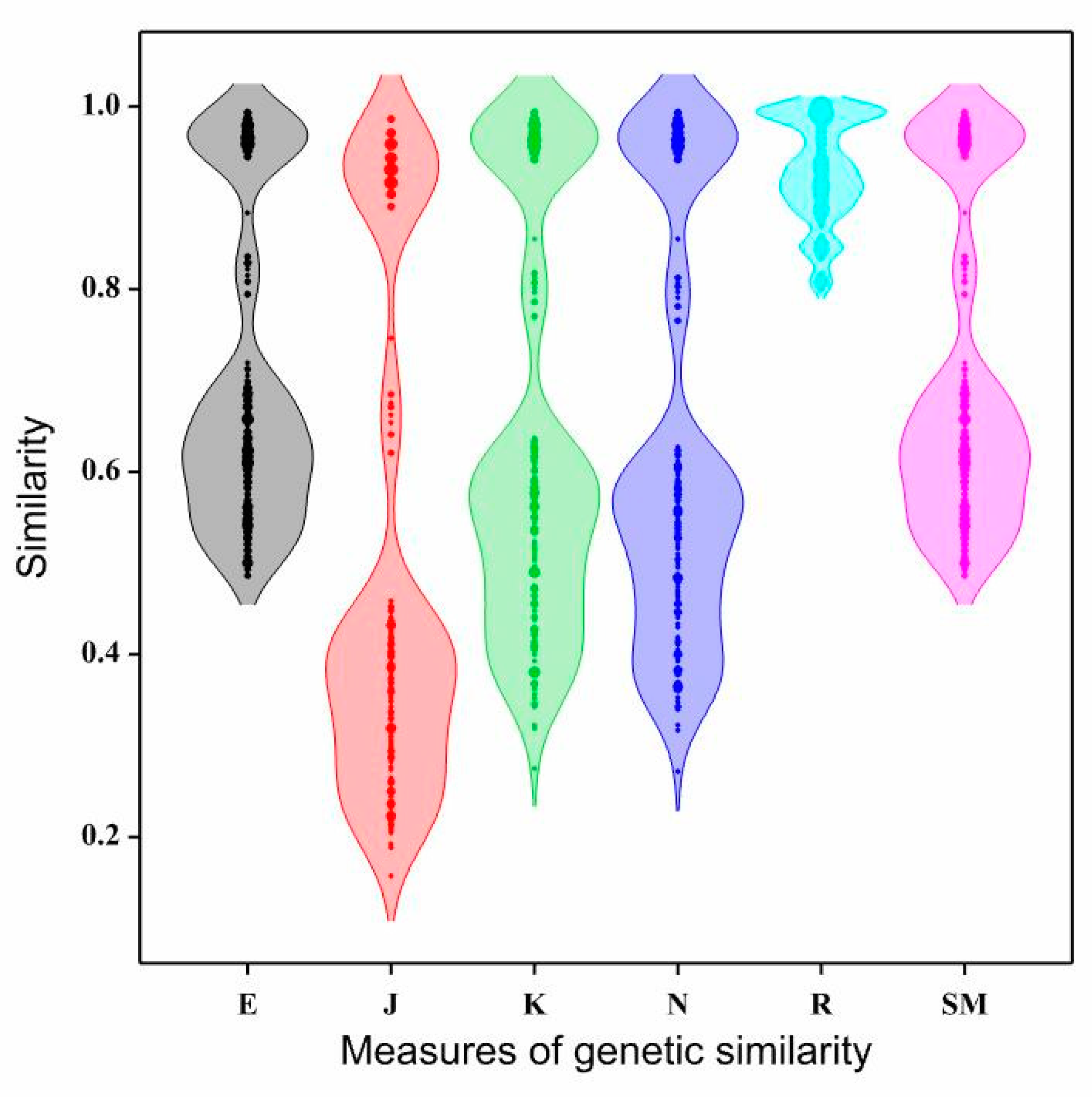
Table 4 presents the statistical characteristics of genetic similarity coefficients obtained using six measures. Genetic similarity values estimated by the Euclidean method ranged from 0.486 (for two pairs of genotypes: SA-BN and SA-BJ) to 0.993 (for three pairs of genotypes: BN×BRc-BN×BJ, BN×SA-BN×BRc, and BN×BT-BN×SA) (
Table 5), with an average of 0.709 (
Table 4). Genetic similarity coefficients calculated using the Euclidean method exhibited moderate variability, with a coefficient of variation of 24.21% (
Figure 1,
Table 4).
The smallest genetic similarity coefficients were obtained for the measure proposed by Jaccard (with an average of 0.508). These coefficients exhibited the highest variability (54.15%) among all measures used (
Table 4,
Figure 1). The lowest genetic similarity according to Jaccard was observed between the pair BJ-SA (0.157), while the highest was between the pair BN×BJ-BN×BRc (0.986) (
Table 5).
Slightly larger genetic similarity coefficients were obtained for the measures proposed by Kulczyński and Nei compared to Jaccard. The results obtained with these two methods were similar in terms of both the values of genetic similarity coefficients (with average values of 0.640 and 0.633 respectively) (
Table 4), and their variability (coefficients of variation were 34.46% and 35.43% respectively) (
Table 4,
Figure 1). The genetic similarity coefficients by Kulczyński ranged from 0.275 (between BJ and SA) to 0.993 (between BN×BJ and BN×BRc) (
Table 6). Meanwhile, genetic similarity calculated using the Nei method ranged from 0.272 (between BJ and SA) to 0.993 (between BN×BJ and BN×BRc) (
Table 6).
Significantly different results were obtained when using the Rogers measure (
Table 4 and
Table 7,
Figure 1). The genetic similarity coefficients exhibited the lowest variability: 6.14% (
Table 4) and ranged from 0.801 (for two genotype pairs: BF-BN×BJ and BF-BN×BRp) to 1.000 (for 23 genotype pairs) (
Table 7). The average value of Rogers coefficients was 0.929 (
Table 4). The minimum value of genetic similarity coefficients estimated by this measure was higher than the average values obtained for the other measures.
