1. Introduction
Plasmodium vivax is the second most important malaria pathogen in the world, being responsible for 2.8% of the 249 million malaria cases in 2022. Outside of Africa, it is the main
Plasmodium species, and it has the widest geographical distribution in the world. It causes significant morbidity in Southeast Asia, North and South America, and the Middle East [
1].
In the Americas, 72% of malaria cases are caused by
P. vivax, and four countries account for 80% of estimated cases in 2022: the Bolivarian Republic of Venezuela (28%), Brazil (27%), Colombia (18%) and Peru (6%) [
1].
In 2023, 142,522 malaria cases were reported in Brazil, of which 99.97% were autochthonous in the states of the legal Amazon region (Acre, Amapá, Amazonas, Maranhão, Mato Grosso, Pará, Rondônia, Roraima, and Tocantins), and 82.6% were caused by
P. vivax. In the same year, 34,555 cases were reported in Roraima, 71% of which were caused by
P. vivax [
2].
To achieve the goal of malaria elimination by 2035, the National Malaria Program (PNCM) envisages only
P. vivax transmission for the last five years after the elimination of malaria transmission by
P. falciparum in 2030 [
3]. However, the unique biology
of P. vivax poses additional challenges to the elimination target, such as relapses caused by hypnozoites, the ability to infect mosquitoes before symptoms appear, and asymptomatic infections in endemic areas [
4].
Adding to this context, the emergence of resistance to chloroquine (CQR), the drug of first choice for the elimination of the blood stages of
P. vivax, may represent a further obstacle to control strategies based on the use of this antimalarial drug [
5,
6]. Indeed, three decades after the emergence of
P. falciparum CQR, such resistance in
P. vivax was first reported in Papua New Guinea in 1980, and subsequent studies have shown an increase in this resistance, accompanied by reports of severe and fatal
P. vivax malaria in these regions [
7,
8].
In addition to Southeast Asia, particularly in Thailand [
9] and Cambodia [
10],
P. vivax CQR has been documented in East Africa, Ethiopia [
11,
12], South America, in part of the Guiana Shield and the Cooperative Republic of Guyana [
13].
In Brazil, CQR was first detected in 1999 in a patient in the city of Manaus in the state of Amazonas [
14]. In the same city, later studies reported a 10.1% failure rate for treatment with CQ in 2007 [
15] and 5.2% in 2014 [
16]. More recently, 1.1% treatment failure following supervised treatment with CQ and primaquine (PQ) was reported in the city of Oiapoque in the state of Amapá, on the border with French Guiana [
17].
The mechanisms of resistance in
P. vivax are still not fully understood, probably also because of the lack of continuous in vitro cultivation methods for this parasite species [
18,
19]. In this context, monitoring with molecular markers has emerged as a practical and cost-effective field tool for antimalarial drug resistance monitoring compared to
in vivo and
in vitro tests [
18].
Molecular markers that can be considered for the prediction of malaria resistance phenotype in
P. vivax have been identified based on the orthologs resistant-related genes in
P. falciparum [
20,
21]. Thus, the multidrug resistance gene 1 of
P. vivax (
pvmdr1) and the CQ resistance transporter gene (
pvcrt-o) are orthologous to the
pfmdr1 and
pfcrt genes in
P. falciparum [
18,
23]. Alterations in the
pvmdr1 sequences are thought to confer CQR by reducing the transport of CQ into the digestive vacuole (DV), where the parasite digests host cell proteins and converts hemoglobin heme to nontoxic hemozoin [
24].
The mutant
pvcrt, in turn, would act as an efflux pump in the active transport of CQ out of the DV and away from its target (the process of converting heme to hemozoin) [
18]. The lysine insertion (AAG) in the first exon (amino acid 10), referred to as the K10 insertion in the
pvcrt-o gene, would be associated with a reduction of half of the maximum inhibitory concentration of CQ (IC50) and has been identified as a possible molecular marker for CQR in
P. vivax [
25,
26]. Concerning
pvmdr1, the amino acid mutations Y976F, F1076L, and T958M have been linked to CQR [
21,
26,
27].
In Roraima, the malaria burden in the state has increased, especially since 2018, due to the increased migration flow from Venezuela and Guyana, together with the boost of illegal mining in the Yanomami indigenous land. In the same period, there has been an increase in hospitalizations and deaths by
P. vivax malaria [
28], and it is known that the clinical severity of malaria could be related to the appearance of CQR [
29].
Since little is known about the P. vivax genotypes of pvmdr1 and pvcrt-o genes circulating in the parasites of Brazilian endemic areas, such as those of Roraima, this study aimed to investigate the polymorphism of these genes in regions with a great influx of people.
2. Materials and Methods
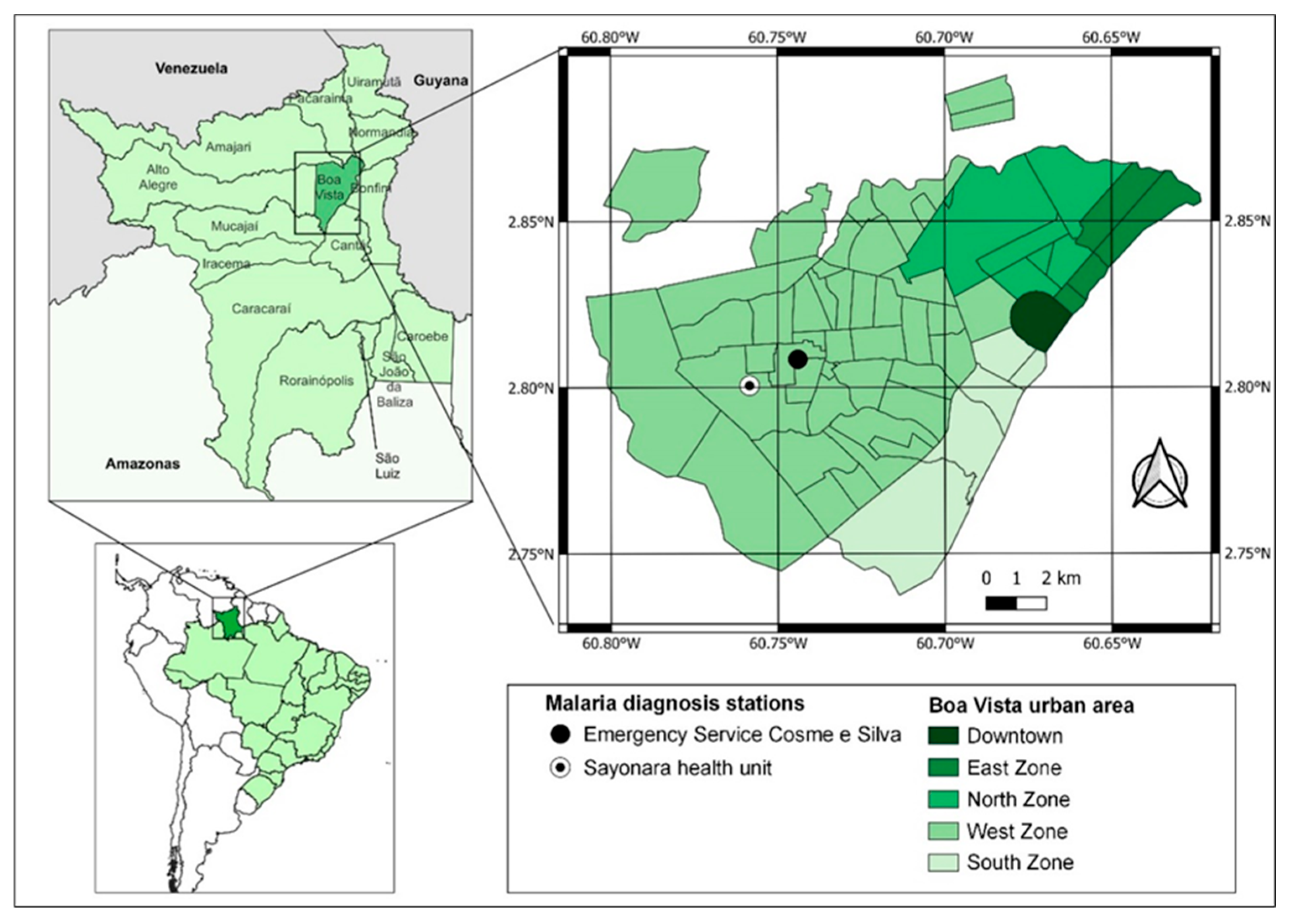
This study was approved by the Research Ethics Committee of the Federal University of Roraima CAAE 24122619.6.0000.5302 (CEP/UFRR, acronym in Portuguese; opinion n. 3,920,373, issued on March 17, 2020). The CEP/UFRR allowed only the inclusion of non-village Indigenous people who speak Brazilian Portuguese and reside in Boa Vista. Additionally, the research project was demanded to be presented to the Kannu Kadan Indigenous Association for obtaining a letter of consent, which was attached to the submission process to the CEP/UFRR. The samples were collected at the Emergency Service Cosme e Silva and Sayonara Health Unit from December 2021 to June 2022, due to the seasonal increase in the number of malaria cases in Roraima during this period. Both health centers are located in the west zone of the city and have the highest number of malaria reports according to the Malaria Epidemiological Surveillance Information System (Sivep-Malaria) (
