Submitted:
30 July 2024
Posted:
01 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
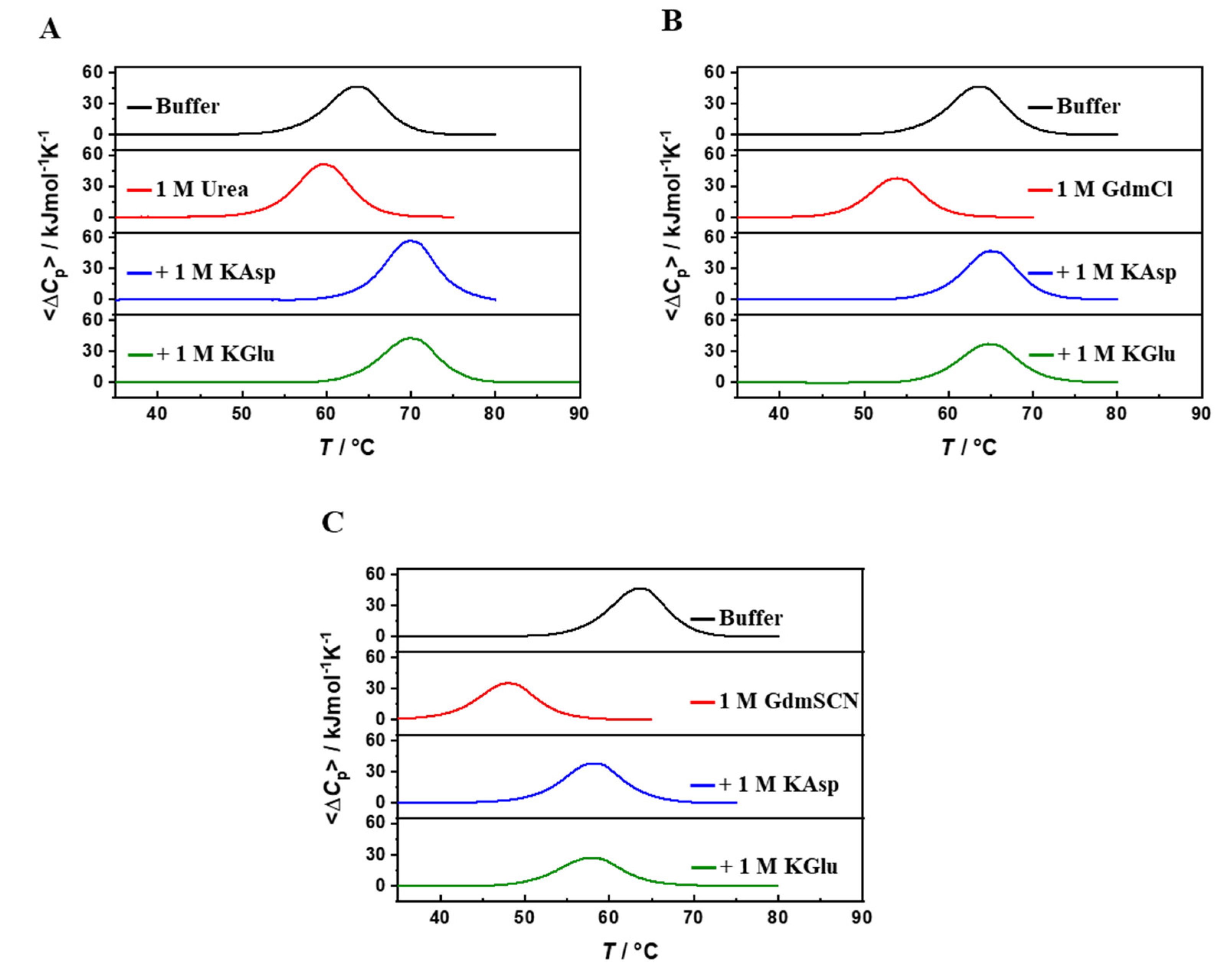
2. DSC Results
3. Theoretical Approach and Its Application
+ [3ξ2/2(1- ξ3)]l + [6ξ1/(1-ξ3)]a·l + [9ξ22/(1-ξ3)2]a·l}
4. Materials and Methods
4.1. Materials
4.2. Density Measurements
4.3. Differential Scanning Calorimetry (DSC)
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hofmeister, F. Zur Lehre von der Wirkung der Salze: Zweite Mittheilung. Arch. Für Exp. Pathol. Pharmakol. 1888, 24, 247–260. [Google Scholar] [CrossRef]
- Von Hippel, P.H.; Wong, K.-Y. On the Conformational Stability of Globular Proteins. J. Biol. Chem. 1965, 240, 3909–3923. [Google Scholar] [CrossRef]
- Baldwin, R.L. How Hofmeister Ion Interactions Affect Protein Stability. Biophys. J. 1996, 71, 2056–2063. [Google Scholar] [CrossRef]
- Schellman, J.A. Protein Stability in Mixed Solvents: A Balance of Contact Interaction and Excluded Volume. Biophys. J. 2003, 85, 108–125. [Google Scholar] [CrossRef]
- Batchelor, J.D.; Olteanu, A.; Tripathy, A.; Pielak, G.J. Impact of Protein Denaturants and Stabilizers on Water Structure. J. Am. Chem. Soc. 2004, 126, 1958–1961. [Google Scholar] [CrossRef]
- Mitra, L.; Smolin, N.; Ravindra, R.; Royer, C.; Winter, R. Pressure Perturbation Calorimetric Studies of the Solvation Properties and the Thermal Unfolding of Proteins in Solution—Experiments and Theoretical Interpretation. Phys. Chem. Chem. Phys. 2006, 8, 1249. [Google Scholar] [CrossRef]
- Godawat, R.; Jamadagni, S.N.; Garde, S. Unfolding of Hydrophobic Polymers in Guanidinium Chloride Solutions. J. Phys. Chem. B 2010, 114, 2246–2254. [Google Scholar] [CrossRef] [PubMed]
- Pegram, L.M.; Record, M.T. Thermodynamic Origin of Hofmeister Ion Effects. J. Phys. Chem. B 2008, 112, 9428–9436. [Google Scholar] [CrossRef] [PubMed]
- Record, M.T.; Guinn, E.; Pegram, L.; Capp, M. Introductory Lecture: Interpreting and Predicting Hofmeister Salt Ion and Solute Effects on Biopolymer and Model Processes Using the Solute Partitioning Model. Faraday Discuss 2013, 160, 9–44. [Google Scholar] [CrossRef]
