Submitted:
31 July 2024
Posted:
01 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
Sample Preparation, Virus Isolation and Sequencing
Phylogenetic Analysis
3. Results
Case Presentation
Assembly and Phylogenetic Position of Imported DENV-1 Genome
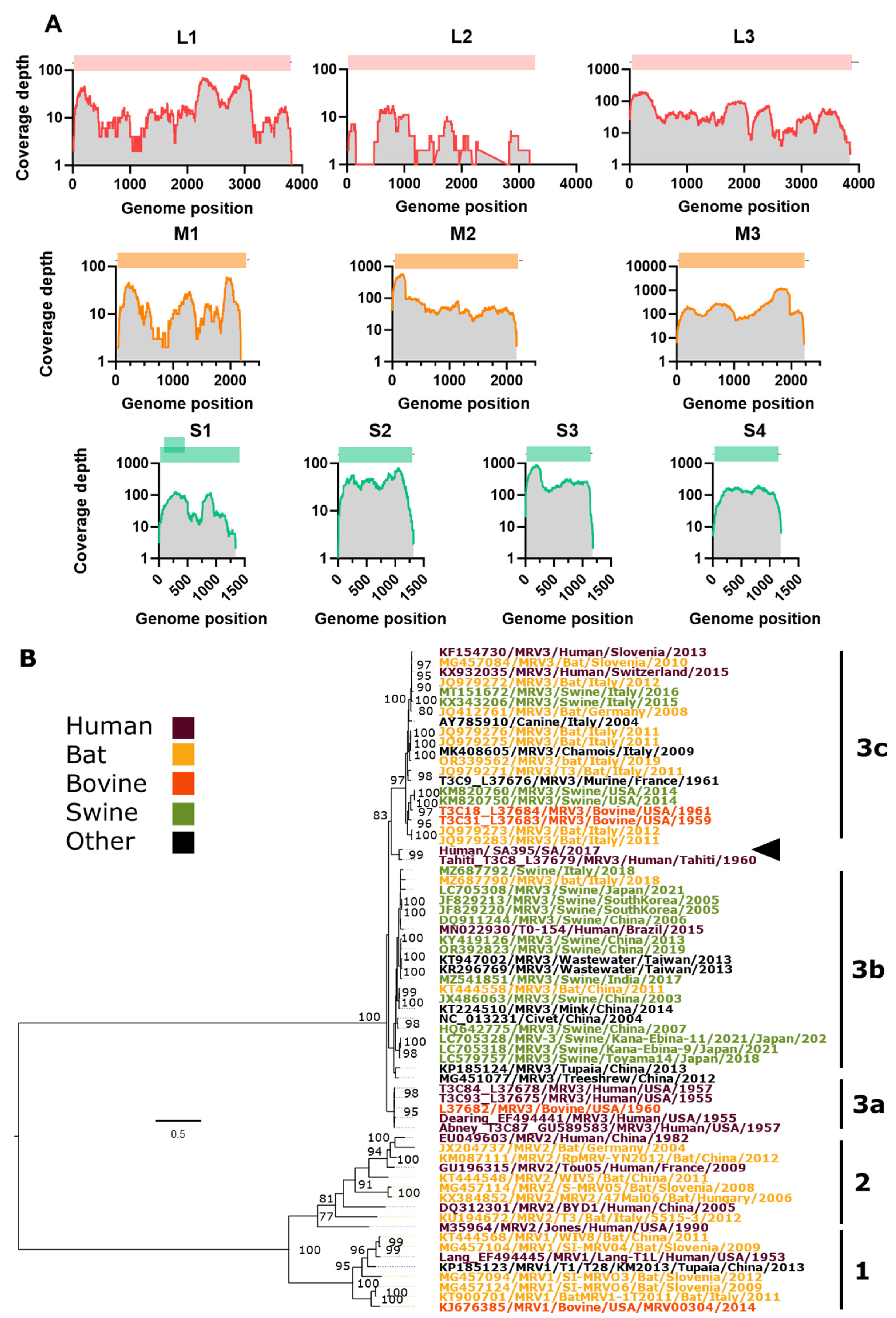
Assembly and Phylogenetic Position of Imported MRV Genome
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; Myers, M.F.; George, D.B.; Jaenisch, T.; Wint, G.R.; Simmons, C.P.; Scott, T.W.; Farrar, J.J.; Hay, S.I. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Zhan, J.; Chen, L.; Chen, H.; Cheng, S. Global, regional, and national dengue burden from 1990 to 2017: A systematic analysis based on the global burden of disease study 2017. EClinicalMedicine 2021, 32, 100712. [Google Scholar] [CrossRef] [PubMed]
- Oishi, K.; Saito, M.; Mapua, C.A.; Natividad, F.F. Dengue illness: clinical features and pathogenesis. J Infect Chemother 2007, 13, 125–133. [Google Scholar] [CrossRef]
- Low, J.G.; Ong, A.; Tan, L.K.; Chaterji, S.; Chow, A.; Lim, W.Y.; Lee, K.W.; Chua, R.; Chua, C.R.; Tan, S.W.; Cheung, Y.B.; Hibberd, M.L.; Vasudevan, S.G.; Ng, L.C.; Leo, Y.S.; Ooi, E.E. The early clinical features of dengue in adults: challenges for early clinical diagnosis. PLoS Negl Trop Dis 2011, 5, e1191. [Google Scholar] [CrossRef]
- Kraemer, M.U.; Sinka, M.E.; Duda, K.A.; Mylne, A.Q.; Shearer, F.M.; Barker, C.M.; Moore, C.G.; Carvalho, R.G.; Coelho, G.E.; Van Bortel, W.; Hendrickx, G.; Schaffner, F.; Elyazar, I.R.; Teng, H.J.; Brady, O.J.; Messina, J.P.; Pigott, D.M.; Scott, T.W.; Smith, D.L.; Wint, G.R.; Golding, N.; Hay, S.I. The global distribution of the arbovirus vectors Aedes aegypti and Ae. albopictus. Elife 2015, 4, e08347. [Google Scholar] [CrossRef]
- Msimang, V.; Kemp, A.; Jansen van Vuren, P.; Weyer, J.; Paweska, J.T. Increased importation of dengue cases into South Africa: a risk for establishment of local endemicity? NICD The National Institute For Communicable Diseases: 01-04-2018, 2018. [Google Scholar]
- Matthijnssens, J.; Attoui, H.; Banyai, K.; Brussaard, C.P.D.; Danthi, P.; Del Vas, M.; Dermody, T.S.; Duncan, R.; Fang, Q.; Johne, R.; Mertens, P.P.C.; Mohd Jaafar, F.; Patton, J.T.; Sasaya, T.; Suzuki, N.; Wei, T. ICTV Virus Taxonomy Profile: Spinareoviridae 2022. J Gen Virol 2022, 103, 11. [Google Scholar] [CrossRef] [PubMed]
- Attoui, H.; Biagini, P.; Stirling, J.; Mertens, P.P.; Cantaloube, J.F.; Meyer, A.; de Micco, P.; de Lamballerie, X. Sequence characterization of Ndelle virus genome segments 1, 5, 7, 8, and 10: evidence for reassignment to the genus Orthoreovirus, family Reoviridae. Biochem Biophys Res Commun 2001, 287, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Ooms, L.S.; Kobayashi, T.; Dermody, T.S.; Chappell, J.D. A post-entry step in the mammalian orthoreovirus replication cycle is a determinant of cell tropism. J Biol Chem 2010, 285, 41604–41613. [Google Scholar] [CrossRef]