Figure 1).
Individuals over the age of 18 who had been diagnosed with P. vivax malaria or mixed malaria (P. vivax + P. falciparum) and had been diagnosed through thick blood smears were included. The non-inclusion of individuals under 18 years old does not represent a limitation of the study because the considerable increase of malaria cases occurred in mining areas where children under 18 years of age do not have access. Individuals who could not read or refused to sign the free and informed consent form (TCLE) were also excluded from the study. After signing the consent form, an epidemiologic questionnaire with questions about the person and malaria was completed.
Blood was collected by venipuncture of 5 ml of peripheral blood. A portion of the blood (approximately 50 microliters) was transferred directly from the syringe to filter paper (Whatman 903 Protein Saver Cards) and the remainder to a Vacutainer tube (Becton, Dickinson & Company) containing EDTA.
All participants were treated according to the National Malaria Control Program (PNCM) protocol for non-severe malaria, which includes the administration of a combination of CQ for 3 days (10 mg/kg on day 1 and 7.5 mg/kg on days 2 and 3) and PQ for 7 days (0.5 mg/kg/day) [
30].
According to the PNCM, cure control should be assessed through the cure verification slide (CVS) on days 3, 7, 14, 21, 28, 42, and 63 after the start of treatment, according to the operational capacity of the local health network, and collections on D3 and D28 should be prioritized for
P. vivax infections. The day on which the diagnosis is made and treatment begins is considered day zero (D0) [
31,
32].
To identify the CVS of the participants who were infected by parasites with target mutations, a search was carried out in Sivep-Malaria for one year before and one year after the date of sample collection. To investigate malaria hospitalization cases after diagnosis, a search was carried out on Sivep-Malaria at the Notification Unit of the Roraima General Hospital, a state reference for severe malaria. The deaths were investigated by a search carried out on the Mortality Information System (SIM).
Relapse or recurrence was considered the reappearance of asexual parasitemia with or without symptoms after treatment due to: i) recrudescence (incomplete clearance of asexual parasites after antimalarial treatment within 28 days); ii) relapse (arising from hypnozoites between 28 and 60 days) or; iii) reinfection (after 60 days) [
31,
32].
The blood samples collected in vacutainer tubes and on filter paper were transported to the Molecular Biology Laboratory (LaBMol) of the Center for Biodiversity Studies (CBio) at the Federal University of Roraima (UFRR). The samples collected in the tubes were centrifuged at 3000 x g for 10 minutes to remove the plasma and the cryopreservation solution glycerolyte 57 (Baxter) was added to the "red blood cell concentrates" (containing leukocytes and platelets) volume by volume (v/v), followed by aliquoting each sample. The aliquots with the cryopreservation solution were stored at - 20°C in racks and packed in individual labeled plastic bags until the deoxyribonucleic acid (DNA) was extracted.
DNA extraction was carried out using the column technique (centrifugation method), using the QIAamp DNA blood mini kit (Qiagen), according to the manufacturer's instructions, from a volume of 500µL of the sample.
The methodology used for the PCR (polymerase chain reaction) of the
pvmdr1 gene was according to the protocol previously described, with the following primers: F: 5'-ATAGTCATGCCCCAGGATTG-3' and R: 5'-ACCGTTGGTCTGGACAAGTAT-3' [
33].
A mixture of PCR reagents was prepared for a final volume of 50 µL, with 26.75 µL of ultrapure water, 6 µL of MgCl2 (25mM), 5 µL of PCR Buffer II (10x), 5 µL of deoxynucleotide triphosphates (dNTPs) (8mM), 1 µL of each primer (10pmol), 0.25 µL of AmpliTaq Gold DNA Polymerase (250U). Finally, 5 µL of DNA was added to each mixture. The PCR conditions in the thermal cycler included: initiation at 95°C for 10 minutes; 40 cycles with denaturation at 94°C for 15 seconds, primer annealing at 60°C for 30 seconds; extension at 72°C for 1 minute and final elongation at 72°C for 7 minutes. The final product was a 762pb fragment amplified for analysis of SNPs T958M, Y976F, and F1076L in the pvmdr1 gene.
The PCR reactions to amplify the
pvcrt-o gene was based on the protocol previously described, with the following primers: F: 5'- AAGAGCCCGTCTAGCCAT CC- 3' and R: 5'- AGTTTCCCTCTACAC CCG-3' [
21]. To the reagent mixture with a final volume of 42 µL, 23.75 µL of ultrapure water, 4 µL of MgCl2 (25mM), 4 µL of PCR Buffer II (10x), 2 µL of deoxynucleotide triphosphates (dNTPs) (8nM), 2 µL of each of the primers (10pmol), 0.25 µL of AmpliTaq Gold DNA Polymerase (250UI) were added. 4 µL of DNA was added to each PCR reagent mixture. The PCR conditions in the thermal cycler were initial heating at 95°C for 10 minutes; followed by 35 cycles with denaturation at 94°C for 30 seconds, primer annealing at 61°C for 1 minute, extension at 72°C for 1 minute and; final elongation at 72°C for 7 minutes. The final product of the PCR reaction (amplicon) generated a fragment of 1186 bp.