Genetic similarity coefficients calculated using the Sokal and Michener measure were identical to the coefficients calculated using the Euclidean method (
Table 5 and
Table 7). The minimum value was twice the minimum value obtained using the Kulczyński or Nei measures, and four times when the Jaccard measure was used for comparison.
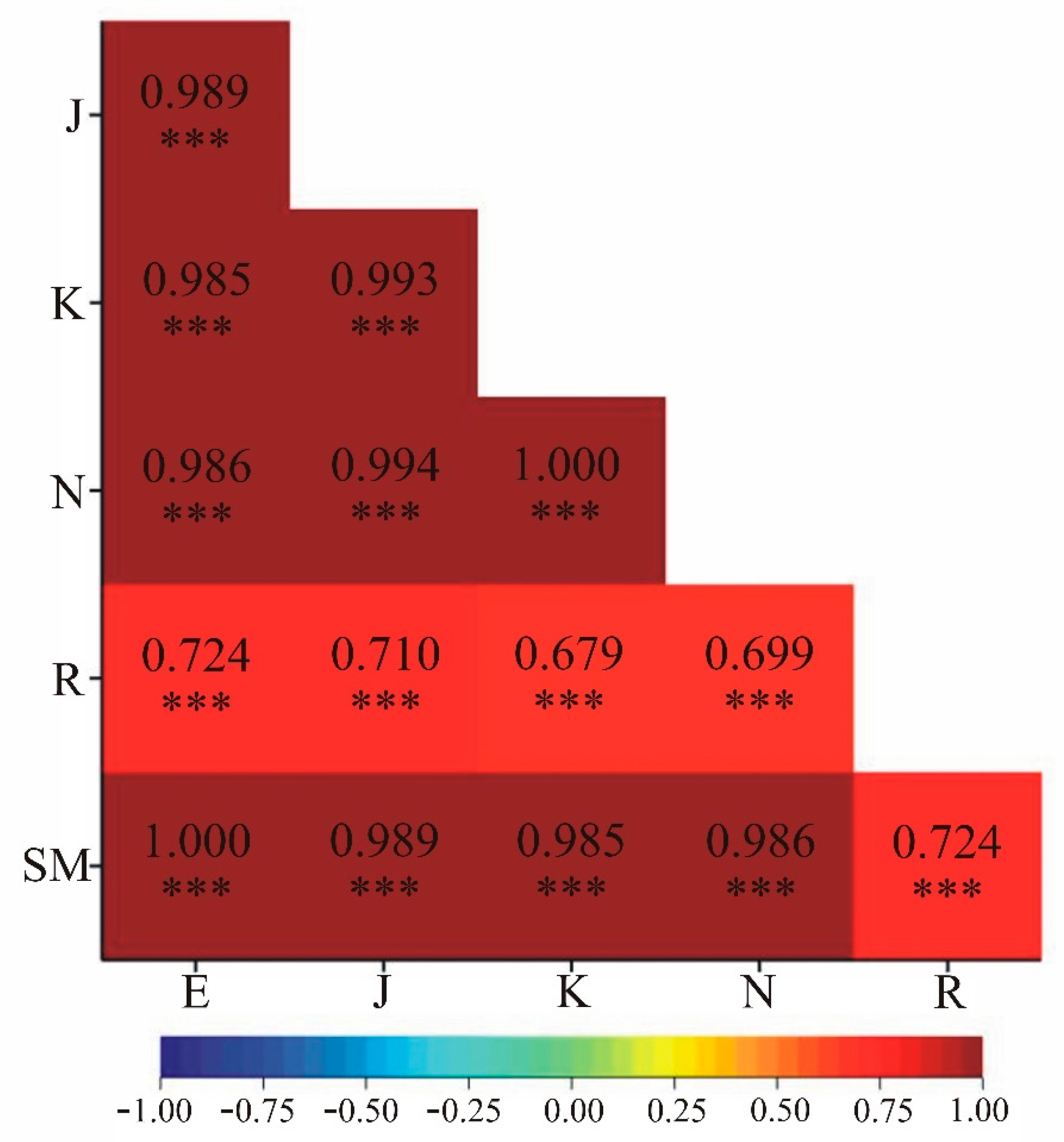
Figure 2 depicts the linear correlation coefficients between genetic similarity values obtained using six measures. The results indicate statistically significant correlations between all applied genetic similarity measures (at
α=0.001 level). It is noteworthy that the genetic similarity coefficients calculated by the Kulczyński and Nei methods show perfect correlation (correlation coefficient was 1.000). A similar result was obtained for Euclidean vs. Sokal and Michener measures (
Figure 2). However, this is due to the identical values obtained from these two methods.