- von Hippel, P.H. Changing the Stability of Macromolecular Surfaces by Manipulating the Aqueous Environment. Biophys. J. 2016, 111, 1817–1820. [Google Scholar] [CrossRef]
- Arakawa, T.; Timasheff, S.N. The Mechanism of Action of Na Glutamate, Lysine HCl, and Piperazine-N,N’-Bis(2-Ethanesulfonic Acid) in the Stabilization of Tubulin and Microtubule Formation. J. Biol. Chem. 1984, 259, 4979–4986. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, R.; Pantel, A.; Cheng, X.; Shkel, I.; Peran, I.; Stenzoski, N.; Raleigh, D.P.; Record, M.T. Positioning the Intracellular Salt Potassium Glutamate in the Hofmeister Series by Chemical Unfolding Studies of NTL9. Biochemistry 2016, 55, 2251–2259. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Guinn, E.J.; Buechel, E.; Wong, R.; Sengupta, R.; Shkel, I.A.; Record, M.T. Basis of Protein Stabilization by K Glutamate: Unfavorable Interactions with Carbon, Oxygen Groups. Biophys. J. 2016, 111, 1854–1865. [Google Scholar] [CrossRef] [PubMed]
- Dill, K.A. Additivity Principles in Biochemistry. J. Biol. Chem. 1997, 272, 701–704. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, S.; Oliva, R.; Graziano, G.; Del Vecchio, P. Counteraction of Denaturant-Induced Protein Unfolding Is a General Property of Stabilizing Agents. Phys. Chem. Chem. Phys. 2018, 20, 29389–29398. [Google Scholar] [CrossRef] [PubMed]
- Cozzolino, S.; Tortorella, A.; Del Vecchio, P.; Graziano, G. General Counteraction Exerted by Sugars against Denaturants. Life 2021, 11, 652. [Google Scholar] [CrossRef] [PubMed]
- Vigorita, M.; Cozzolino, S.; Oliva, R.; Graziano, G.; Del Vecchio, P. Counteraction Ability of TMAO toward Different Denaturing Agents. Biopolymers 2018, 109, e23104. [Google Scholar] [CrossRef] [PubMed]
- Graziano, G. On the Molecular Origin of Cold Denaturation of Globular Proteins. Phys. Chem. Chem. Phys. 2010, 12, 14245. [Google Scholar] [CrossRef]
- Graziano, G. On the Mechanism of Cold Denaturation. Phys Chem Chem Phys 2014, 16, 21755–21767. [Google Scholar] [CrossRef]