To obtain the target sequences of the isolates, the PCR products were purified using the Wizard® Kit, according to the manufacturer's instructions. The sequencing reaction was performed using the Big DyeTM Terminator Cycle Sequencing Ready Reaction version 3.1 kit (Applied Biosystems) and 3.2 pmol of forward and reverse primers were used separately in the reactions.
To investigate SNPs in the pvmdr1 and pvcrt-o genes, the sense and antisense sequences of the samples and the reference sequence were aligned and analyzed with a ClustalW multiple sequence aligner in BioEdit software version 7.7.1 (North Carolina State University, Raleigh, USA). The Salvador 1 (Sal-1) strain was used as the reference sequence (GenBank Accession No. AF314649.1 for pvcrt-o and No. AY571984.1 for pvmdr1).
The information from the questionnaires and the results of the laboratory analysis were entered and tabulated in the Excel program (Microsoft Office®). The maps were drawn up using the QGIS program version 3.28.10. Mining areas in Roraima were obtained from Mapbiomas [
34]. Geopolitical limits of Brazil and Indigenous Lands were accessed on the IBGE website [
35].
3. Results
Samples were collected from 164 participants, of whom 153 had P. vivax and 11 (7%) had mixed malaria (P. vivax + P. falciparum).
The participants ranged in age from 18 to 67, and 82% (135/164) were men. The most commonly reported symptoms were fever 91.5% (150/164), headache 86% (141/164), chills 67% (110/164), abdominal pain 55% (90/164), and nausea/vomiting 43% (70/164). Detailed epidemiological information on these patients has recently been published [
36].
Regarding the main activity carried out in the 15 days before the onset of symptoms, 96% (157/164) of the participants reported gold mining, mainly in the municipalities of Alto Alegre, with 76% (120/157) of the participants, and Mucajaí with 18.5% (29/157) of them. Agriculture accounted for 3% (5/164) of the participants, with 40% (2/5) in the municipality of Mucajaí and 20% (1/5) in Alto Alegre, Cantá, and Caroebe municipalities. Hunting/fishing and tourism, both with a percentage of 0.6% (1/164), occurred in participants from the municipalities of Alto Alegre and Mucajaí, respectively (
Table 1).
The
pvcrt-o gene was amplified in 94% (154/164) of the samples and 99% (151/154) of the amplified products were sequenced. Of all the samples sequenced for the
pvcrt-o gene, 87% (131/151) were identical to the Sal 1 strain, which was used as a wild-type reference for CQ-sensitive parasites. The lysine insertion (codon AAG) at position 10, called the K10 insertion, was identified in 13% (20/151) of the sequenced samples. The K10 insertion was detected in 25% (1/4) of the samples from participants engaged in agriculture, 100% (1/1) of those engaged in hunting/fishing, and 12% (18/145) of those engaged in illegal mining (
Table 2). Despite the small number of samples, the K10 insertion was more frequent in prospectors (18) than in farmers (1) or hunters/fishermen (1).
The two participants who carried parasites with a K10 insertion in the
pvcrt-o gene performed farming activities (1) and hunting/fishing activities (1) in the municipalities of Mucajaí and Alto Alegre, respectively, while the three farmer participants who carried parasites without a K10 insertion were probably infected in the municipalities of Cantá, Caroebe, and Mucajaí. The only participant who reported tourism carried parasites without a K10 insertion in the
pvcrt-o gene, and the probable place of infection was the municipality of Mucajaí (
Table 1 and
Table 2).
Of all the participants who reported mining activities in the Yanomami indigenous area of Roraima, the municipality of Alto Alegre ranked first, with 14% (15/109) of the parasites having a K10 insertion. The second-ranked municipality was Mucajaí, where 7% (2/28) of
P. vivax samples showed K10 insertion. The only sample from a mining site in the municipality of Amajarí had no K10 insertion (
Table 1 and
Table 2).
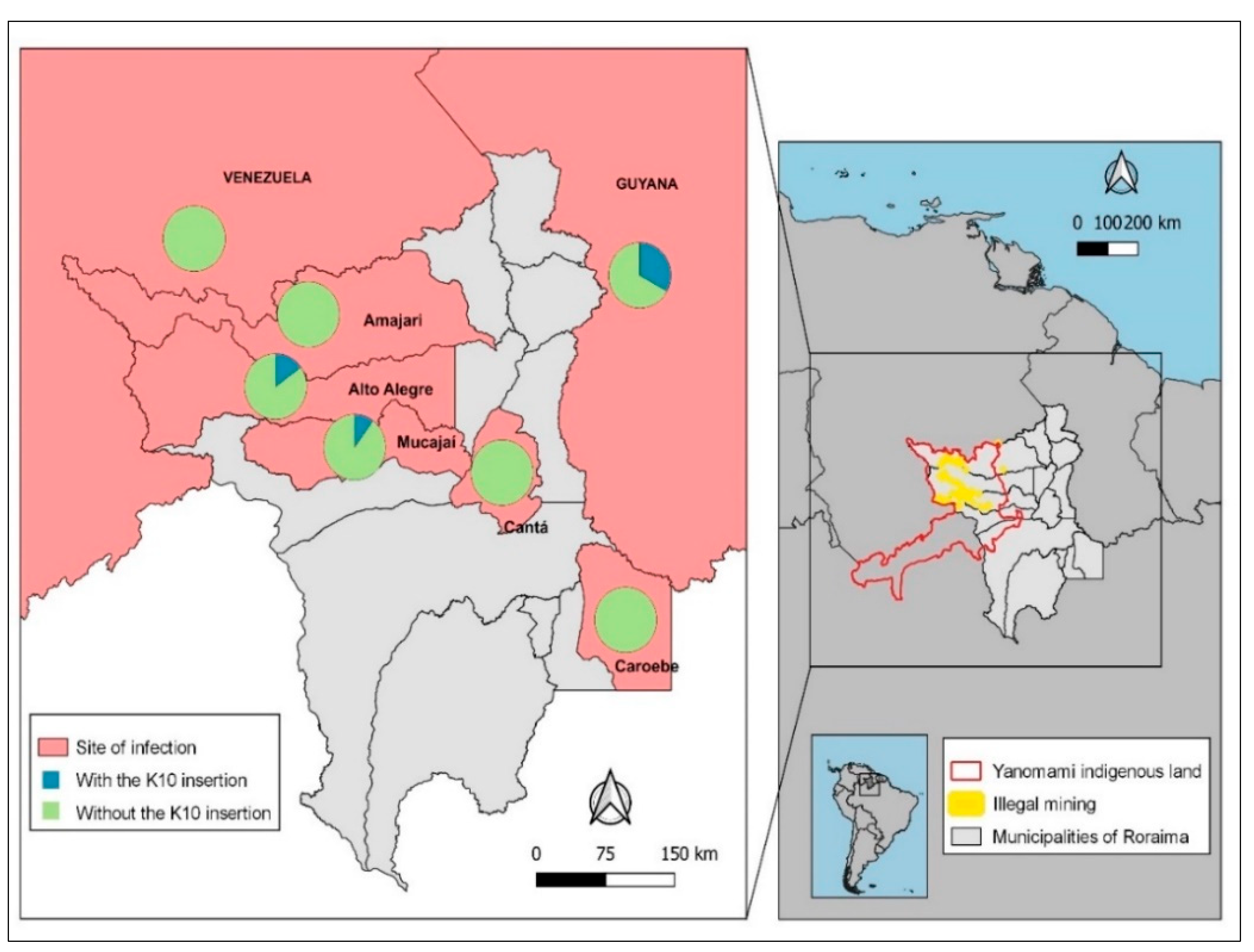
Among the 110 individuals infected in Alto Alegre, 15% (16/110) had parasites carrying a K10 insertion. Of all samples with probable infection in Mucajaí, 10% (3/31) had a K10 insertion. No K10 insertion was detected in the four samples from the municipalities of Amajarí, Cantá and Caroebe. K10 was also not detected in samples of the four participants infected in Venezuela, but it was noted in 1/3 of the samples from participants infected in Guyana (
Figure 2). In short, the K10 mutant was present in Alto Alegre, Mucajaí and Guyana and seems to be more fixed in Alto Alegre.