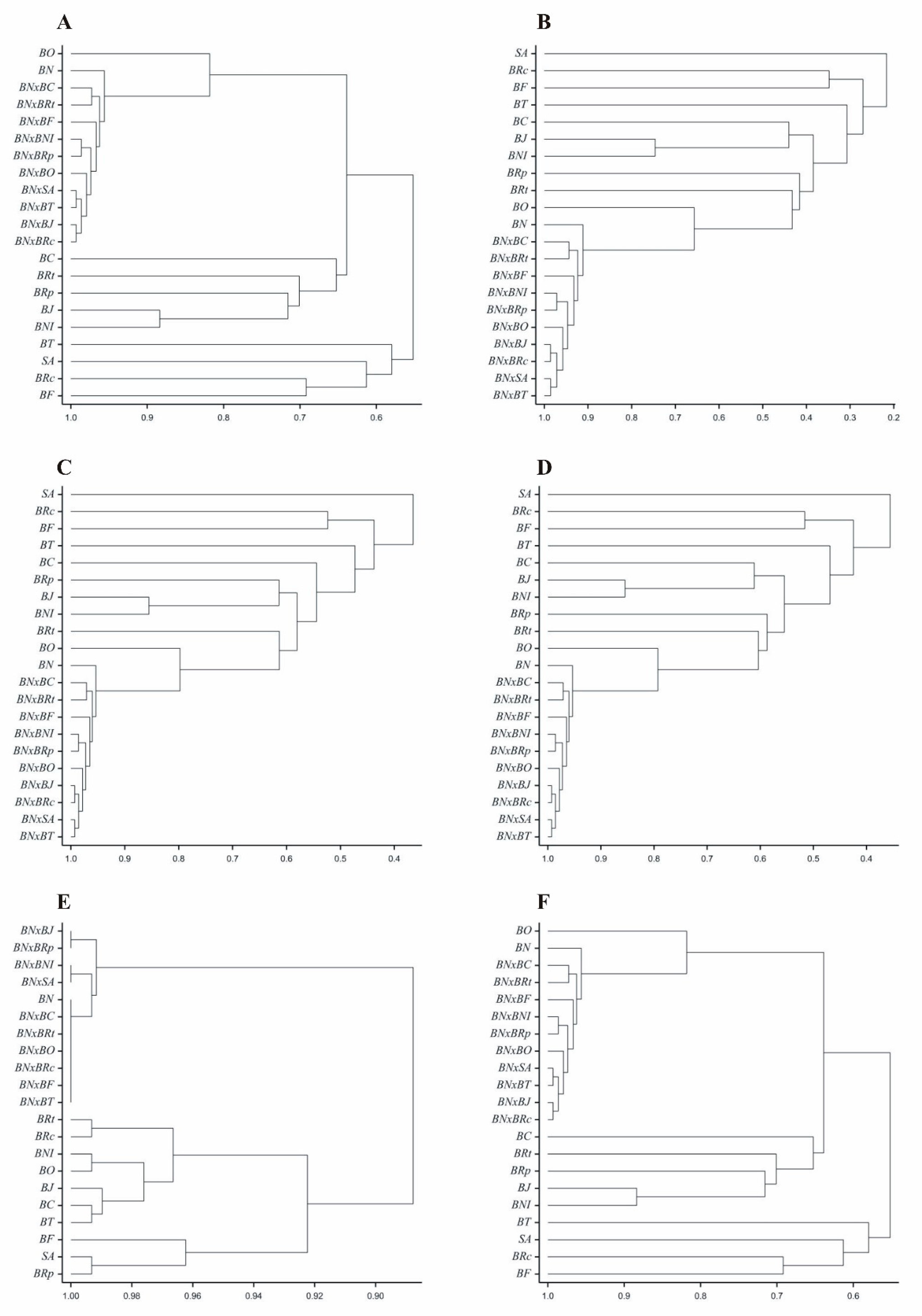
The obtained genetic similarity coefficients were used for hierarchical clustering of objects using the average linkage method. The results of the clustering are presented in the form of dendrograms for respective measures (
Figure 3). Clustering of genotypes based on genetic similarity coefficients according to the Euclidean measure (
Figure 3A) and Sokal and Michener measure (
Figure 3F) was identical, due to identical similarity values obtained from these two methods. Clustering of genotypes based on genetic similarity coefficients according to the Kulczyński measure (
Figure 3C) and Nei measure (
Figure 3D) was nearly identical. Very similar clustering of objects to the aforementioned two measures was obtained based on Jaccard coefficients (
Figure 3B). It is noteworthy that based on genetic similarity coefficients estimated by all six measures, the examined rapeseed genotypes were clustered similarly into two clusters. One of these clusters comprised ten rapeseed hybrids, and the other comprised their eleven parental forms.
4. Discussion
An important element in the accurate assessment of genetic similarity in plant material is both the choice of molecular analysis technique, the number of molecular markers used in the study, and the appropriate statistical measure to estimate relationships between studied genotypes [
62]. Genetic variability among studied objects is defined based on the frequency of DNA polymorphism obtained using different types of molecular markers. Various measures can be applied in estimating genetic similarity/distance based on results obtained from molecular markers. In studies of
Brassica crops, the Nei and Li measure [
63] has been most commonly used for Indian mustard (
B. juncea) [
64], cabbage (
B. oleracea) [
65], and rapeseed (
B. napus) [
66]. The Jaccard measure has been applied in studies on rapeseed [
67] and cabbage [
68,
69], among others. To a lesser extent in rapeseed studies, the Sokal and Michener measure has been utilized [
70]. The Rogers measure has been used to assess genetic variability in crops such as maize (
Zea mays L.) [
71,
72] and sorghum (
Sorghum bicolor L.) [
73].
In the literature on the subject, little attention has been devoted to theoretical comparisons of genetic similarity measures. Lamboy [
74] compared the Nei and Jaccard measures, pointing out Nei's measure as less biased for analyzing closely related organisms. Five of the genetic similarity measures used in the present study (excluding the Rogers measure) yielded very similar results. However, determining the best measure is not feasible. Nearly identical clustering of genotypes suggests that any of the proposed methods would be suitable for the overall characterization of the studied materials. Mohammadi and Prasanna [
62] highlight Rogers' genetic distance measure as useful for estimating genetic similarity based on co-dominant molecular markers, where the amplification product can be equated with alleles. This is corroborated by Gauthier et al. [
75] in their study of European maize populations, where both Rogers' measure utilizing allele frequencies and Nei and Li's measure based on a binary system (where 1 indicates allele presence and 0 absence) were applied. Similarly, Lombard et al. [
76] demonstrated the utility of Rogers' measure in assessing homogeneity within three rapeseed varieties, assuming the tested material was homozygous and each marker was treated as a diallelic locus. Similar findings were reported by Lee et al. [
77] and Dudley et al. [
78] for homozygous maize breeding lines, as well as by Jordan et al. [
73] for sorghum lines analyzed using co-dominant markers.
The distinctly different similarity coefficient values obtained using the Rogers method stem from the fact that its calculation principle differs from the other proposed methods. Rogers' method considers the number of alleles present only in one of the compared genotypes, whereas the other methods are based on the number of alleles present simultaneously in both compared genotypes. Previous studies on generated data [
79] as well as real data [
80] indicate a lack of significant correlation between Rogers' genetic similarity measure and the other measures. The significant correlation obtained in this study may be a result of a large number of markers obtained, significantly exceeding the number of genotype observations considered in the aforementioned studies. Furthermore, it is noteworthy that each of the six applied measures yielded identical two clusters (hybrids and their parental forms), as a consequence of clustering using the UPGMA method. Rogers’ GS coefficients also distinguished these two groups, despite significantly different values compared to those obtained by the other methods.