- Pica, A.; Graziano, G. Shedding Light on the Extra Thermal Stability of Thermophilic Proteins. Biopolymers 2016, 105, 856–863. [Google Scholar] [CrossRef]
- Reiss, H. Scaled Particle Methods in the Statistical Thermodynamics of Fluids. In Advances in Chemical Physics; Prigogine, I., Ed.; Wiley, 1965; Vol. 9, pp. 1–84 ISBN 978-0-470-69910-2.
- Lebowitz, J.L.; Helfand, E.; Praestgaard, E. Scaled Particle Theory of Fluid Mixtures. J. Chem. Phys. 1965, 43, 774–779. [Google Scholar] [CrossRef]
- Royer, C.A. Revisiting Volume Changes in Pressure-Induced Protein Unfolding. Biochim. Biophys. Acta BBA - Protein Struct. Mol. Enzymol. 2002, 1595, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Chalikian, T.V. Volumetric Properties of Proteins. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 207–235. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.R.; Makhatadze, G.I. Molecular Determinant of the Effects of Hydrostatic Pressure on Protein Folding Stability. Nat. Commun. 2017, 8, 14561. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Richards, F.M. The Interpretation of Protein Structures: Estimation of Static Accessibility. J. Mol. Biol. 1971, 55, 379–IN4. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Janin, J.; Lesk, A.M.; Chothia, C. Interior and Surface of Monomeric Proteins. J. Mol. Biol. 1987, 196, 641–656. [Google Scholar] [CrossRef]
- Creamer, T.P.; Srinivasan, R.; Rose, G.D. Modeling Unfolded States of Peptides and Proteins. Biochemistry 1995, 34, 16245–16250. [Google Scholar] [CrossRef] [PubMed]
- Kell, G.S. Density, Thermal Expansivity, and Compressibility of Liquid Water from 0.Deg. to 150.Deg.. Correlations and Tables for Atmospheric Pressure and Saturation Reviewed and Expressed on 1968 Temperature Scale. J. Chem. Eng. Data 1975, 20, 97–105. [Google Scholar] [CrossRef]
- Sorenson, J.M.; Hura, G.; Glaeser, R.M.; Head-Gordon, T. What Can X-Ray Scattering Tell Us about the Radial Distribution Functions of Water? J. Chem. Phys. 2000, 113, 9149–9161. [Google Scholar] [CrossRef]