The
pvmdr1 gene was sequenced in 80 samples. Mutations in the entire fragment were analyzed, including the 3 codons (T958
M, F1076
L, and Y976
F) potentially associated with the phenotype of RCQ in
P. vivax. The T958
M mutation (
MYF haplotype), in other words, threonine replaced by methionine at codon 958, was found in 92.5% (74/80) of the sequenced samples, while the wild-type TYF haplotype was identified in 7.5% (6/80) of the samples. The F1076
L and Y976
F mutations were absent in all the samples (
Table 3).
When relating the sequenced samples of the
pvmdr1 gene and the main activity carried out by the patient in the 15 days before the onset of symptoms, the only participant who reported agriculture carried a parasite with the single mutation
MYF haplotype (958M) from the municipality of Caroebe. The only participant who reported hunting/fishing carried a parasite with the wild TYF haplotype from the municipality of Alto Alegre. Concerning gold mining, 94% (73/78) of the participants carried parasites with the
MYF haplotype from the municipality of Alto Alegre (55), Mucajaí and the neighboring country Venezuela (1) and 6% (5/78) with the wild TYF haplotype from Alto Alegre (3) and Mucajaí (2) (
Table 3 and
Figure 4).
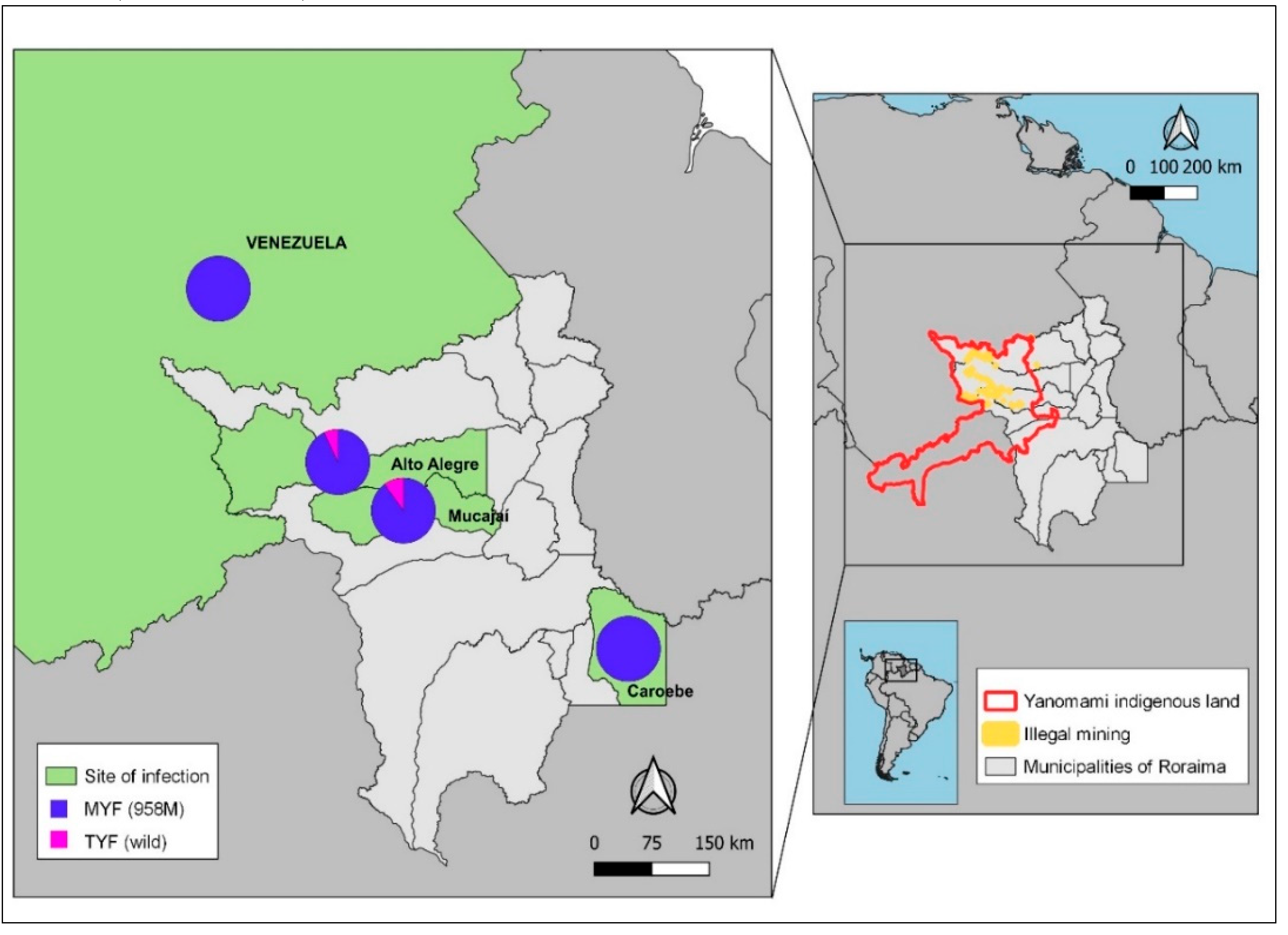
The distribution of the
pvmdr1 gene haplotypes by probable site of infection reveals that the
MYF haplotype is predominant in the municipalities of Alto Alegre (93%; 55/59) and Mucajaí (90%). The only sample from the municipality of Caroebe and neighboring Venezuela also had the
MYF haplotype. The wild TYF haplotype was only present in a small number of samples from Mucajaí (11%) and Alto Alegre (5%) (
Figure 3).