Duarte et al. [
81] obtained dendrograms for bean varieties with the same structure using UPGMA clustering based on Jaccard's and Nei's coefficients. Lombard et al. [
70] applied genetic similarity (GS) coefficients Jaccard (J), Sokal and Michener (SM), and modified Sokal and Michener (MSM) to compare 83 spring and winter rapeseed varieties, whose diversity was assessed using AFLP. All tested similarity coefficients showed significant correlations (
r=0.96 for J and MSM; 0.97 for SM and MSM; and 0.98 for J and SM at the significance level of 0.001). Genetic similarity calculated using these three coefficients allowed for very similar estimation of relationships among the studied varieties.
Author Contributions
Conceptualization, J.B.; methodology, J.B., J.N., A.J. and J.S.; software, J.B.; validation, J.B., J.N., A.J. and J.S.; formal analysis, J.B.; investigation, J.B. and J.N.; resources, J.B., J.N. and J.S.; data curation, J.N., A.J. and J.S.; writing—original draft preparation, J.B. and J.N.; writing—review and editing, J.B., J.N., A.J. and J.S.; visualization, J.B.; supervision, J.B.; project administration, J.B.; funding acquisition, J.B. All authors have read and agreed to the published version of the manuscript.
Figure 1.
Density plots of coefficients of genetic similarity of interspecific Brassicaceae hybrids and their parental forms using six measures of genetic similarity; E – Euclidean, J – Jaccard, K – Kulczyński, N – Nei, R – Rogers, SM – Sokal and Michener.
Figure 1.
Density plots of coefficients of genetic similarity of interspecific Brassicaceae hybrids and their parental forms using six measures of genetic similarity; E – Euclidean, J – Jaccard, K – Kulczyński, N – Nei, R – Rogers, SM – Sokal and Michener.
Figure 2.
Linear correlation coefficients between pairs of measures of genetic similarity of the studied interspecific Brassicaceae hybrids and their parental forms. E – Euclidean, J – Jaccard, K – Kulczyński, N – Nei, R – Rogers, SM – Sokal and Michener.
Figure 2.
Linear correlation coefficients between pairs of measures of genetic similarity of the studied interspecific Brassicaceae hybrids and their parental forms. E – Euclidean, J – Jaccard, K – Kulczyński, N – Nei, R – Rogers, SM – Sokal and Michener.
Figure 3.
Dendrograms of genetic similarity of 21 Brassicaceae genotypes constructed using six measures based on 146 ISSR markers. A – Euclidean, B – Jaccard, C – Kulczyński, D – Nei, E - Rogers, F – Sokal and Michener.
Figure 3.
Dendrograms of genetic similarity of 21 Brassicaceae genotypes constructed using six measures based on 146 ISSR markers. A – Euclidean, B – Jaccard, C – Kulczyński, D – Nei, E - Rogers, F – Sokal and Michener.
Table 1.
Plant material used in this study for the analysis of genetic similarity based on ISSR markers.
Table 1.
Plant material used in this study for the analysis of genetic similarity based on ISSR markers.
| Maternal form |
Paternal forms |
Hybrids of F2 generation |
|
Brassica napus (BN) |
Brassica carinata (BC) |
B. napus × B. carinata (BNxBC) |
|
Brassica rapa ssp. trilocularis (BRt) |
B. napus × B. rapa ssp. trilocularis (BNxBRt) |
|
Brassica rapa ssp. chinensis (BRc) |
B. napusx × B. rapa ssp. chinensis (BNxBRc) |
|
Brassica fruticulosa (BF) |
B. napus × B. fruticulosa (BNxBF) |
|
Brassica rapa ssp. pekinensis (BRp) |
B. napus × B. rapa ssp. pekinensis (BNxBRp) |
|
Brassica nigra (BNI) |
Brassica napus × Brassica nigra (BNxBNI) |
|
Brassica juncea (BJ) |
Brassica napus × Brassica juncea (BNxBJ) |
|
Sinapis alba (SA) |
Brassica napus × Sinapis alba (BNxSA) |
|
Brassica tournefortii (BT) |
Brassica napus × Brassica rapa ssp. tournefortii (BNxBT) |
|
Brassica oleracea var. alboglabra (BO) |
Brassica napus × Brassica oleracea var. alboglabra (BNxBO) |
Table 2.
PCR amplification profiles for tested primers.
Table 2.
PCR amplification profiles for tested primers.
| Reaction profile |
Primers |
Parameters |
Temperature |
Number of cycles |
Source |
| 1 |
mstg 1, mstg 4, mstg 12, mstg 13, mstg 38 |
Initial denaturation |
94°C 30 s |
x1 |
[52] |
| Denaturation |
94°C 30 s |
x40 |
| Primers annealing |
60°C* 30 s |
| Elongation |
72°C 1 min |
| Final elongation |
72°C 5 min |
x1 |
| 2 |
UBC812, UBC840, UBC845 |
Initial denaturation |
95°C 2 min |
x1 |
[53] |
| Denaturation |
93°C 20 s |
x40 |
| Primers annealing |
52°C* 1 min |
| Elongation |
72°C 20 s |
| Final elongation |
72°C 6 min |
x1 |
| 3 |
UBC891, ISSR-840, ISSR-856, ISSR-857, ISSR-887, ISSR-888, ISSR-889, ISSR-890, scm51, P1, P2, P4, P5 |
Initial denaturation |
94°C 5 min |
x1 |
[54] |
| Denaturation |
94°C 45 s |
x35 |
| Primers annealing |
50°C* 45 s |
| Elongation |
72°C 90 s |
| Final elongation |
72°C 7 min |
x1 |
Table 3.
List of ISSR primers used in this study.
Table 3.
