1. Introduction
Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) is a chronic complex disorder classified by the WHO with the ICD-11 8E49 code as a postviral fatigue syndrome [
1]. It is characterized by debilitating fatigue that is not alleviated by rest, generalized pain, sleep disturbances, cognitive impairments, and a variety of other symptoms that can severely impact daily functioning [
2]. Diagnosis of ME/CFS is based on the clinical assessment of nonspecific symptoms and frequently overlaps with Fibromyalgia (FM) (ICD-11 MG30.0 for chronic primary pain) [
3]. FM is a chronic disorder characterized by low pain threshold, stiffness and tenderness in the muscles of neck, usually accompanied by fatigue, memory loss and sleep disturbances [
4,
5]. Their frequent co-diagnosis and the lack of biomarkers for either condition, have prompted the exploration of both common and unique molecular factors [
6,
7,
8,
9] that may explain their frequent joint appearance and aid in differential diagnosis.
The absence of reliable biomarkers is partly due to the limited understanding of the disease, as the precise etiology of ME/CFS and FM remain elusive. Nevertheless, a growing body of evidence suggests that viral infections may play a pivotal role in the onset and progression of the disease [
10,
11,
12,
13,
14]. Active viral infections may cause immunological disturbances like autoimmunity or immunological dysfunction [
15,
16,
17] and trigger dramatic changes in the epigenetic landscape [
18], leading to viral reactivation of exogenous [
19,
20,
21] or endogenous retroviruses [
22]. This may contribute to the chronic immune activation and dysregulation observed in ME/CFS patients [
23], as well as the persistence and exacerbation of symptoms.
Up to date, no single infectious agent has been associated with ME/CFS, as reviewed by Rasa et al., [
14]. Conflicting research results and experimental limitations like heterogeneous ME/CFS cohorts, high prevalence of persistent viral infection in the general population, and different methodological approaches, among other, may have hindered this finding. In a previous work, we identified reactivation of human endogenous retroviral sequences in a subgroup of ME/CFS patients whose expression correlated with an altered immunological profile [
9]. Those features allowed for perfect discrimination of that ME/CFS subgroup, suggesting alterations in patients’ epigenetic landscape.
The aim of this study was to identify candidate exogenous viral RNA sequences that may associate with the HERV activation profile and correlated immunological disturbances observed in the described subset of ME/CFS patients. To address this question, we performed a comprehensive analysis through microarray technology of viral RNA sequences present in the exact same RNA samples analyzed in our previous study, focusing on increased viral load in the subgroup of ME/CFS patients exhibiting altered HERV profiles as compared to FM, co-diagnosed cases and healthy controls. Our findings revealed significant overexpression of the Torque Teno Mini Virus 9 (TTMV9) in the immune cells of this subgroup of ME/CFS patients compared to the other study groups. The identification of TTMV9 and its association with a particular subset of ME/CFS cases provides new insights into the viral mechanisms that may underlie this complex disorder and opens avenues for further research into, diagnostic markers, targeted treatments, and prevention programs.
2. Materials and Methods
2.1. Participant Recruitment
This cross-sectional observational study was approved by the Public Health Research Ethics Committee DGSP-CSISP of Valencia, num. 20190301/12, Valencia, Spain. The study included a total of 34 female patients from local patient associations (National Biobank Registry Ref. 0006024) who were clinically diagnosed with ME/CFS (n=8) according to the Canadian (4) and International Consensus (5) criteria, with FM (n=10) according to the 1990 (7) and 2011 (3, 6) American College of Rheumatology (ACR) criteria for FM, or both, ME/CFS and FM (n=16), hereafter referred to as co-diagnosed group. A healthy control individuals’ population-matched for age and BMI consisting of 9 female donors was also included in the study (National Biobank Registry Ref. 0006034). Patients with health problems other than FM and ME/CFS were excluded. Individuals with any similar o related pathology, including a medical history of chronic pain and/or fatigue, or serious health complications, were excluded from control group, as well as medicated healthy controls. Written informed consent was obtained from all study participants.