- Ichikawa, A.; Katayama, M.; Lai, H.; Wataru, S.; Takenaka-Uema, A.; Horimoto, T.; Murakami, S. Isolation and genetic characterization of a mammalian orthoreovirus from Vespertilio sinensis in Japan. Arch Virol 2023, 168, 165. [Google Scholar] [CrossRef]
- Lim, M.C.; Wang, Y.F.; Huang, S.W.; Yang, J.Y.; Wang, J.R. High Incidence of Mammalian Orthoreovirus Identified by Environmental Surveillance in Taiwan. PLoS One 2015, 10, e0142745. [Google Scholar] [CrossRef]
- Sedmak, G.; Bina, D.; Macdonald, J.; Couillard, L. Nine-year study of the occurrence of culturable viruses in source water for two drinking water treatment plants and the influent and effluent of a Wastewater Treatment Plant in Milwaukee, Wisconsin (August 1994 through July 2003). Appl Environ Microbiol 2005, 71, 1042–1050. [Google Scholar] [CrossRef] [PubMed]
- Jansen van Vuren, P.; Wiley, M.; Palacios, G.; Storm, N.; McCulloch, S.; Markotter, W.; Birkhead, M.; Kemp, A.; Paweska, J.T. Isolation of a Novel Fusogenic Orthoreovirus from Eucampsipoda africana Bat Flies in South Africa. Viruses 2016, 8, 65. [Google Scholar] [CrossRef]
- Chua, K.B.; Voon, K.; Crameri, G.; Tan, H.S.; Rosli, J.; McEachern, J.A.; Suluraju, S.; Yu, M.; Wang, L.F. Identification and characterization of a new orthoreovirus from patients with acute respiratory infections. PLoS One 2008, 3, e3803. [Google Scholar] [CrossRef] [PubMed]
- Chua, K.B.; Voon, K.; Yu, M.; Keniscope, C.; Abdul Rasid, K.; Wang, L.F. Investigation of a potential zoonotic transmission of orthoreovirus associated with acute influenza-like illness in an adult patient. PLoS One 2011, 6, e25434. [Google Scholar] [CrossRef]
- Giordano, M.O.; Martinez, L.C.; Isa, M.B.; Ferreyra, L.J.; Canna, F.; Pavan, J.V.; Paez, M.; Notario, R.; Nates, S.V. Twenty year study of the occurrence of reovirus infection in hospitalized children with acute gastroenteritis in Argentina. Pediatr Infect Dis J 2002, 21, 880–882. [Google Scholar] [CrossRef] [PubMed]
- Mikuletic, T.; Steyer, A.; Kotar, T.; Zorec, T.M.; Poljak, M. A novel reassortant mammalian orthoreovirus with a divergent S1 genome segment identified in a traveler with diarrhea. Infect Genet Evol 2019, 73, 378–383. [Google Scholar] [CrossRef]