Regarding mining activities, the parasite haplotype was the
MYF in the only sample from Venezuela. In parasites transmitted in mining Yanomami indigenous areas of the Alto Alegre, 95% (55/58) of the samples carried the
MYF haplotype. Similarly, in infections from the Mucajaí, the MYF haplotype predominated (89%; 17/19) (
Figure 3).
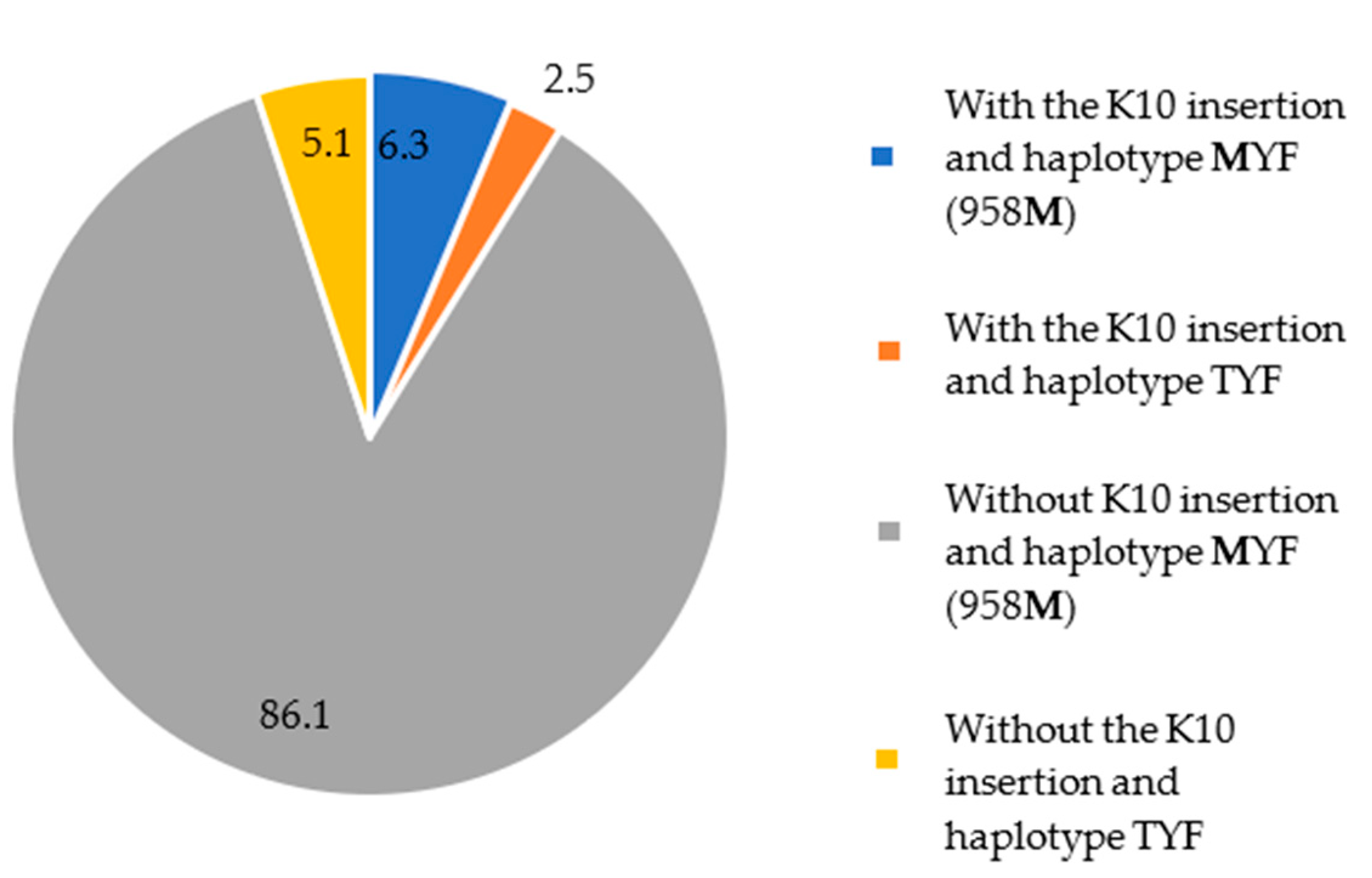
A total of 79 samples were sequenced for both the pvcrt-o and pvmdr1 genes. In addition to parasites with a single mutation in pvcrt-o or pvmdr1, five patients were infected with parasites carrying double mutants: insertion of K10 in the pvcrt-o gene and MYF (958M) haplotype in the pvmdr1 gene. These patients reported mining activities in the 15 days preceding symptoms in the municipalities of Mucajaí (1) and Alto Alegre (4).
These data showed that around 86% (68/79) of the parasite samples carried the
MYF haplotype and lacked the K10 insertion and clearly show that the
MYF mutant parasites are present in all studied municipalities, in contrast to the K10 insertion (
Figure 4).
No case notifications in the 28 days preceding or following the date of sample collection of the 18 patients carrying parasites with the pvcrt-o K10 insertion were registered in Sivep-Malaria, showing, therefore, no recrudescence episodes. However, probable cases of relapse in 11% (2/18) and reinfections in 39% (7/18) of those who reported mining activities were recorded (
Table 4).
None of these participants were hospitalized or died during the study. The use of antimalarials as prophylaxis was narrated by only one participant with mining in the municipality of Mucajaí (
Table 4).
We also searched for registers in Sivep-Malaria of participants carrying parasites with the MYF haplotype of the pvmdr1 gene within 28 days before or after the date of sample collection, and no cases in the recrudescence period were observed in these patients. However, among them, the miner patients had 12% (9/73) cases of probable relapses and 29% (21/73) of reinfections. There were no records of hospitalization or death among these participants. Only 12% (9/73) of the participants with mining activities in the municipalities of Alto Alegre (7) and Mucajaí (2) reported use of antimalarials as prophylaxis.
4. Discussion
The emergence of CQR in
P. vivax presents a significant challenge for the elimination of malaria in Brazil by 2035. This species is responsible for the largest malaria burden in the legal Amazon. Moreover, the complex biology of
P. vivax and the limited availability of laboratory research tools make it difficult to identify cases of antimalarial resistance in this parasite. Monitoring molecular markers to identify mutations related to antimalarial resistance over time can provide essential information to identify effective treatment policies and help determine the change of first-line drugs according to the local reality [
22].
Detecting
P. vivax CQR is complex due to the difficulty in distinguishing whether relapses or recurrences of the disease are due to relapse (related to hypnozoites), recrudescence (related to antimalarial resistance), or reinfections [
7]. This difficulty is greater when monitoring occurs in individuals who carry out mining activities, as they return often to the place of infection. The illegal mining sites in Roraima are located in isolated areas of forest in the Yanomami Indigenous land, which comprises the municipalities of Amajarí, Alto Alegre, Mucajaí, Iracema and Caracaraí. In these locations, the miners have no access to the Brazilian Health Unic System (SUS) healthcare diagnosis network [
28]. When they travel to Boa Vista for malaria diagnosis and treatment, the gold miners return to the miners immediately after receiving diagnosis and antimalarial drugs, making it impossible to monitor the clinical efficacy of CQ through the negativity of parasitemia (CVS).