2.2. Blood Sample Collection, Processing and Storage
After a 12h overnight fasting and medication withdrawal, whole blood was extracted from participants following the protocol described in [
9]. Briefly, up to 10 mL of whole blood were collected via venipuncture in K2EDTA tubes (Becton Dickinson, Franklin Lakes, NJ, USA) and processed within 2 hours. Peripheral blood mononuclear cells (PBMCs) were isolated by dilution at 1:1 (v/v) ratio in phosphate-buffered saline solution (PBS) with layering on top of 1 volume of Ficoll-Paque Premium (GE Healthcare, Chicago, IL, USA) and separation by density centrifugation at 500× g for 30 minutes (20°C, brakes off). The PBMC layer was washed with PBS, contaminant erythrocytes removed, and cells adjusted to a final concentration of 10
7 cells/mL in freezing medium (90 % FBS, 10 % DMSO), aliquoted, and deeply frozen in liquid nitrogen until use.
2.3. RNA Extraction, Processing and Analysis by High-Density Microarray
Total RNA from PBMCs was extracted using RNeasy Mini Kit (Qiagen, MD, USA) according to the manufacturer’s instructions. RNA quality was assessed using Agilent TapeStation 4200 (Agilent). All RNA samples had an RNA Integrity number (RIN) above seven. To evaluate the presence of foreign viral RNA sequences, cDNA was synthesized and amplified from 45 ng of RNA using the Ovation Pico WTA System V2 kit (Nugen) according to manufacturer’s instructions. The resulting amplified ssDNA was purified using the QIAquick purification kit (Qiagen, MD, USA). Total DNA concentration was measured with NanoDrop 2000 spectrophotometer (Thermo Scientific) and quality was assessed on the Bioanalyzer 2100. Purified ssDNA was fragmented, biotin-labeled, hybridized and read according to the protocol described in detail in [
9] by Sampled (Piscataway, NJ 08854, USA). The custom high-density HERV-V3 microarrays [
24] were used to detect the presence of 289 exogenous infectious virus, including: (i) dsDNA viruses (103): adenovirus, herpesvirus, polyomavirus, papillomavirus; (ii) RT viruses (82): ALV, BFV, ELL, FFV, FLV, FIV, MLV, HIV, HTVL, SIV, KoRV, etc; (iii) ssDNA viruses (31): Torque virus, and (iv) ssRNA viruses (75): hepatitis C, dengue, hantaviruses, influenza, coronavirus (Full list is provided with the MTA to users).
2.4. Bioinformatic Analysis
All bioinformatic analysis were performed with RStudio software version 4.2.1. Microarray CEL files were processed and analyzed using R oligo package [
25]. Data were normalized, adjusted for background noise, and summarized using the RMA (Robust Multi-Array) algorithm. Differential expression (DE) analysis was performed using limma R package [
26], considering differentially expressed those probes with a “Benjamin-Hochberg” (BH) adjusted
p value < 0.05 and an absolute log2 fold-change>1. Plots were represented with ggplot2 R package [
27]. Correlation analyses were performed with the package Hmisc v5.1. ME/CFS diagnosis variable was binarized in order to be compared with continuous variables. Functional analysis by gene ontology of biological processes was performed with ShinyGO 8.0 [
28]. Digital cytometry analysis was performed with CIBERSORTx [
29] software on normalized non-logarithmic gene expression data for all samples. The default LM22 leukocyte gene signature matrix [
30] was used as a reference. Response operating characteristic (ROC) analysis with area under curve (AUC) were used to assess the discriminating capacity of the variable and were performed with the R package pROC version 1.18.5 [
31].
2.4. Statistical Analysis
Continuous variables were presented as mean ± standard deviation All statistical analyses were done in R v4.2.1. Data distributions were tested for normality. Normally distributed data were tested using two-tailed unpaired Student’s t-tests; non-normal data were analyzed with non-parametric statistical test, as detailed.