- Rosa, U.A.; Ribeiro, G.O.; Villanova, F.; Luchs, A.; Milagres, F.A.P.; Komninakis, S.V.; Tahmasebi, R.; Lobato, M.; Brustulin, R.; Chagas, R.T.D.; Abrao, M.; Soares, C.; Tinker, R.J.; Pandey, R.P.; Raj, V.S.; Sabino, E.C.; Deng, X.; Delwart, E.; Costa, A.C.D.; Leal, E. First identification of mammalian orthoreovirus type 3 by gut virome analysis in diarrheic child in Brazil. Sci Rep 2019, 9, 18599. [Google Scholar] [CrossRef] [PubMed]
- Steyer, A.; Gutierrez-Aguire, I.; Kolenc, M.; Koren, S.; Kutnjak, D.; Pokorn, M.; Poljsak-Prijatelj, M.; Racki, N.; Ravnikar, M.; Sagadin, M.; Fratnik Steyer, A.; Toplak, N. High similarity of novel orthoreovirus detected in a child hospitalized with acute gastroenteritis to mammalian orthoreoviruses found in bats in Europe. J Clin Microbiol 2013, 51, 3818–3825. [Google Scholar] [CrossRef] [PubMed]
- Lewandowska, D.W.; Capaul, R.; Prader, S.; Zagordi, O.; Geissberger, F.D.; Kugler, M.; Knorr, M.; Berger, C.; Gungor, T.; Reichenbach, J.; Shah, C.; Boni, J.; Zbinden, A.; Trkola, A.; Pachlopnik Schmid, J.; Huber, M. Persistent mammalian orthoreovirus, coxsackievirus and adenovirus co-infection in a child with a primary immunodeficiency detected by metagenomic sequencing: a case report. BMC Infect Dis 2018, 18, 33. [Google Scholar] [CrossRef] [PubMed]
- Ouattara, L.A.; Barin, F.; Barthez, M.A.; Bonnaud, B.; Roingeard, P.; Goudeau, A.; Castelnau, P.; Vernet, G.; Paranhos-Baccala, G.; Komurian-Pradel, F. Novel human reovirus isolated from children with acute necrotizing encephalopathy. Emerg Infect Dis 2011, 17, 1436–1444. [Google Scholar] [CrossRef]
- Tyler, K.L.; Barton, E.S.; Ibach, M.L.; Robinson, C.; Campbell, J.A.; O’Donnell, S.M.; Valyi-Nagy, T.; Clarke, P.; Wetzel, J.D.; Dermody, T.S. Isolation and molecular characterization of a novel type 3 reovirus from a child with meningitis. J Infect Dis 2004, 189, 1664–1675. [Google Scholar] [CrossRef] [PubMed]
- Lelli, D.; Beato, M.S.; Cavicchio, L.; Lavazza, A.; Chiapponi, C.; Leopardi, S.; Baioni, L.; De Benedictis, P.; Moreno, A. First identification of mammalian orthoreovirus type 3 in diarrheic pigs in Europe. Virol J 2016, 13, 139. [Google Scholar] [CrossRef] [PubMed]
- Naglic, T.; Rihtaric, D.; Hostnik, P.; Toplak, N.; Koren, S.; Kuhar, U.; Jamnikar-Ciglenecki, U.; Kutnjak, D.; Steyer, A. Identification of novel reassortant mammalian orthoreoviruses from bats in Slovenia. BMC Vet Res 2018, 14, 264. [Google Scholar] [CrossRef] [PubMed]
- Kohl, C.; Lesnik, R.; Brinkmann, A.; Ebinger, A.; Radonic, A.; Nitsche, A.; Muhldorfer, K.; Wibbelt, G.; Kurth, A. Isolation and characterization of three mammalian orthoreoviruses from European bats. PLoS One 2012, 7, e43106. [Google Scholar] [CrossRef] [PubMed]