List of ISSR primers used in this study.
| ISSR primer |
Sequence 5’-3’ |
Annealing temperature [°C] |
| mstg 1 |
(TA)3(TG)9
|
53.9 |
| mstg 4 |
(TA)3(TG)10
|
60 |
| |
|
|
| mstg 12 |
(TG)7
|
54 |
| mstg 13 |
(TG)7CG(TG)2
|
59.7 |
| |
|
|
| mstg 38 |
(TG)3TT(TG)5
|
53.9 |
| |
|
|
| UBC835 |
(AG)8YC |
51.5 |
| UBC843 |
(CT)8RA |
48.1 |
| UBC889 |
DBD(AC)7
|
51.5 |
| UBC891 |
HVH(TG)7
|
51.5 |
| scm 51 |
(AGG)5
|
52.8 |
| P1 |
GAG(CAA)5
|
49.7 |
| P2 |
CTG(GT)8
|
56.5 |
| P4 |
(AG)8GTG |
52.8 |
| P5 |
(GA)8ACC |
51.5 |
| |
|
|
| ISSR-840 |
(GA)8YT |
48.4 |
| ISSR-856 |
(GA)8YA |
48.4 |
| ISSR-857 |
(GA)8YG |
55.3 |
| ISSR-887 |
DVD(TC)7
|
47.8 |
| ISSR-888 |
BDB(CA)7
|
48.4 |
| ISSR-889 |
DBD(AC)7
|
48.1 |
| ISSR-890 |
VHV(GT)7
|
48.4 |
Table 4.
Statistical characteristics of genetic similarity of interspecific Brassicaceae hybrids F2 generation and their parental forms.
Table 4.
Statistical characteristics of genetic similarity of interspecific Brassicaceae hybrids F2 generation and their parental forms.
| Genetic similarity measure |
Minimal value |
Mean value |
Maximum value |
Coefficient of variability [in %] |
| Euclidean |
0.486 |
0.709 |
0.993 |
24.21 |
| Jaccard |
0.157 |
0.508 |
0.986 |
54.15 |
| Kulczyński |
0.275 |
0.640 |
0.993 |
34.46 |
| Nei |
0.272 |
0.633 |
0.993 |
35.43 |
| Rogers |
0.801 |
0.929 |
1.000 |
6.14 |
| Sokal and Michener |
0.486 |
0.709 |
0.993 |
24.21 |
Table 5.
Genetic similarity coefficients between the examined genotypes calculated using the Euclidean measure (above diagonal) and Jaccard measure (below diagonal).
Table 5.
Genetic similarity coefficients between the examined genotypes calculated using the Euclidean measure (above diagonal) and Jaccard measure (below diagonal).
| Genotype |
BN |
BC |
BRt |
BJ |
BNI |
BO |
BRc |
SA |
BRp |
BF |
BT |
BNxBC |
BNxBRt |
BNxBJ |
BNxBNI |
BNxBO |
BNxBRc |
BNxSA |
BNxBRp |
BNxBF |
BNxBT |
| BN |
1.000 |
0.596 |
0.658 |
0.603 |
0.610 |
0.808 |
0.541 |
0.486 |
0.644 |
0.534 |
0.562 |
0.959 |
0.959 |
0.952 |
0.952 |
0.945 |
0.959 |
0.952 |
0.966 |
0.959 |
0.959 |
| BC |
0.359 |
1.000 |
0.582 |
0.692 |
0.699 |
0.610 |
0.658 |
0.548 |
0.637 |
0.637 |
0.500 |
0.637 |
0.637 |
0.603 |
0.603 |
0.596 |
0.596 |
0.603 |
0.616 |
0.610 |
0.610 |
| BRt |
0.419 |
0.282 |
1.000 |
0.685 |
0.706 |
0.644 |
0.555 |
0.555 |
0.712 |
0.589 |
0.521 |
0.671 |
0.671 |
0.678 |
0.678 |
0.671 |
0.685 |
0.692 |
0.664 |
0.658 |
0.685 |
| BJ |
0.370 |
0.430 |
0.410 |
1.000 |
0.884 |
0.616 |
0.623 |
0.486 |
0.712 |
0.630 |
0.589 |
0.616 |
0.616 |
0.637 |
0.623 |
0.644 |
0.630 |
0.623 |
0.623 |
0.616 |
0.630 |
| BNI |
0.387 |
0.450 |
0.449 |
0.746 |
1.000 |
0.610 |
0.658 |
0.521 |
0.719 |
0.623 |
0.582 |
0.610 |
0.623 |
0.630 |
0.603 |
0.610 |
0.623 |
0.616 |
0.616 |
0.610 |
0.623 |
| BO |
0.641 |
0.337 |
0.366 |
0.349 |
0.352 |
1.000 |
0.527 |
0.541 |
0.699 |
0.589 |
0.589 |
0.795 |
0.795 |
0.829 |
0.829 |
0.836 |
0.836 |
0.829 |
0.815 |
0.808 |
0.822 |
| BRc |
0.287 |
0.367 |
0.235 |
0.329 |
0.383 |
0.233 |
1.000 |
0.575 |
0.596 |
0.692 |
0.555 |
0.555 |
0.541 |
0.507 |
0.534 |
0.514 |
0.514 |
0.521 |
0.548 |
0.541 |
0.527 |
| SA |
0.211 |
0.214 |
0.207 |
0.157 |
0.205 |
0.221 |
0.225 |
1.000 |
0.569 |
0.651 |
0.527 |
0.500 |
0.500 |
0.493 |
0.507 |
0.500 |
0.500 |
0.507 |
0.493 |
0.514 |
0.500 |
| BRp |
0.381 |
0.321 |
0.409 |
0.425 |
0.446 |
0.413 |
0.253 |
0.192 |
1.000 |
0.671 |