3. Results
3.1. Clinical Characteristics of Participants
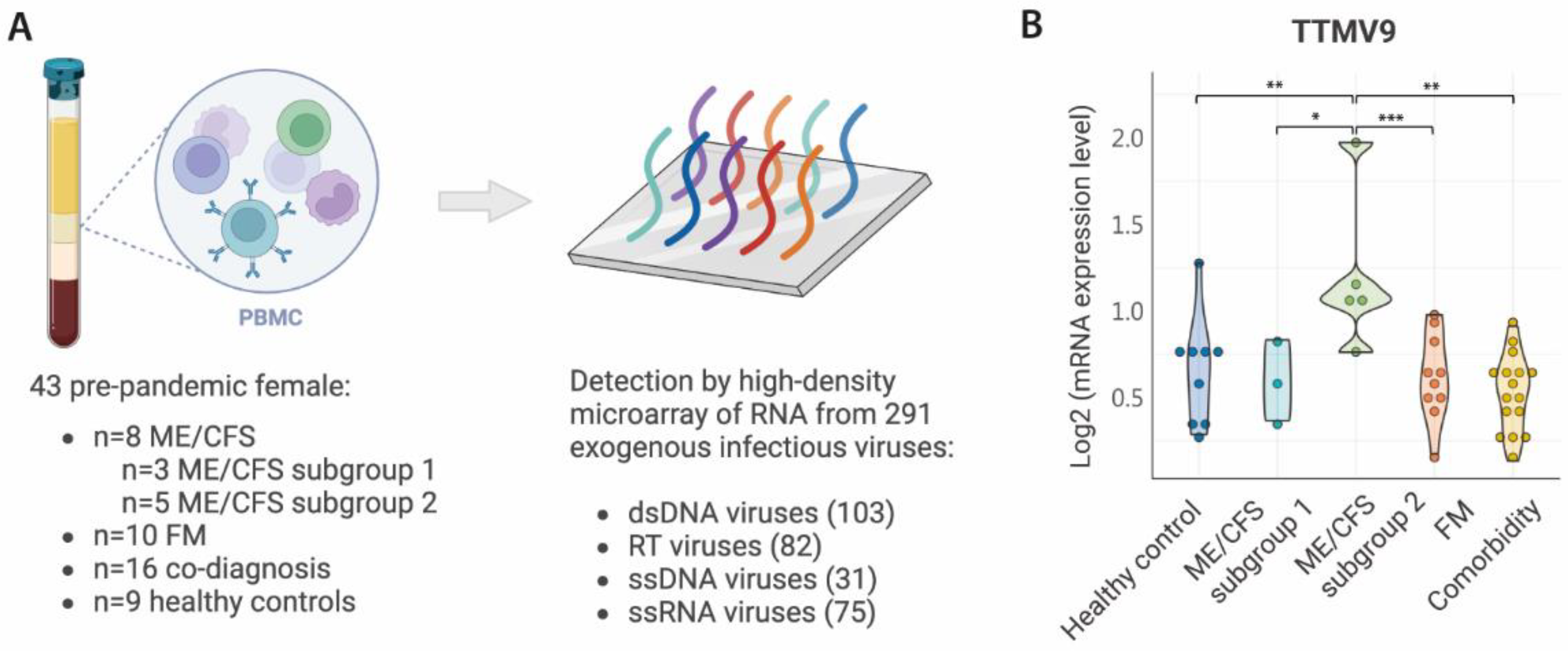
The study included 43 pre-pandemic female participants aged 42 to 58 years, with cases: n=8 ME/CFS, n=10 FM, n=16 ME/CFS + FM (co-diagnosed), and n=9 matched healthy controls. According to a previous study on the same cohort, ME/CFS cases were stratified into two subgroups based on gene and human endogenous retrovirus (HERV) expression, ME/CFS subgroup 1 (n=3) and ME/CFS subgroup 2 (n=5) [
9]. Patient symptoms were assessed through the Fibromyalgia Impact Questionnaire (FIQ) [
32], Multi Fatigue Inventory (MFI) for general fatigue [
33], and Short-Form-36 Health Survey (SF-36) [
34] for quality of life. Itemized questionnaire scores can be accessed in the reference study published by our group [
9]. Significant differences were only found between ME/CFS subgroup 2 and co-diagnosed groups for MFI General fatigue (19 ± 1.7 vs 13.6 ± 3.2, respectively,
p=0.035). Nonetheless, a tendency on lower scores with
p<0.1 could be observed for other items of the MFI questionnaire in ME/CFS subgroup 2, suggesting worse health status for the patients included in the group.