- Graziano, G. Water: Cavity Size Distribution and Hydrogen Bonds. Chem. Phys. Lett. 2004, 396, 226–231. [Google Scholar] [CrossRef]
- Ben-Amotz, D.; Willis, K.G. Molecular Hard-Sphere Volume Increments. J. Phys. Chem. 1993, 97, 7736–7742. [Google Scholar] [CrossRef]
- Pauling, L. The Nature of the Chemical Bond and the Structure of Molecules and Crystals: An Introduction to Modern Structural Chemistry; 3. ed., 17. print.; Cornell Univ. Press: Ithaca, NY, 2010; ISBN 978-0-8014-0333-0. [Google Scholar]
- Baxa, M.C.; Haddadian, E.J.; Jumper, J.M.; Freed, K.F.; Sosnick, T.R. Loss of Conformational Entropy in Protein Folding Calculated Using Realistic Ensembles and Its Implications for NMR-Based Calculations. Proc. Natl. Acad. Sci. 2014, 111, 15396–15401. [Google Scholar] [CrossRef] [PubMed]
- Sharp, K.A.; O’Brien, E.; Kasinath, V.; Wand, A.J. On the Relationship between NMR-Derived Amide Order Parameters and Protein Backbone Entropy Changes: Measurement of Protein Backbone Entropy by NMR. Proteins Struct. Funct. Bioinforma. 2015, 83, 922–930. [Google Scholar] [CrossRef] [PubMed]
- Fogolari, F.; Corazza, A.; Fortuna, S.; Soler, M.A.; VanSchouwen, B.; Brancolini, G.; Corni, S.; Melacini, G.; Esposito, G. Distance-Based Configurational Entropy of Proteins from Molecular Dynamics Simulations. PLOS ONE 2015, 10, e0132356. [Google Scholar] [CrossRef] [PubMed]
- Rose, G.D. Reframing the Protein Folding Problem: Entropy as Organizer. Biochemistry 2021, 60, 3753–3761. [Google Scholar] [CrossRef] [PubMed]
- Rose, G.D. Protein Folding - Seeing Is Deceiving. Protein Sci. 2021, 30, 1606–1616. [Google Scholar] [CrossRef] [PubMed]
- Rees, D.C.; Robertson, A.D. Some Thermodynamic Implications for the Thermostability of Proteins. Protein Sci. 2001, 10, 1187–1194. [Google Scholar] [CrossRef]
- Zangi, R.; Zhou, R.; Berne, B.J. Urea’s Action on Hydrophobic Interactions. J. Am. Chem. Soc. 2009, 131, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Lindgren, M.; Westlund, P.-O. On the Stability of Chymotrypsin Inhibitor 2 in a 10 M Urea Solution. The Role of Interaction Energies for Urea-Induced Protein Denaturation. Phys. Chem. Chem. Phys. 2010, 12, 9358. [Google Scholar] [CrossRef]