0.658 |
0.658 |
0.671 |
0.678 |
0.678 |
0.685 |
0.685 |
0.692 |
0.664 |
0.658 |
0.685 |
| BF |
0.236 |
0.293 |
0.221 |
0.290 |
0.295 |
0.250 |
0.348 |
0.261 |
0.294 |
1.000 |
0.658 |
0.562 |
0.548 |
0.527 |
0.541 |
0.534 |
0.534 |
0.541 |
0.541 |
0.548 |
0.548 |
| BT |
0.319 |
0.207 |
0.214 |
0.302 |
0.307 |
0.310 |
0.244 |
0.188 |
0.342 |
0.315 |
1.000 |
0.548 |
0.562 |
0.555 |
0.582 |
0.575 |
0.562 |
0.569 |
0.569 |
0.562 |
0.562 |
| BNxBC |
0.917 |
0.405 |
0.435 |
0.385 |
0.387 |
0.620 |
0.301 |
0.223 |
0.398 |
0.264 |
0.305 |
1.000 |
0.973 |
0.952 |
0.966 |
0.945 |
0.959 |
0.966 |
0.980 |
0.959 |
0.973 |
| BNxBRt |
0.917 |
0.405 |
0.435 |
0.385 |
0.402 |
0.620 |
0.287 |
0.223 |
0.415 |
0.250 |
0.319 |
0.944 |
1.000 |
0.952 |
0.966 |
0.945 |
0.959 |
0.966 |
0.980 |
0.959 |
0.973 |
| BNxBJ |
0.904 |
0.370 |
0.447 |
0.411 |
0.413 |
0.675 |
0.258 |
0.221 |
0.427 |
0.233 |
0.316 |
0.904 |
0.904 |
1.000 |
0.973 |
0.980 |
0.993 |
0.986 |
0.959 |
0.966 |
0.980 |
| BNxBNI |
0.903 |
0.363 |
0.441 |
0.389 |
0.376 |
0.671 |
0.277 |
0.226 |
0.420 |
0.239 |
0.337 |
0.930 |
0.930 |
0.944 |
1.000 |
0.980 |
0.980 |
0.986 |
0.986 |
0.966 |
0.980 |
| BNxBO |
0.890 |
0.359 |
0.435 |
0.416 |
0.387 |
0.684 |
0.260 |
0.223 |
0.432 |
0.236 |
0.333 |
0.890 |
0.890 |
0.958 |
0.957 |
1.000 |
0.986 |
0.980 |
0.966 |
0.959 |
0.973 |
| BNxBRc |
0.917 |
0.359 |
0.452 |
0.400 |
0.402 |
0.684 |
0.260 |
0.223 |
0.432 |
0.236 |
0.319 |
0.917 |
0.917 |
0.986 |
0.957 |
0.971 |
1.000 |
0.993 |
0.966 |
0.973 |
0.986 |
| BNxSA |
0.903 |
0.363 |
0.458 |
0.389 |
0.391 |
0.671 |
0.263 |
0.226 |
0.438 |
0.239 |
0.323 |
0.930 |
0.930 |
0.971 |
0.971 |
0.957 |
0.986 |
1.000 |
0.973 |
0.966 |
0.993 |
| BNxBRp |
0.931 |
0.385 |
0.430 |
0.396 |
0.398 |
0.654 |
0.298 |
0.221 |
0.410 |
0.247 |
0.330 |
0.958 |
0.958 |
0.918 |
0.971 |
0.931 |
0.931 |
0.944 |
1.000 |
0.966 |
0.980 |
| BNxBF |
0.917 |
0.374 |
0.419 |
0.385 |
0.387 |
0.641 |
0.287 |
0.237 |
0.398 |
0.250 |
0.319 |
0.917 |
0.917 |
0.931 |
0.930 |
0.917 |
0.944 |
0.930 |
0.931 |
1.000 |
0.973 |
| BNxBT |
0.917 |
0.374 |
0.452 |
0.400 |
0.402 |
0.662 |
0.274 |
0.223 |
0.432 |
0.250 |
0.319 |
0.944 |
0.944 |
0.958 |
0.957 |
0.944 |
0.971 |
0.986 |
0.958 |
0.944 |
1.000 |
Table 6.
Genetic similarity coefficients between the examined genotypes calculated using the Kulczyński measure (above diagonal) and Nei measure (below diagonal).
Table 6.
Genetic similarity coefficients between the examined genotypes calculated using the Kulczyński measure (above diagonal) and Nei measure (below diagonal).
| Genotype |
BN |
BC |
BRt |
BJ |
BNI |
BO |
BRc |
SA |
BRp |
BF |
BT |
BNxBC |
BNxBRt |
BNxBJ |
BNxBNI |
BNxBO |
BNxBRc |
BNxSA |
BNxBRp |
BNxBF |
BNxBT |
| BN |
1.000 |
0.534 |
0.601 |
0.545 |
0.561 |
0.786 |
0.455 |
0.362 |
0.572 |
0.408 |
0.490 |
0.957 |
0.957 |
0.950 |
0.949 |
0.942 |
0.957 |
0.949 |
0.964 |
0.957 |
0.957 |
| BC |
0.528 |
1.000 |
0.441 |
0.602 |
0.621 |
0.505 |
0.538 |
0.356 |
0.489 |
0.465 |
0.342 |
0.582 |
0.582 |
0.546 |
0.537 |
0.534 |
0.534 |
0.537 |
0.563 |
0.550 |
0.550 |
| BRt |
0.590 |
0.440 |
1.000 |
0.583 |
0.622 |
0.537 |
0.381 |
0.345 |
0.582 |
0.368 |
0.352 |
0.617 |
0.617 |
0.630 |
0.621 |
0.617 |
0.634 |
0.638 |
0.613 |
0.601 |
0.634 |
| BJ |
0.540 |
0.602 |
0.582 |
1.000 |
0.855 |
0.517 |
0.497 |
0.275 |
0.602 |
0.461 |
0.464 |
0.561 |
0.561 |
0.589 |
0.564 |
0.593 |
0.577 |
0.564 |