3.2. Increased Levels of TTMV9 in PBMCs of a Subgroup of ME/CFS Cases
To identify exogenous viral RNA potentially causing the activation of HERVs in a subgroup of ME/CFS cases, we analyzed by high-density microarray the expression levels in PBMC of up to 289 viruses belonging to eleven families (
Figure 1A): (i) Retro-transcribing viruses:
Hepadnaviridae, Retroviridae; (ii) ssRNA viruses: ssRNA-positive-strand viruses with no DNA stage,
Coronaviridae; (iii) dsDNA viruses:
Mastadenovirus, Chordopoxvirinae, Herpesviridae, Alphapapillomavirus, Betapapillomavirus, Gammapapillomavirus; and (iv) ssDNA viruses:
Anelloviridae.
Overall analysis of viral RNA by family did not reveal any significant changes associated with ME/CFS subgroup 2 compared to the others. Differences were only detected for the
Coronaviridae, Gammapapillomavirus and
Retroviridae families, and only between FM and ME/CFS subgroup 2 (
Figure S1). Individual analysis of the viruses from each of these families revealed that only for some viruses these differences were confirmed (
Figure S2-S4). As we aimed to identify viral RNA sequences discriminating the ME/CFS subgroup 2 from all the other study groups, we independently compared the expression of each of the 289 viral sequences included in the microarray. Surprisingly, only one virus, the Torque Teno Mini Virus 9 (TTMV9) from the family
Anelloviridae, was specific for the subgroup 2 of ME/CFS patients, with levels above all the other study groups (
Figure 1B). Interestingly, TTMV9 was the only member of the
Anelloviridae family with increased RNA levels in this disease group (