Thus, to identify the cure and whether there had been any progression to the severe form of the disease, resulting in hospitalization or death, we consulted the SUS Health Information Systems (SIS), the Mortality Information System (SIM) and SIVEP-Malaria. This search proved to be an important strategy for investigation integrating research with local surveillance and minimizing SUS costs for monitoring the patients.
Few single nucleotide polymorphisms (SNPs) have been reported in the
pvcrt gene, unlike
pfcrt its ortholog in
P. falciparum. The most common polymorphism in the
pvcrt gene is a lysine insertion (codon AAG) at position 10 (K10), which was proposed to be associated to CQR [
22,
27,
37].
In this study, we found a K10 insertion in the pvcrt-o gene in 13% (20/151) of the sequenced parasite samples, and no recrudescence, hospitalization, or death episodes in the patients infected with parasites carrying this mutation was noticed. Interestingly, the K10 insertion was only identified in parasites from locations with intense mining activity, such as Alto Alegre and Mucajaí, in Roraima and the neighboring country of Guyana, suggesting that this mutant is becoming established in these areas because of an intense flow of individuals from different areas and possibly reflecting the greater diversity of P. vivax in areas with greater transmission.
In previous studies with patients from Acre, Amazonas, Amapá, Pará, Rondônia and Roraima, the K10 insertion was not also associated with CQR [
38]. The same was true in Manaus, where the K10 insertion was not related to in vitro
P. vivax resistance to CQ [
39], and in French Guiana, where no polymorphism in
pvcrt-o and
pvmdr1 genes was identified in CQR parasites [
40].
In Southeast Asia parasites, the K10 insertion has been observed in prevalences ranging from 9.4% in India to 72% in Myanmar. However, no association between the presence of K10 insertion and in vitro
P. vivax resistance to CQ was shown [
41,
42,
43]. In fact, unlike what happens with
P. falciparum, polymorphisms in
pvcrt do not seem to be good molecular markers for monitoring CQR [
38,
40].
Regarding the
pvmdr1 gene, in patients carrying parasites with the T958M mutation (92%), no recrudescence or progression to severe malaria resulting in hospitalizations and deaths was identified through research into the search platforms. This finding is supported by other studies which have shown that the T958M mutation allele is a majority in parasite populations in endemic areas of Brazil, French Guiana, Asia, Pakistan, Afghanistan, Sri Lanka, Nepal, Sudan, São Tomé and Ecuador and that its presence is not associated with CQR [
18,
21,
38,
40,
41,
44,
45,
46,
47].
The double mutation haplotype with amino acid changes in Y976F and F1076L, in particular, has already been cited as a possible marker of resistance to CQ [
33,
47]. However, this haplotype can also be found in regions with no reported cases of CQR, making its association with drug resistance uncertain and, consequently, its applicability as a marker of chemoresistance of
P. vivax to CQ unprovable [
38,
47].
In the present study, no haplotypes with a double mutation profile (T958M + F1076L or Y976F + F1076L) were found in Roraima, Venezuela, or Guyana. Double mutants have not been previously identified in Roraima, although they have been found in patients from the state of Amazonas who responded well to treatment with CQ [
38].
Parasites from French Guiana may have double T958M/F1076L and triple T958M/Y976F/F1076L mutated haplotypes in the
pvmdr1 gene [
45]. However, the association between clinical response to the drug and/or in vitro susceptibility has never been demonstrated.
These data seem to suggest that the presence of these mutations in the pvcrt-o and pvmdr1 genes are due to the remarkable genetic diversity of P. vivax and that these polymorphisms have no implications for the phenotype of CQR parasites.
Over two decades of research into the resistance of
P. vivax to antimalarials using molecular markers orthologous to
P. falciparum, the findings have shown no or a weak relationship with resistance to CQ, potentially leading to false conclusions that could have an impact on national policy for the treatment of the disease [
22].
There are marked differences in the topologies and number of SNPs in the
crt-o and
mdr1 genes between
P. vivax and
P. falciparum, which reinforces the idea that other genes may be involved in the CQR phenotype in
P. vivax. Therefore, understanding the molecular mechanisms of antimalarial resistance in
P. vivax and investigating candidate genes to monitor CQR through ex vivo assays and sequencing could help identify genes other than these
P. falciparum orthologs [
22,
38].
Despite the comparable selection pressure from the massive use of CQ,
P. vivax CQR was only reported in 1989, whereas in
P. falciparum it has been evident since the late 1950s. This can be explained by the differences in genetic determinants and molecular mechanisms of CQR in
P. falciparum and
P. vivax parasites [
48,
49] beyond the lower parasite biomass, the gametocytes production at the beginning of the infection, and the recurrence of hepatic hypnozoites. These conditions allow the parasite to be transmitted before the start of treatment or after the concentration of the drug has decreased. Furthermore, it has been suggested that the use of PQ may have the potential to reduce the transmission of CQR parasites [
50].
Understanding the evolutionary and population dynamics of antimalarial resistance in
P. vivax will be crucial for strengthening molecular surveillance, both to identify when these alleles arise and to understand how they move through and between populations [
22,
51]. This is probably especially true in the state of Roraima, where the dynamics of migratory flows and mining activities make malaria elimination an even more challenging goal.
Similar to Brazil, French Guiana is experiencing intense gold mining and human migration between countries in the Guiana Shield. This raises concerns about the spread of CQR
P. vivax isolates, making surveillance and detection of resistant parasites critical [
45].
5. Conclusions
The results of this study corroborate that mutations in the pvcrt-o and pvmdr-1 genes have no predictive potential of the CQR P. vivax phenotype in Brazilian endemic areas. The molecular mechanisms of antimalarial resistance in P. vivax and the difference in the evolutionary dynamics of the P. vivax and P. falciparum populations suggest that molecular markers associated with P. vivax chemoresistance to CQ may lie beyond the P. falciparum orthologues.
Author Contributions
Conceptualization: JAB, MFFC, FG.; methodology: JAB, DSS, RAF, LTQ, MFFC, FG; software: JAC, AC; validation: JAB, DSS, RAF, LTQ, MFFC, FG; formal analysis: JAB, DSS, RAF, LTQ, MFFC, FG, NKAOM; investigation: JAB, DSS, MFFC; resources: MFFC, CTDR, FG; data curation: JAB, DSS, AC; writing—original draft preparation: JAC, MFFC, FG; writing—review and editing: JAB, DSS, RAF, LTQ, MFFC, FG, NKAOM, CTDR; visualization: MFFC, FG, CDTR; supervision: MFFC, FG; project administration: JAB, MFFC, FG; funding acquisition: MFFC.
Funding
This work was funded by the National Council for Scientific and Technological Development (CNPq), the Department of Science and Technology in Health/Ministry of Health (DECIT/MS), the Oswaldo Cruz Foundation (Fiocruz), the Carlos Chagas Filho Foundation for Research Support in the State of Rio de Janeiro (FAPERJ), the Health Surveillance Secretariat/Ministry of Health (SVS/MS) and the Federal University of Roraima (UFRR). MFFC and CTDR are supported by CNPq, Brazil, through a Research Productivity Grant, and are "Cientistas do Nosso Estado" by Faperj.