0.573 |
0.561 |
0.577 |
| BNI |
0.558 |
0.621 |
0.620 |
0.855 |
1.000 |
0.521 |
0.556 |
0.346 |
0.626 |
0.472 |
0.471 |
0.561 |
0.576 |
0.588 |
0.549 |
0.561 |
0.576 |
0.565 |
0.573 |
0.561 |
0.576 |
| BO |
0.781 |
0.504 |
0.536 |
0.517 |
0.521 |
1.000 |
0.380 |
0.368 |
0.593 |
0.413 |
0.474 |
0.770 |
0.770 |
0.812 |
0.807 |
0.818 |
0.818 |
0.807 |
0.797 |
0.786 |
0.802 |
| BRc |
0.446 |
0.537 |
0.381 |
0.495 |
0.554 |
0.378 |
1.000 |
0.369 |
0.405 |
0.524 |
0.393 |
0.472 |
0.455 |
0.419 |
0.441 |
0.422 |
0.422 |
0.424 |
0.469 |
0.455 |
0.438 |
| SA |
0.348 |
0.353 |
0.343 |
0.272 |
0.340 |
0.362 |
0.367 |
1.000 |
0.323 |
0.415 |
0.319 |
0.380 |
0.380 |
0.378 |
0.383 |
0.380 |
0.380 |
0.383 |
0.378 |
0.399 |
0.380 |
| BRp |
0.552 |
0.485 |
0.580 |
0.596 |
0.617 |
0.585 |
0.404 |
0.323 |
1.000 |
0.457 |
0.513 |
0.590 |
0.608 |
0.622 |
0.612 |
0.626 |
0.626 |
0.630 |
0.605 |
0.590 |
0.626 |
| BF |
0.382 |
0.454 |
0.362 |
0.449 |
0.455 |
0.400 |
0.516 |
0.414 |
0.455 |
1.000 |
0.490 |
0.447 |
0.428 |
0.406 |
0.411 |
0.408 |
0.408 |
0.411 |
0.425 |
0.428 |
0.428 |
| BT |
0.484 |
0.342 |
0.352 |
0.464 |
0.470 |
0.474 |
0.393 |
0.317 |
0.510 |
0.479 |
1.000 |
0.474 |
0.490 |
0.487 |
0.510 |
0.507 |
0.490 |
0.493 |
0.503 |
0.490 |
0.490 |
| BNxBC |
0.957 |
0.576 |
0.607 |
0.556 |
0.558 |
0.766 |
0.463 |
0.365 |
0.569 |
0.418 |
0.468 |
1.000 |
0.971 |
0.950 |
0.964 |
0.942 |
0.957 |
0.964 |
0.979 |
0.957 |
0.971 |
| BNxBRt |
0.957 |
0.576 |
0.607 |
0.556 |
0.574 |
0.766 |
0.446 |
0.365 |
0.586 |
0.400 |
0.484 |
0.971 |
1.000 |
0.950 |
0.964 |
0.942 |
0.957 |
0.964 |
0.979 |
0.957 |
0.971 |
| BNxBJ |
0.950 |
0.540 |
0.618 |
0.583 |
0.585 |
0.806 |
0.410 |
0.362 |
0.598 |
0.378 |
0.480 |
0.950 |
0.950 |
1.000 |
0.971 |
0.979 |
0.993 |
0.986 |
0.957 |
0.964 |
0.979 |
| BNxBNI |
0.949 |
0.532 |
0.612 |
0.560 |
0.547 |
0.803 |
0.433 |
0.368 |
0.591 |
0.385 |
0.504 |
0.964 |
0.964 |
0.971 |
1.000 |
0.978 |
0.978 |
0.985 |
0.986 |
0.964 |
0.978 |
| BNxBO |
0.942 |
0.528 |
0.607 |
0.587 |
0.558 |
0.813 |
0.413 |
0.365 |
0.603 |
0.382 |
0.500 |
0.942 |
0.942 |
0.978 |
0.978 |
1.000 |
0.986 |
0.978 |
0.964 |
0.957 |
0.971 |
| BNxBRc |
0.957 |
0.528 |
0.623 |
0.571 |
0.574 |
0.813 |
0.413 |
0.365 |
0.603 |
0.382 |
0.484 |
0.957 |
0.957 |
0.993 |
0.978 |
0.986 |
1.000 |
0.993 |
0.964 |
0.971 |
0.986 |
| BNxSA |
0.949 |
0.532 |
0.628 |
0.560 |
0.563 |
0.803 |
0.417 |
0.368 |
0.609 |
0.385 |
0.488 |
0.964 |
0.964 |
0.986 |
0.985 |
0.978 |
0.993 |
1.000 |
0.971 |
0.964 |
0.993 |
| BNxBRp |
0.964 |
0.556 |
0.602 |
0.567 |
0.569 |
0.791 |
0.459 |
0.362 |
0.581 |
0.396 |
0.496 |
0.978 |
0.978 |
0.957 |
0.986 |
0.964 |
0.964 |
0.971 |
1.000 |
0.964 |
0.979 |
| BNxBF |
0.957 |
0.544 |
0.590 |
0.556 |
0.558 |
0.781 |
0.446 |
0.383 |
0.569 |
0.400 |
0.484 |
0.957 |
0.957 |
0.964 |
0.964 |
0.957 |
0.971 |
0.964 |
0.964 |
1.000 |
0.971 |
| BNxBT |
0.957 |
0.544 |
0.623 |
0.571 |
0.574 |
0.797 |
0.430 |
0.365 |
0.603 |
0.400 |
0.484 |
0.971 |
0.971 |
0.978 |
0.978 |
0.971 |
0.986 |
0.993 |
0.978 |
0.971 |
1.000 |
Table 7.
Genetic similarity coefficients between the examined genotypes calculated using the Rogers measure (above diagonal) and Sokal and Michener measure (below diagonal).
Table 7.
Genetic similarity coefficients between the examined genotypes calculated using the Rogers measure (above diagonal) and Sokal and Michener measure (below diagonal).