Figure S5) and would have been overlooked if the analysis had been conducted only at the family level.
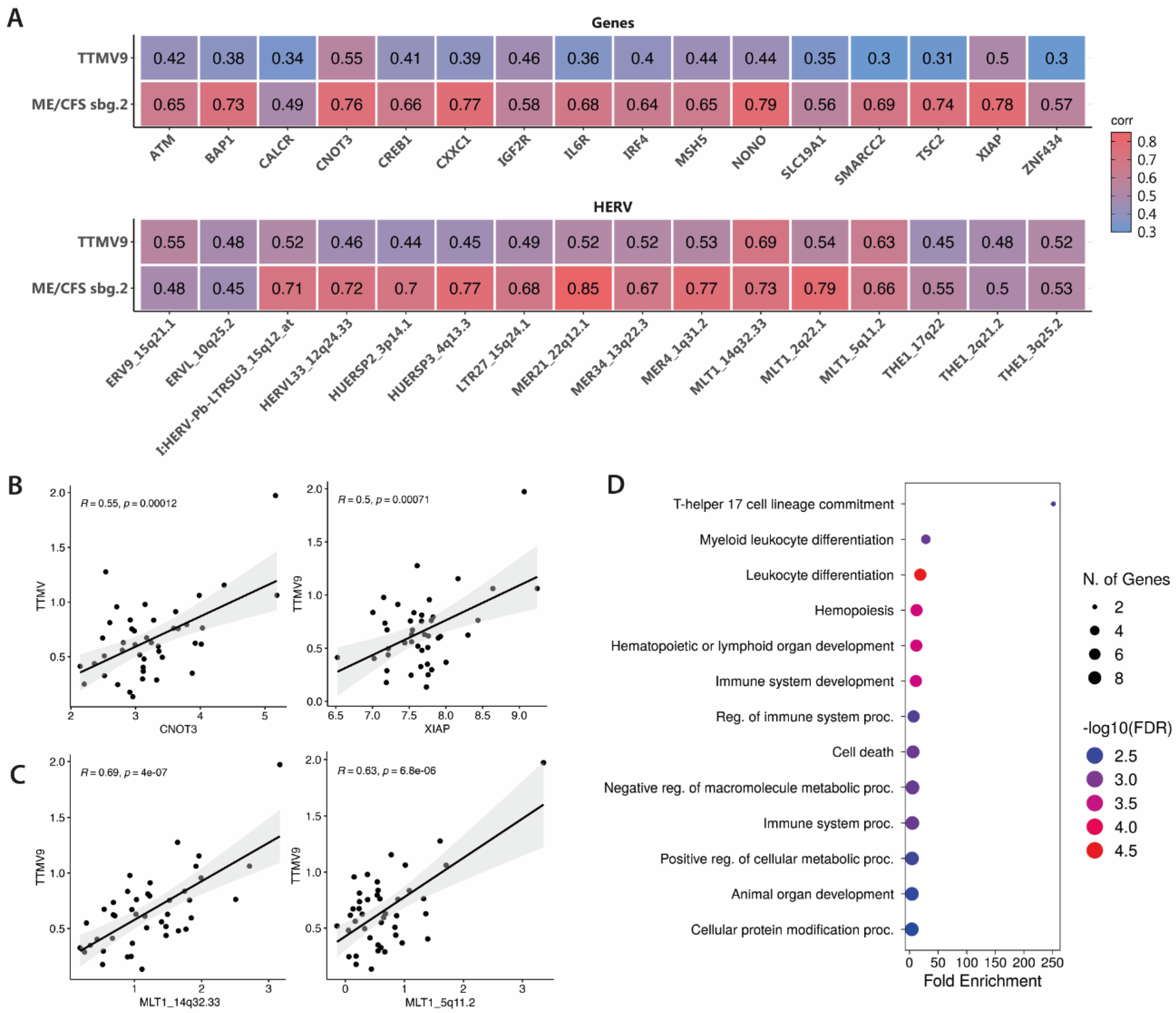
3.3. TTMV9 Levels Correlate with DE HERVs and Genes
To understand if increased viral RNA of TTMV9 associates with the molecular fingerprint previously identified in the same subgroup of ME/CFS patients [
9], we searched for potential correlations between the levels of TTMV9 and the ME/CFS subgroup 2-associated HERV and gene signatures [
9] (
Figure 2A). Moderate correlations with TTMV9 levels were observed for top DE genes, especially for
CNOT3 (
R=0.55,
p=0.00012) and
XIAP (X-linked inhibitor of apoptosis protein) (
R=0.5,
p=0.00071) (
Figure 2A, 2B). Interestingly, the genes showing the highest correlation with TTMV9 levels were also strongly correlated with ME/CFS diagnosis for subgroup 2 (as previously published, [
9])(
R=0.76,
p<0.0001 and
R=0.78,
p<0.0001, respectively;
Figure 2A), indicating potential involvement of TTMV9 levels with disease and gene expression in this patient subgroup. Similarly, HERV levels highly correlating with ME/CFS diagnosis for subgroup 2 [
9], showed moderate to strong associations with TTMV9 expression, e.g., MLT1_14q32.33 (
R=0.69,
p=0.00007), located within the lncRNA
LINC02298, and MLT1_5q11.2 (
R=0.63,
p<0.0001), 25 kbp away from the gene
COX6C (
Figure 2A, 2C). Functional analysis of the genes significantly correlated with TTMV9 levels revealed enrichment for the pathway involved in the commitment of CD4 T cells towards T-helper 17 cells and other leukocyte differentiation events (
Figure 2D).