Data Availability Statement
Sequences were deposited in Genbank™ with pvmdr1 accession number PP693801- PP693880 and pvcrt-o accession number PP681704 - PP681857.
Acknowledgments
We would like to thank all the participants who agreed to take part in this study. Hugo Almeida for your support in collecting samples. Jener Franco and José Carlos Nascimento for transporting the team to the field. For their logistical support, we would like to thank the Boa Vista Municipal Malaria Coordination, the General Health Surveillance Coordination, the Epidemiological Surveillance Department and the State Malaria Control Center. To the students and technical staff at LaBMol/CBio/UFRR and the Malaria Research Laboratory/Fiocruz for their support in laboratory analyses and to the microscopists in the malaria rooms at the Sayonara Health Unit and Cosme e Silva Emergency Room.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- World malaria report 2023. Geneva: World Health Organization; 2023. License: ISBN 978-92-4-008617-3.
- Brazil. Ministry of Health. Data for citizens from the Sivep-Malaria, Sinan and E- SUS-VS data sources, for notifications in Brazil from 2007 to 2023. Available online: https://public.tableau.com/app/profile/mal.ria.brasil/viz/Dadosparacidado_20 1925_03_2020/Incio. Accessed on 10 Nov 2023.
- Brazil. Ministry of Health. Health Surveillance Secretariat. Department of Immunization and Communicable Diseases. Eliminate Malaria Brazil: National Malaria Elimination Plan. Brasília, 2022. 60pp.
- Adams JH, Mouller I. The Biology of Plasmodium vivax. In: Malaria: Biology in the Era of Eradication. Cold Spring Harbor, New York, 2017, pp. 43-54.
- Commons RJ, Thriemer K, Humphreys G, et al. The Vivax Surveyor: Online mapping database for Plasmodium vivax clinical trials. International Journal for parasitology. Drugs and Drug Resistance,. [CrossRef]
- Negreiros S, Farias S, Viana GM, et al. Efficacy of Chloroquine and Primaquine for the Treatment of Uncomplicated Plasmodium vivax Malaria in Cruzeiro do Sul, Brazil. Am J Trop Med Hyg., 1068. [CrossRef]
- Rieckmann KH, Davis DR, Hutton DC. Plasmodium vivax resistance to chloroquine? Lancet, 1183. [CrossRef]
- Price RN, von Seidlein L, Valecha N, Nosten F, Baird JK, White NJ. Global extent of chloroquine-resistant Plasmodium vivax: a systematic review and meta- analysis. Lancet Infect Dis, 982–91. [CrossRef]
- Phyo AP, Lwin KM, Price RN, et al. Dihydroartemisininpiperaquine versus chloroquine in the treatment of Plasmodium vivax malaria in Thailand: a randomized controlled trial. Clin Infect Dis, 977–84.
- Leang R, Barrette A, Mey Bouth D, et al. Effi cacy of dihydroartemisinin- piperaquine for the treatment of uncomplicated Plasmodium falciparum and Plasmodium vivax in Cambodia, 2008-2010. Antimicrob Agents Chemother.
- Yohannes AM, Teklehaimanot A, Bergqvist Y, Ringwald P. Confi rmed vivax resistance to chloroquine and eff ectiveness of artemether-lumefantrine for the treatment of vivax malaria in Ethiopia. Am J Trop Med Hyg, 84.
- Ketema T, Getahun K, Bacha K. Therapeutic effi cacy of chloroquine for treatment of Plasmodium vivax malaria cases in Halaba district, South Ethiopia. Parasit Vectors, 4–46.
- Phillips EJ, Keystone JS, Kain KC. Failure of combined chloroquine and high-dose primaquine therapy for Plasmodium vivax malaria acquired in Guyana, South America. Clin Infect Dis, 1171.
- Alecrim MG, Alecrim W, Macêdo V. Plasmodium vivax resistance to chloroquine (R2) and mefloquine (R3) in Brazilian Amazon region. Rev Soc Bras Med Trop.,. [CrossRef]
- De-Santana-Filho FS, Arcanjo AR, Chehuan YM, Costa MR, Martinez-Espinosa FE, Vieira JL, et. al. Chloroquine-resistant Plasmodium vivax, Brazilian Amazon. Emerg Infect Dis., 1125. [CrossRef] [PubMed]
- Marques MM, Costa MR, Santana Filho FS, Vieira JL, Nascimento MT, Brasil LW, et al. Plasmodium vivax chloroquine resistance and anemia in the western Brazilian Amazon. Antimicrob Agents Chemother, 342–7.
- Gomes LR, Lavigne A, Brasil P, Peterka CL, Ménard D, Daniel-Ribeiro CT, Ferreira-da-Cruz MF. Lack of quadruple and quintuple mutant alleles associated with sulfadoxine-pyrimethamine resistance in Plasmodium vivax isolates from Brazilian endemic areas. Mem Inst Oswaldo Cruz,. [CrossRef]
- Ferreira, MU, Nobrega-de-Sousa, T, Rangel, GW, Johansen, IC, Corder, RM, Ladeia-Andrade, S, et. al. Monitoring Plasmodium vivax resistance to antimalarials: Persisting challenges and future directions. International journal for parasitology. Drugs and drug resistance. 2021, 15, 9–24. [Google Scholar] [CrossRef]
- Ibrahim A, Manko E, Dombrowski JG, Campos M, Benavente ED, Nolder D, et. al. Population-based genomic study of Plasmodium vivax malaria in seven Brazilian states and across South America. Lancet Reg Health Am. [CrossRef]
- Sánches, PO. Genetic diversity in Plasmodium vivax populations: analysis of neutral genetic markers and genes potentially associated with drug resistance. 50 p. Thesis (Doctorate in Sciences). University of São Paulo. 2010.