- Colombino, E.; Lelli, D.; Canziani, S.; Quaranta, G.; Guidetti, C.; Leopardi, S.; Robetto, S.; De Benedictis, P.; Orusa, R.; Mauthe von Degerfeld, M.; Capucchio, M.T. Main causes of death of free-ranging bats in Turin province (North-Western Italy): gross and histological findings and emergent virus surveillance. BMC Vet Res 2023, 19, 200. [Google Scholar] [CrossRef]
- Qin, P.; Li, H.; Wang, J.W.; Wang, B.; Xie, R.H.; Xu, H.; Zhao, L.Y.; Li, L.; Pan, Y.; Song, Y.; Huang, Y.W. Genetic and pathogenic characterization of a novel reassortant mammalian orthoreovirus 3 (MRV3) from a diarrheic piglet and seroepidemiological survey of MRV3 in diarrheic pigs from east China. Vet Microbiol 2017, 208, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Thimmasandra Narayanappa, A.; Sooryanarain, H.; Deventhiran, J.; Cao, D.; Ammayappan Venkatachalam, B.; Kambiranda, D.; LeRoith, T.; Heffron, C.L.; Lindstrom, N.; Hall, K.; Jobst, P.; Sexton, C.; Meng, X.J.; Elankumaran, S. A novel pathogenic Mammalian orthoreovirus from diarrheic pigs and Swine blood meal in the United States. mBio 2015, 6, e00593–15. [Google Scholar] [CrossRef] [PubMed]
- Beaty, B.J.; Calisher, C.H.; Shope, R.E. Arboviruses. In: Diagnostic procedures for viral, rickettsial, and chlamydial infections, 7th ed. Washington (DC): American Public Health Association; 1995. p. 189–212.
- Jansen van Vuren, P.; Parry, R.; Khromykh, A.A.; Paweska, J.T. A 1958 Isolate of Kedougou Virus (KEDV) from Ndumu, South Africa, Expands the Geographic and Temporal Range of KEDV in Africa. Viruses 2021, 13, 7. [Google Scholar] [CrossRef]
- Djikeng, A.; Halpin, R.; Kuzmickas, R.; Depasse, J.; Feldblyum, J.; Sengamalay, N.; Afonso, C.; Zhang, X.; Anderson, N.G.; Ghedin, E.; Spiro, D.J. Viral genome sequencing by random priming methods. BMC Genomics 2008, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Parry, R.; James, M.E.; Asgari, S. Uncovering the Worldwide Diversity and Evolution of the Virome of the Mosquitoes Aedes aegypti and Aedes albopictus. Microorganisms 2021, 9, 8. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nature biotechnology 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Hill, V.; Cleemput, S.; Fonseca, V.; Tegally, H.; Brito, A.F.; Gifford, R.; Tran, V.T.; Kien, D.T.H.; Huynh, T.; Yacoub, S.; Dieng, I.; Ndiaye, M.; Balde, D.; Diagne, M.M.; Faye, O.; Salvato, R.; Wallau, G.L.; Gregianini, T.S.; Godinho, F.M.S.; Vogels, C.B.F.; Breban, M.I.; Leguia, M.; Jagtap, S.; Roy, R.; Hapuarachchi, C.; Mwanyika, G.; Giovanetti, M.; Alcantara, L.C.J.; Faria, N.R.; Carrington, C.V.F.; Hanley, K.A.; Holmes, E.C.; Dumon, W.; de Oliveira, T.; Grubaugh, N.D. A new lineage nomenclature to aid genomic surveillance of dengue virus. medRxiv 2024, 2024.05.16.24307504.