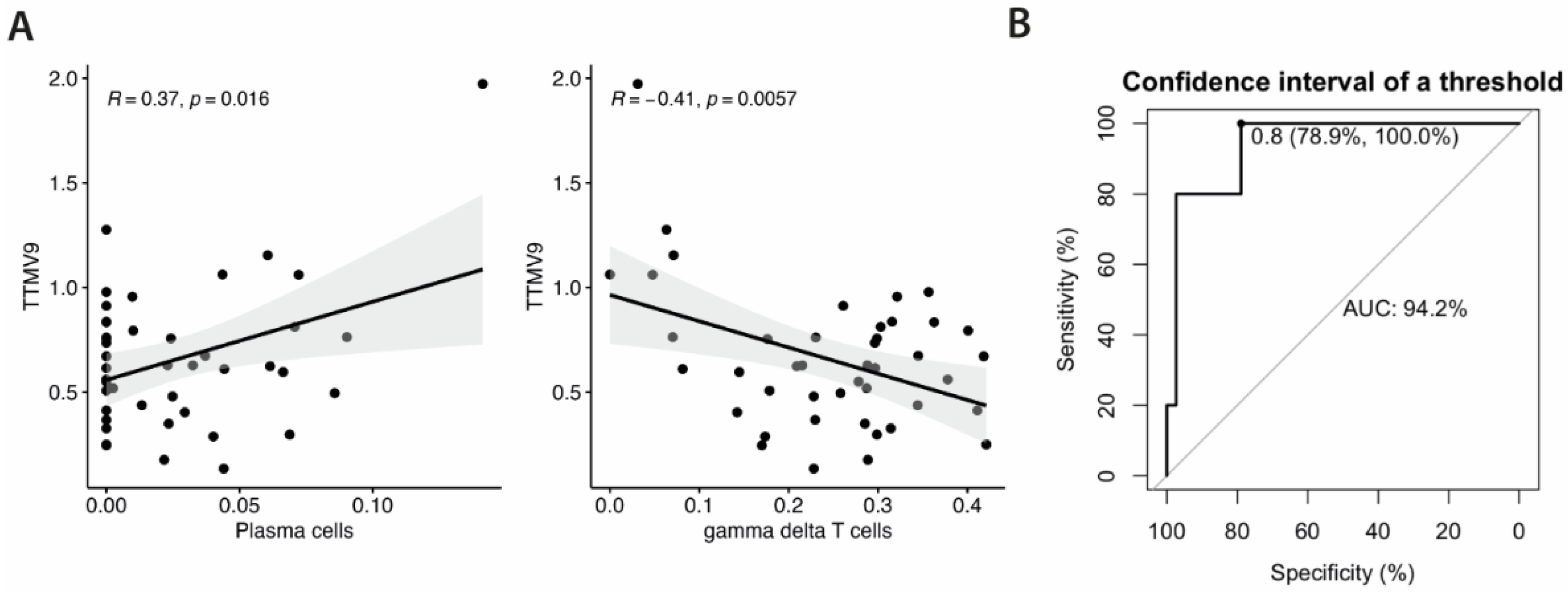
3.3. TTMV9 Levels Correlate with Plasma Cell and Gamma Delta T Cell Population Levels and Discriminate ME/CFS Subgroup 2 from all Other Study Groups
Given the correlation of TTMV9 levels with genes involved in T cell differentiation pathways, and the previously reported association between ME/CFS subgroup 2- HERV fingerprint and certain immune cell populations [
9], we sought to investigate the relationship between TTMV9 levels and immune cell proportions. To this aim, we studied potential correlations of the frequencies of each cell type with TTMV9 RNA levels, using CIBERSORTx [
29]. Interestingly enough, significant correlations were only found for plasma cells (
R=0.37,
p=0.016) and γδ T cells (
R=-0.41,
p=0.0057) (
Figure 3A), coinciding with the already reported for HERV fingerprints in this ME/CFS subgroup [
9], suggesting selective effects on these cell types by the virus.