- Paladino, A.; Balasco, N.; Graziano, G.; Vitagliano, L. A Protein Data Bank Survey of Multimodal Binding of Thiocyanate to Proteins: Evidence for Thiocyanate Promiscuity. Int. J. Biol. Macromol. 2022, 208, 29–36. [Google Scholar] [CrossRef]
- Paladino, A.; Balasco, N.; Vitagliano, L.; Graziano, G. A Structure-Based Mechanism for the Denaturing Action of Urea, Guanidinium Ion and Thiocyanate Ion. Biology 2022, 11, 1764. [Google Scholar] [CrossRef] [PubMed]
- Paladino, A.; Vitagliano, L.; Graziano, G. The Action of Chemical Denaturants: From Globular to Intrinsically Disordered Proteins. Biology 2023, 12, 754. [Google Scholar] [CrossRef] [PubMed]
- Collins, K.D. Sticky Ions in Biological Systems. Proc. Natl. Acad. Sci. 1995, 92, 5553–5557. [Google Scholar] [CrossRef]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to Measure and Predict the Molar Absorption Coefficient of a Protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef] [PubMed]
- Foglia, F.; Mandrich, L.; Pezzullo, M.; Graziano, G.; Barone, G.; Rossi, M.; Manco, G.; Del Vecchio, P. Role of the N-Terminal Region for the Conformational Stability of Esterase 2 from Alicyclobacillus Acidocaldarius. Biophys. Chem. 2007, 127, 113–122. [Google Scholar] [CrossRef]
- Zhou, Y.; Hall, C.K.; Karplus, M. The Calorimetric Criterion for a Two-State Process Revisited. Protein Sci. Publ. Protein Soc. 1999, 8, 1064–1074. [Google Scholar] [CrossRef]
- Chan, H.S. Modeling Protein Density of States: Additive Hydrophobic Effects Are Insufficient for Calorimetric Two-State Cooperativity. Proteins Struct. Funct. Genet. 2000, 40, 543–571. [Google Scholar] [CrossRef]
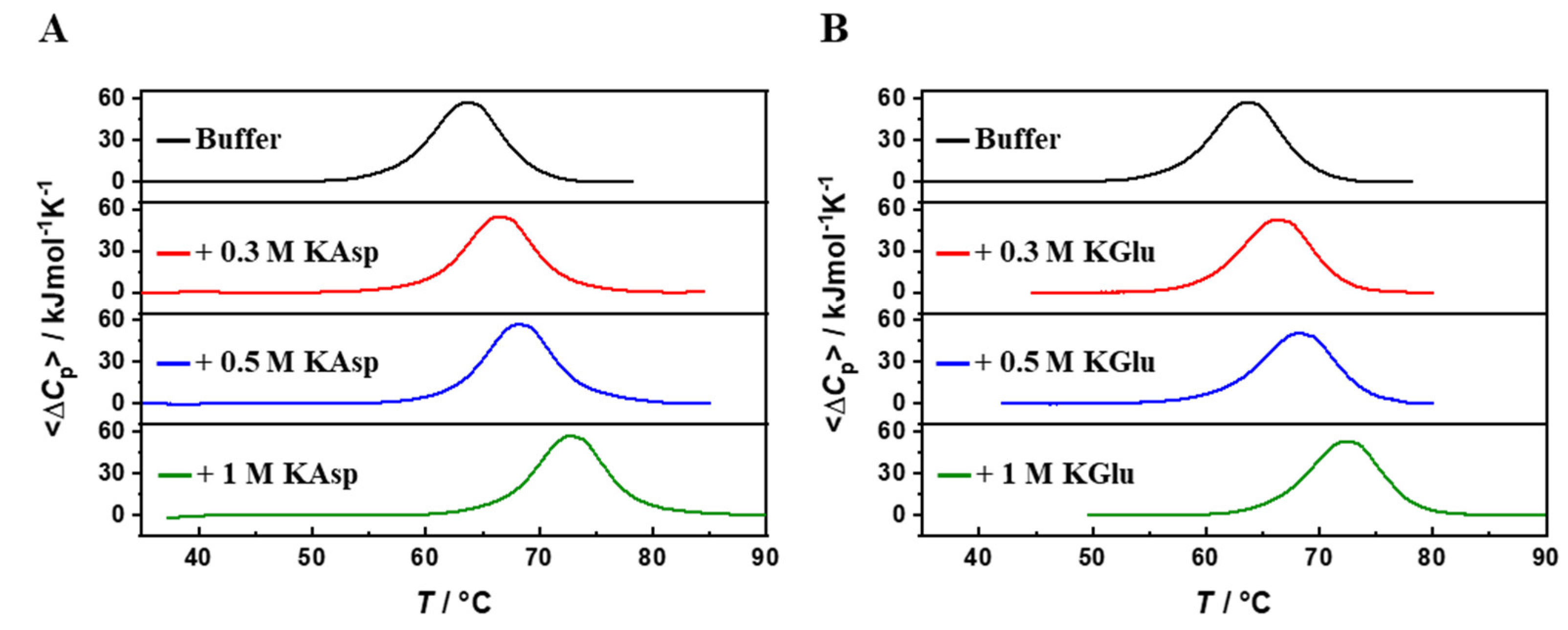
| Td/(°C) | ΔHd(Td) / kJ mol-1 | CU | ΔGd(25°C) / kJ mol-1 | |
|---|---|---|---|---|
| H2O | 63.5 | 464 | 0.99 | 38 |
| 0.3 M KAsp | 66.4 | 455 | 0.99 | 39 |
| 0.5 M KAsp | 68.2 | 480 | 0.97 | 42 |