- Huang, F. , Li, S., Tian, P. et al. Genetic polymorphisms in genes associated with drug resistance in Plasmodium vivax parasites from northeastern Myanmar. Malar J. [CrossRef]
- Buyon LE, Elsworth B, Duraisingh MT. The molecular basis of antimalarial drug resistance in Plasmodium vivax. Int J Parasitol Drugs Drug Resist, 23–37. [CrossRef]
- Cui L, Mharakurwa S, Ndiaye D, Rathod PK, Rosenthal PJ. Antimalarial Drug Resistance: Literature Review and Activities and Findings of the ICEMR Network. Am J Trop Med Hyg, 93. [CrossRef]
- Veiga, M. , Dhingra, S., Henrich, P. et al. Globally prevalent PfMDR1 mutations modulate Plasmodium falciparum susceptibility to artemisinin-based combination therapies. Nat Commun, 2016, v. 7, p. 11553. [CrossRef]
- Melo GC, Monteiro WM, Siqueira AM, Silva SR, Magalhães BM, Alencar AC, Kuehn A, del Portillo HA, Fernandez-Becerra C, Lacerda MV. Expression levels of pvcrt-o and pvmdr-1 are associated with chloroquine resistance and severe Plasmodium vivax malaria in patients of the Brazilian Amazon. PLoS One. [CrossRef]
- Stanley P, Rajkumari N, Sivaradjy M. Molecular detection of antimalarial resistance in Plasmodium vivax isolates from a tertiary care setting in Puducherry. Indian J Med Microbiol, 1004. [CrossRef]
- Tantiamornkul K, Pumpaibool T, Piriyapongsa J, Culleton R, Lek-Uthai U. The prevalence of molecular markers of drug resistance in Plasmodium vivax from the border regions of Thailand in 2008 and 2014. Int J Parasitol Drugs Drug Resist, 229–237. [CrossRef]
- De-Aguiar-Barros J, Granja F, Pequeno P, Marchesini P, Ferreira da Cruz MF. Gold miners increase malaria transmission in indigenous territories of Roraima state, Brazil. Malar J, 21. [CrossRef]
- Baird, JK. Neglect of Plasmodium vivax malaria. Trends Parasitol. 2007, v. 23, p. 533-9. [CrossRef]
- Brazil. Ministry of Health. Health Surveillance Secretariat. Department of Immunization and Communicable Diseases. Guide to malaria treatment in Brazil. 2nd ed. Brasília: 2020.
- Brazil. Ministry of Health. Health Surveillance Secretariat. Malaria. In: Guia de Vigilância em Saúde, Brasília; 2023. 921-953.
- Simões LR, Alves ER Jr, Ribatski-Silva D, Gomes LT, Nery AF, Fontes CJ. Factors associated with recurrent Plasmodium vivax malaria in Porto Velho, Rondônia State, Brazil, 2009. Cad Saude Publica. 1403. [CrossRef]
- Brega S, Meslin B, De Monbrison F, Severini C, Gradoni L; Udomsangpetch, R., et. al. Identification of the Plasmodium vivax Mdr-like Gene (Pvmdr1) and Analysis of Single-Nucleotide Polymorphisms among Isolates from Different Areas of Endemicity. J. Infect. Dis.
- MapBiomas Project – Collection 7.0 of the 2021 Annual Series of Land Use and Coverage Maps of Brazil. Available in: https://brasil.mapbiomas.org/colecoes-mapbiomas/. Accessed on, 2023.
- IBGE - Brazilian Institute of Geography and Statistics. Vectors of geopolitical limits, of Indigenous Lands, Available in: https://www.ibge.gov.br/geociencias/downloads-geociencias.html. Accessed on: 01/09/2023.
- Barros JA, Granja F, Silva DS, Citó AC, Peterka C, Ferreira-da-Cruz MF. A snapshot of a representative Brazilian state of illegal mining in indigenous areas during the era of malaria elimination. Cad de Saude Publica 2024, v 40. Available in: https://cadernos.ensp.fiocruz.br/ojs/index.php/csp/article/view/8629. [CrossRef]
- Nyunt MH, Han JH, Wang B, Aye KM, Aye KH, Lee SK, Htut Y, et al. Clinical and molecular surveillance of drug resistant vivax malaria in Myanmar (2009-2016). Malar J, 16. [CrossRef]
- Abreu-Fernandes, R.d.; Almeida-de-Oliveira, N.K.; de Lavigne Mello, A.R.; Queiroz, L.T.d.; Barros, J.d.A.; Baptista, B.D.O.; et al. Are pvcrt-o and pvmdr1 Gene Mutations Associated with Plasmodium vivax Chloroquine-Resistant Parasites? Biomedicines,. [CrossRef]
- Silva, S.R. , Almeida, A.C.G., da Silva, G.A.V. et al. Chloroquine resistance is associated to multi-copy pvcrt-o gene in Plasmodium vivax malaria in the Brazilian Amazon. Malar J. [CrossRef]
- Musset L, Heugas C, Naldjinan R, Blanchet D, Houze P, Abboud P, et al. Emergence of Plasmodium vivax resistance to chloroquine in French Guiana. Antimicrob Agents Chemother. [CrossRef]
- Zhao, Yan, et al. "Molecular surveillance for drug resistance markers in Plasmodium vivax isolates from symptomatic and asymptomatic infections at the China-Myanmar border." Malaria Journal, 2020, v. 19, p. 1-12. [CrossRef]
- Tantiamornkul K, Pumpaibool T, Piriyapongsa J, Culleton R, Lek-Uthai U. The prevalence of molecular markers of drug resistance in Plasmodium vivax from the border regions of Thailand in 2008 and 2014. Int J Parasitol Drugs Drug Resist. [CrossRef]
- Rungsihirunrat K, Muhamad P, Chaijaroenkul W, Kuesap J, Na-Bangchang K. Plasmodium vivax drug resistance genes; Pvmdr1 and Pvcrt-o polymorphisms in relation to chloroquine sensitivity from a malaria endemic area of Thailand. Korean J Parasitol. 2015, v. 53, p. 43-49. [CrossRef]
- Gomes MS, Vieira JL, Machado RL, Nacher M, Stefani A, Musset L, et. al. Efficacy in the treatment of malaria by Plasmodium vivax in Oiapoque, Brazil, on the border with French Guiana: the importance of control over external factors. Malar J 14, 402 (2015). [CrossRef]
- Faway, E. , Musset, L., Pelleau, S. et al. Plasmodium vivax multidrug resistance-1 gene polymorphism in French Guiana. Malar J, 2016,. [CrossRef]
- Orjuela-Sánchez, Pamela, et al. "Analysis of single-nucleotide polymorphisms in the crt-o and mdr1 genes of Plasmodium vivax among chloroquine-resistant isolates from the Brazilian Amazon region." Antimicrob Agents Chemother. 2009, v. 53, p. 3561-4. [CrossRef]
- Schousboe ML, Ranjitkar S, Rajakaruna RS, Amerasinghe PH, Morales F, Pearce R, et. al. Multiple Origins of Mutations in the mdr1 Gene--A Putative Marker of Chloroquine Resistance in P. vivax. PLoS Negl Trop Dis. 2015, v. 9. [CrossRef]
- Spotin, M. , Mahami-Oskouei E., Ahmadpour M., Parsaei A., Rostami S., et. al. Global assessment of genetic paradigms of Pvmdr1 mutations in chloroquine-resistant Plasmodium vivax isolates. Trans. R. Soc. Trop. Med. Hyg.,. [CrossRef]
- Nomura T, Carlton JM, Baird JK, del Portillo HA, Fryauff DJ, Rathore D, et al. Evidence for Different Mechanisms of Chloroquine Resistance in 2 Plasmodium Species That Cause Human Malaria. J Infect Dis, 1653. [CrossRef]
- Escalante, A.A. , Cepeda, A.S. & Pacheco, M.A. Why are Plasmodium vivax and Plasmodium falciparum so different? A tale of two clades and their species diversities. Malar J. [CrossRef]
- Schneider KA, Escalante AA. "Fitness components and natural selection: why are there different patterns on the emergence of drug resistance in Plasmodium falciparum and Plasmodium vivax?" Malaria journal, 2013, v. 12, p. 1-11. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).