Lastly, to assess the discriminating power of TTMV9 RNA levels and its potential as biomarker of the ME/CFS subgroup 2, we performed receiver operating characteristics (ROC) analysis. The area under the curve (AUC) was above 0.9, reaching 100% sensitivity and 78.9% specificity, indicative of good capacity to discriminate this subgroup of ME/CFS cases not only from healthy subjects but also from FM and co-diagnosed cases.
4. Discussion
The use of high-density microarrays containing probe sets with capacity to detect active transcription for 289 exogenous infectious viruses has allowed us to rule out differential expression of EBV, herpes, enterovirus, or other in our ME/CFS cohort. A relevant aspect since many have been formerly detected in other cohorts [
35]. It has also allowed us to study the correlation of differential active infection of the TTMV virus with HERV and immune gene profiles. Main limitations of the study being the reduced number of cases per study group, and the research of viral sequences not detectable by our microarrays, particularly some previously reported for ME/CFS, including CMV, Borna disease virus or Parvovirus 19 [
35], or the effects by coinfections with bacteria or other microorganisms.
Torque Teno Mini Viruses (TTMV) are non-enveloped and circular ssDNA viruses belonging to the genus
Betatorquevirus from the family
Anelloviridae [
36]. There are multiple species of TTMV, among which we can find TTMV9 [
36]. Anelloviruses (AV) are a major component of the human virome and they are ubiquitously present in the human population across different tissues in the human body [
37]. Interestingly, the “anellome” of each individual is unique, being comprised of multiple AV genera and species in different ratios [
38]. Despite being acquired at an early stage of life, no pathogenic role has been attributed to their presence. However, high AV titers are considered an indicator of weak immune competence [
39] and may potentially reflect immunocompromised status of some ME/CFS patients. Furthermore, AV load has been shown to be altered in different pathologies, including diverse cancer types or pathologies of the immune system, diabetes, intestinal bowel disease, or in persistent infections such as hepatitis or AIDS [
40,
41]. Perhaps ME/CFS, or at least a subset of cases, similarly to our subgroup 2, will be added to this list of chronic pathologies once these findings get validated at the epidemiological level. In support of compromised immunocompetence of these patients, the few statistically significant differences found in this study also involved increased presence in ME/CFS group 2 (e.g., human papilloma virus 5).
The study of TTMV has been hampered by the lack of in vitro culture systems. However, recent advances on the structure of Anellovirus particles indicates that the strategy they use for immune evasion is to expose highly diverse epitopes acting as immune decoys that elicit weak immune responses, while hiding conserved regions, which permits multiple strains to coexist in an individual [
42]. This brings up the question as to what may be different in the TTMV9 from other TTMV species to become prominently active in this subgroup of ME/CFS patients to cause or sustain disease. A possibility may come from inclined instability of the conserved domains, perhaps leading to autoimmunity through molecular mimicry events, as described for lupus and the HRES-1 endogenous retrovirus encoded autoantigen [
43,
44]. Continuation studies pursuing serological reactivity titrations of these patients’ blood against MMTV9, as well as against the human proteome, should clarify this possibility.
The finding of increased numbers of plasma cells with increased levels of TTMV9 RNA, could just reflect response to infection, perhaps at a chronic stage in these patients. However, in addition to humoral immunity, other roles in hematopoiesis and neuro-inflammation have been attributed to this cell type offering other potential interventions [
45]. On another end, reduced ϒδT cell numbers may indicate increased susceptibility to infections in these subjects.
Through the identified correlations of increased TTMV9 levels and DE genes, and the association of the latter with patient symptoms, as previously published [
9], it can be inferred that increased levels of TTMV9, may directly or indirectly relate with increased “General fatigue” scores, as measured by the MFI instrument, and with worsening of “Role Emotional” and “Mental health” domains of the SF-36 questionnaire. In addition, the finding of this viral strain in the blood and cerebrospinal fluid of a pediatric case of encephalitis [
46] seems to set a pathogenic spectrum for the virus involving the CNS. Although the relationship between TTMV9 and CNOT3 or XIAP levels could be cause, consequence or even coincidence (unlikely according to statistic parameters), it should be mentioned that altered levels of CNOT3 and other CNOT members or XIAP triggered by infections have been described [
47,
48]. In addition to potential biomarker value, the understanding of the mechanisms leading to increased XIAP in ME/CFS, either due by activation of TTMV or not, suggests potential benefit of embelin, a plant-derived benzoquinone with anti-oxidant and anti-inflammatory properties targeting XIAP [
49].