- Vilsker, M.; Moosa, Y.; Nooij, S.; Fonseca, V.; Ghysens, Y.; Dumon, K.; Pauwels, R.; Alcantara, L.C.; Vanden Eynden, E.; Vandamme, A.M.; Deforche, K.; de Oliveira, T. Genome Detective: an automated system for virus identification from high-throughput sequencing data. Bioinformatics 2019, 35, 871–873. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, V.; Libin, P.J.K.; Theys, K.; Faria, N.R.; Nunes, M.R.T.; Restovic, M.I.; Freire, M.; Giovanetti, M.; Cuypers, L.; Nowe, A.; Abecasis, A.; Deforche, K.; Santiago, G.A.; Siqueira, I.C.; San, E.J.; Machado, K.C.B.; Azevedo, V.; Filippis, A.M.B.; Cunha, R.V.D.; Pybus, O.G.; Vandamme, A.M.; Alcantara, L.C.J.; de Oliveira, T. A computational method for the identification of Dengue, Zika and Chikungunya virus species and genotypes. PLoS Negl Trop Dis 2019, 13, e0007231. [Google Scholar] [CrossRef] [PubMed]
- Siew, Z.Y.; Loh, A.; Segeran, S.; Leong, P.P.; Voon, K. Oncolytic Reoviruses: Can These Emerging Zoonotic Reoviruses Be Tamed and Utilized? DNA Cell Biol 2023, 42, 289–304. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol Biol Evol 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Bohl, J.A.; Lay, S.; Chea, S.; Ahyong, V.; Parker, D.M.; Gallagher, S.; Fintzi, J.; Man, S.; Ponce, A.; Sreng, S.; Kong, D.; Oliveira, F.; Kalantar, K.; Tan, M.; Fahsbender, L.; Sheu, J.; Neff, N.; Detweiler, A.M.; Yek, C.; Ly, S.; Sath, R.; Huch, C.; Kry, H.; Leang, R.; Huy, R.; Lon, C.; Tato, C.M.; DeRisi, J.L.; Manning, J.E. Discovering disease-causing pathogens in resource-scarce Southeast Asia using a global metagenomic pathogen monitoring system. Proc Natl Acad Sci U S A 2022, 119, e2115285119. [Google Scholar] [CrossRef] [PubMed]
- Poltep, K.; Phadungsombat, J.; Nakayama, E.E.; Kosoltanapiwat, N.; Hanboonkunupakarn, B.; Wiriyarat, W.; Shioda, T.; Leaungwutiwong, P. Genetic Diversity of Dengue Virus in Clinical Specimens from Bangkok, Thailand, during 2018-2020: Co-Circulation of All Four Serotypes with Multiple Genotypes and/or Clades. Trop Med Infect Dis 2021, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Dermody, T.S.; Nibert, M.L.; Bassel-Duby, R.; Fields, B.N. Sequence diversity in S1 genes and S1 translation products of 11 serotype 3 reovirus strains. J Virol 1990, 64, 4842–4850. [Google Scholar] [CrossRef]
- He XiaoMing He, X.; Yao HuoChun Yao, H.; Zhang HongBiao Zhang, H.; Lin Tao Lin, T.; Yuan ShiShan Yuan, S.; Long JinXue Long, J.; Ding Chan Ding, C. Isolation and identification of a strain of porcine reovirus serotype 1 in China. 2013, 43, 22–29.