| Genotype |
BN |
BC |
BRt |
BJ |
BNI |
BO |
BRc |
SA |
BRp |
BF |
BT |
BNxBC |
BNxBRt |
BNxBJ |
BNxBNI |
BNxBO |
BNxBRc |
BNxSA |
BNxBRp |
BNxBF |
BNxBT |
| BN |
1.000 |
0.911 |
0.890 |
0.918 |
0.938 |
0.932 |
0.884 |
0.843 |
0.849 |
0.808 |
0.904 |
1.000 |
1.000 |
0.993 |
0.993 |
1.000 |
1.000 |
0.993 |
0.993 |
1.000 |
1.000 |
| BC |
0.596 |
1.000 |
0.980 |
0.993 |
0.973 |
0.980 |
0.973 |
0.932 |
0.938 |
0.897 |
0.993 |
0.911 |
0.911 |
0.904 |
0.918 |
0.911 |
0.911 |
0.918 |
0.904 |
0.911 |
0.911 |
| BRt |
0.658 |
0.582 |
1.000 |
0.973 |
0.952 |
0.959 |
0.993 |
0.952 |
0.959 |
0.918 |
0.986 |
0.890 |
0.890 |
0.884 |
0.897 |
0.890 |
0.890 |
0.897 |
0.884 |
0.890 |
0.890 |
| BJ |
0.603 |
0.692 |
0.685 |
1.000 |
0.980 |
0.986 |
0.966 |
0.925 |
0.932 |
0.890 |
0.986 |
0.918 |
0.918 |
0.911 |
0.925 |
0.918 |
0.918 |
0.925 |
0.911 |
0.918 |
0.918 |
| BNI |
0.610 |
0.699 |
0.706 |
0.884 |
1.000 |
0.993 |
0.945 |
0.904 |
0.911 |
0.870 |
0.966 |
0.938 |
0.938 |
0.932 |
0.945 |
0.938 |
0.938 |
0.945 |
0.932 |
0.938 |
0.938 |
| BO |
0.808 |
0.610 |
0.644 |
0.616 |
0.610 |
1.000 |
0.952 |
0.911 |
0.918 |
0.877 |
0.973 |
0.932 |
0.932 |
0.925 |
0.938 |
0.932 |
0.932 |
0.938 |
0.925 |
0.932 |
0.932 |
| BRc |
0.541 |
0.658 |
0.555 |
0.623 |
0.658 |
0.527 |
1.000 |
0.959 |
0.966 |
0.925 |
0.980 |
0.884 |
0.884 |
0.877 |
0.890 |
0.884 |
0.884 |
0.890 |
0.877 |
0.884 |
0.884 |
| SA |
0.486 |
0.548 |
0.555 |
0.486 |
0.521 |
0.541 |
0.575 |
1.000 |
0.993 |
0.966 |
0.938 |
0.843 |
0.843 |
0.836 |
0.849 |
0.843 |
0.843 |
0.849 |
0.836 |
0.843 |
0.843 |
| BRp |
0.644 |
0.637 |
0.712 |
0.712 |
0.719 |
0.699 |
0.596 |
0.569 |
1.000 |
0.959 |
0.945 |
0.849 |
0.849 |
0.843 |
0.856 |
0.849 |
0.849 |
0.856 |
0.843 |
0.849 |
0.849 |
| BF |
0.534 |
0.637 |
0.589 |
0.630 |
0.623 |
0.589 |
0.692 |
0.651 |
0.671 |
1.000 |
0.904 |
0.808 |
0.808 |
0.801 |
0.815 |
0.808 |
0.808 |
0.815 |
0.801 |
0.808 |
0.808 |
| BT |
0.562 |
0.500 |
0.521 |
0.589 |
0.582 |
0.589 |
0.555 |
0.527 |
0.658 |
0.658 |
1.000 |
0.904 |
0.904 |
0.897 |
0.911 |
0.904 |
0.904 |
0.911 |
0.897 |
0.904 |
0.904 |
| BNxBC |
0.959 |
0.637 |
0.671 |
0.616 |
0.610 |
0.795 |
0.555 |
0.500 |
0.658 |
0.562 |
0.548 |
1.000 |
1.000 |
0.993 |
0.993 |
1.000 |
1.000 |
0.993 |
0.993 |
1.000 |
1.000 |
| BNxBRt |
0.959 |
0.637 |
0.671 |
0.616 |
0.623 |
0.795 |
0.541 |
0.500 |
0.671 |
0.548 |
0.562 |
0.973 |
1.000 |
0.993 |
0.993 |
1.000 |
1.000 |
0.993 |
0.993 |
1.000 |
1.000 |
| BNxBJ |
0.952 |
0.603 |
0.678 |
0.637 |
0.630 |
0.829 |
0.507 |
0.493 |
0.678 |
0.527 |
0.555 |
0.952 |
0.952 |
1.000 |
0.986 |
0.993 |
0.993 |
0.986 |
1.000 |
0.993 |
0.993 |
| BNxBNI |
0.952 |
0.603 |
0.678 |
0.623 |
0.603 |
0.829 |
0.534 |
0.507 |
0.678 |
0.541 |
0.582 |
0.966 |
0.966 |
0.973 |
1.000 |
0.993 |
0.993 |
1.000 |
0.986 |
0.993 |
0.993 |
| BNxBO |
0.945 |
0.596 |
0.671 |
0.644 |
0.610 |
0.836 |
0.514 |
0.500 |
0.685 |
0.534 |
0.575 |
0.945 |
0.945 |
0.980 |
0.980 |
1.000 |
1.000 |
0.993 |
0.993 |
1.000 |
1.000 |
| BNxBRc |
0.959 |
0.596 |
0.685 |
0.630 |
0.623 |
0.836 |
0.514 |
0.500 |
0.685 |
0.534 |
0.562 |
0.959 |
0.959 |
0.993 |
0.980 |
0.986 |
1.000 |
0.993 |
0.993 |
1.000 |
1.000 |
| BNxSA |
0.952 |
0.603 |
0.692 |
0.623 |
0.616 |
0.829 |
0.521 |
0.507 |
0.692 |
0.541 |
0.569 |
0.966 |
0.966 |
0.986 |
0.986 |
0.980 |
0.993 |
1.000 |
0.986 |
0.993 |
0.993 |
| BNxBRp |
0.966 |
0.616 |
0.664 |
0.623 |
0.616 |
0.815 |
0.548 |
0.493 |
0.664 |
0.541 |
0.569 |
0.980 |
0.980 |
0.959 |
0.986 |
0.966 |
0.966 |
0.973 |
1.000 |
0.993 |
0.993 |
| BNxBF |
0.959 |
0.610 |
0.658 |
0.616 |
0.610 |
0.808 |
0.541 |
0.514 |
0.658 |
0.548 |
0.562 |
0.959 |
0.959 |
0.966 |
0.966 |
0.959 |
0.973 |
0.966 |
0.966 |
1.000 |
1.000 |
| BNxBT |
0.959 |
0.610 |
0.685 |
0.630 |
0.623 |
0.822 |
0.527 |
0.500 |
0.685 |
0.548 |
0.562 |
0.973 |
0.973 |
0.980 |
0.980 |
0.973 |
0.986 |
0.993 |
0.980 |
0.973 |
1.000 |