| 1 M KAsp | 72.7 | 482 | 0.99 | 44 |
| 0.3 M KGlu | 66.3 | 485 | 0.97 | 42 |
| 0.5 M KGlu | 68.2 | 455 | 0.99 | 39 |
| 1 M KGlu | 72.2 | 480 | 0.97 | 44 |
| 1 M urea | 59.7 | 440 | 1.0 | 34 |
| 1 M KAsp + 1 M urea | 69.9 | 460 | 0.99 | 41 |
| 1 M KGlu + 1 M urea | 68.8 | 420 | 0.98 | 35 |
| 1 M GdmCl | 53.9 | 383 | 0.97 | 25 |
| 1 M KAsp + 1 M GdmCl | 64.9 | 400 | 0.99 | 31 |
| 1 M KGlu + 1 M GdmCl | 64.7 | 378 | 0.98 | 29 |
| 0.5 M GdmSCN | 47.9 | 320 | 0.96 | 17 |
| 1 M KAsp + 0.5 M GdmSCN | 58.0 | 352 | 0.96 | 24 |
| 1 M KGlu + 0.5 M GdmSCN | 57.9 | 360 | 0.97 | 25 |
| ρ / g L-1 | [H2O] / M | σ / Å | ξ3 | < σ> / Å | |
| H2O | 997 | 55.3 | 2.80 | 0.383 | 2.80 |
| 1 M KAsp | 1085 | 50.7 | 6.06 & 2.66 | 0.427 | 2.86 |
| 1 M K Glu | 1088 | 50.1 | 6.37 & 2.66 | 0.434 | 2.87 |
| 1 M urea | 1013 | 52.9 | 4.64 | 0.398 | 2.83 |
| 1 M KAsp + 1 M urea | 1099 | 48.2 | 6.06 & 2.66 & 4.64 | 0.441 | 2.90 |
| 1 M KGlu + 1 M urea | 1104 | 47.7 | 6.37 & 2.66 & 4.64 | 0.449 | 2.90 |
| 1 M GdmCl | 1024 | 51.5 | 4.70 & 3.62 | 0.404 | 2.85 |
| 1 M KAsp + 1 M GdmCl | 1109 | 46.7 | 6.06 & 2.66 & 4.70 & 3.62 | 0.447 | 2.91 |
| 1 M KGlu + 1 M GdmCl | 1112 | 46.1 | 6.37 & 2.66 & 4.70 & 3.62 | 0.458 | 2.92 |
| 0.5 M GdmSCN | 1011 | 52.8 | 4.70 & 3.94 | 0.392 | 2.83 |
| 1 M KAsp + 0.5 M GdmSCN | 1095 | 48.0 | 6.06 & 2.66 & 4.70 & 3.94 | 0.434 | 2.89 |
| 1 M KGlu + 0.5 M GdmSCN | 1101 | 47.5 | 6.37 & 2.66 & 4.70 & 3.62 | 0.442 | 2.90 |
| ΔGC(N) | ΔGC(D) | ΔΔGC | ΔΔGC’ | |
| H2O | 1072 | 1875 | 803 | - |
| 1 M urea | 1111 | 1942 | 831 | 28 |
| 1 M GdmCl | 1135 | 1984 | 849 | 46 |
| 0.5 M GdmSCN | 1085 | 1897 | 812 | 9 |
| 1 M KAsp | 1203 | 2102 | 899 | 96 |
| 1 M KAsp + 1 M urea | 1245 | 2176 | 931 | 128 |
| 1 M KAsp + 1 M GdmCl | 1269 | 2217 | 948 | 145 |
| 1 M KAsp + 0.5 M GdmSCN | 1207 | 2109 | 902 | 99 |
| 1 M KGlu | 1218 | 2128 | 910 | 107 |
| 1 M KGlu + 1 M urea | 1268 | 2215 | 947 | 144 |
| 1 M KGlu + 1 M GdmCl | 1323 | 2311 | 988 | 185 |
| 1 M KGlu + 0.5 M GdmSCN | 1228 | 2147 | 919 | 116 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