- Wang, L.; Fu, S.; Cao, L.; Lei, W.; Cao, Y.; Song, J.; Tang, Q.; Zhang, H.; Feng, Y.; Yang, W.; Liang, G. Isolation and identification of a natural reassortant mammalian orthoreovirus from least horseshoe bat in China. PLoS One 2015, 10, e0118598. [Google Scholar] [CrossRef] [PubMed]
- Lo, V.T.; Yoon, S.W.; Noh, J.Y.; Jang, S.S.; Na, W.; Song, D.; Jeong, D.G.; Kim, H.K. Characterization of replication and variations in genome segments of a bat reovirus, BatMRV/B19-02, by RNA-seq in infected Vero-E6 cells. Arch Virol 2022, 167, 2133–2142. [Google Scholar] [CrossRef]
- Rosen, L.; Hovis, J.F.; Mastrota, F.M.; Bell, J.A.; Huebner, R.J. Observations on a newly recognized virus (Abney) of the reovirus family. Am J Hyg 1960, 71, 258–265. [Google Scholar] [PubMed]
- Hrdy, D.B.; Rosen, L.; Fields, B.N. Polymorphism of the migration of double-stranded RNA genome segments of reovirus isolates from humans, cattle, and mice. J Virol 1979, 31, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, H.; Wang, J.; Wang, S.; Wu, J.; Zhou, Y.; Wang, X.; Luo, F.; Tu, X.; Chen, Q.; Huang, Y.; Ju, W.; Peng, X.; Rao, J.; Wang, L.; Jiang, N.; Ai, J.; Zhang, W. Emergence and Autochthonous Transmission of Dengue Virus Type I in a Low-Epidemic Region in Southeast China. Front Cell Infect Microbiol 2021, 11, 638785. [Google Scholar] [CrossRef] [PubMed]
- Kemp, A.; Jupp, P.G. Potential for dengue in South Africa: mosquito ecology with particular reference to Aedes aegypti. J Am Mosq Control Assoc 1991, 7, 574–583. [Google Scholar] [PubMed]
- Jupp, P.G.; Kemp, A. The potential for dengue in South Africa: vector competence tests with dengue 1 and 2 viruses and 6 mosquito species. Trans R Soc Trop Med Hyg 1993, 87, 639–643. [Google Scholar] [CrossRef] [PubMed]

| Date of blood collection | Case # | Age | Sex | Symptoms | Clinical features and complications |
|---|---|---|---|---|---|
| 17-July-2017 | SA395 | 46 | M | Onset (date not recorded): Fever (38.7°C) without rash, headache, severe body aches | No blood hematology test results were reported. The patients were not hospitalized. No previous history of dengue or yellow fever vaccination. All patients reported mosquito bites. |
| SA396 | 8 | F | Onset (date not recorded): Fever (38.5°C) without rash, severe body aches, nausea | ||
| SA397 | 45 | F | Onset (16/07/2017): Fever (39°C) without rash, severe body aches, nausea |
| Segment Protein |
Nucleotide (nt), Amino acid (aa) length |
Closest strain, GenbankID, (%) | MRV Serotype |
Location, Date | Host | Ref |
|---|---|---|---|---|---|---|
| L1 λ3 protein |
3822nt 1267aa |
SHR-A JX415466 88.57% |
MRV-1 | China, 2011 |
Pig | [43] |
| L2 λ2 protein |
2992/3918nt (76.3%) 993/1299aa (76.4%) |
AP-151 MN022938 93.99% |
MRV-3 | Brazil, 2015 |
Human | [18] |
| L3 λ1 protein |
3901nt 1275aa |
RpMRV-YN2012 KM087107 92.72% |
MRV-2 | Yunnan province, China, 2012 | Rhinolophus pusillus (bat) | [44] |
| M1 μ2 protein |
2295nt 736aa |
BatMRV/B19-02 MW582625 96.19% |
MRV-1 | Jeju Island, South Korea, 2019 | Miniopterus schreibersii (bat) | [45] |
| M2 μ1 protein |
2203nt 708aa |
Abney T3C87 GU589581 93.64% |
MRV-3 | Washington, D.C, USA, 1957 | Human | [46] |
| M3 μNS protein |
2241nt 721aa |
BatMRV/B19-02 MW582627 96.90% |
MRV-1 | Jeju Island, South Korea, 2019 | Miniopterus schreibersii (bat) | [45] |
| S1 σ1/σ1s protein |
1380nt σ1—455aa σ1s—120aa |
Tahiti L37679 93.57% |
MRV-3 | Tahiti, French Polynesia, 1960 | Human | [42] |
| S2 σ2 protein |
1323nt 427aa |
Lang L19774 98.11% |
MRV-1 | Ohio, USA, 1953 | Human | [47] |
| S3 σNS protein |
1198nt 366aa |
BatMRV/B19-02 MW582630 96.79% |
MRV-1 | Jeju Island, South Korea, 2019 | Miniopterus schreibersii (bat) | [45] |
| S4 σ3 protein |
1196nt 365aa |
SI-MRV07 MG999585 92.47% |
MRV-3 | Slovenia, 2017* | Human | [17] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