Lastly, potential effects of TTMV9 through HERV elements are more difficult to envision as their functions are mainly unknown. However, molecular effects through the associated competitive endogenous (ceRNA) of miR-140-3p-GPRIN1 axis of LINC02298 [
50], or through cytochrome c oxidase subunit 6C (COX6C) function, as found in other neurological diseases [
51], may be still possible.
5. Conclusions
In summary, this study identifies overexpression of TTMV9 viral RNA correlating with activation of particular DE genes and HERV elements, coinciding with increased plasma and decreased ϒ
T cell numbers, in a subgroup of ME/CFS patients, a phenotype characterized by worsening of symptoms, mainly involving fatigue and emotional-mental health status. The mechanism behind this defined subgroup phenotype, as detailed by this and previous work of our group [
9], seems to deserve further exploration for the advancement of proper patient classification and the development of personalized medicine programs to effectively treat ME/CFS.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org, Figure S1: Virus families represented in the HERV-V3 microarray; Figure S2: Detailed analysis of viruses belonging to the Coronaviridae family; Figure S3: Detailed analysis of viruses belonging to the Gammapapillomavirus family; Figure S4: Detailed analysis of viruses belonging to the Retroviridae family; Figure S5: Detailed analysis of viruses belonging to the Anelloviridae family.
Author Contributions
Conceptualization and methodology KGO. and EO.; patient recruitment and diagnosis EMM; investigation, and formal analysis KGO;. writing—original draft preparation, KGO and EO; writing—review and editing, all authors.; supervision and project administration, EO; funding acquisition, KGO and EO. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by an ME Research UK (SCIO charity number SC036942) grant, and by Generalitat Valenciana CIAICO/2021/103 grant to EO. KG-O is supported by the Generalitat Valenciana ACIF2021/179 grant.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the Public Health Research Ethics Committee DGSP-CSISP of Valencia, núm. 20190301/12, Valencia, Spain.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, or previous publications, as indicated. Further inquiries can be directed to the corresponding author/s.
Acknowledgments
We particularly thank all the patients who participated in the study and Dr. Vicente Serra (Umivale,Valencia, Spain) for his help in the recruitment of healthy volunteers. We also acknowledge bioMérieux and Sampled for assisting us in the authorization and analysis of samples by microarray.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Harrison, J.E.; Weber, S.; Jakob, R.; Chute, C.G. ICD-11: An International Classification of Diseases for the Twenty-First Century. BMC Méd. Inform. Decis. Mak. 2021, 21, 206. [Google Scholar] [CrossRef] [PubMed]
- Bateman, L.; Bested, A.C.; Bonilla, H.F.; Chheda, B.V.; Chu, L.; Curtin, J.M.; Dempsey, T.T.; Dimmock, M.E.; Dowell, T.G.; Felsenstein, D.; et al. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome: Essentials of Diagnosis and Management. Mayo Clin. Proc. 2021, 96, 2861–2878. [Google Scholar] [CrossRef]
- Castro-Marrero, J.; Faro, M.; Aliste, L.; Sáez-Francàs, N.; Calvo, N.; Martínez-Martínez, A.; Sevilla, T.F. de; Alegre, J. Comorbidity in Chronic Fatigue Syndrome/Myalgic Encephalomyelitis: A Nationwide Population-Based Cohort Study. Psychosomatics 2017, 58, 533–543. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, F.; Smythe, H.A.; Yunus, M.B.; Bennett, R.M.; Bombardier, C.; Goldenberg, D.L.; Tugwell, P.; Campbell, S.M.; Abeles, M.; Clark, P.; et al. The American College of Rheumatology 1990 Criteria for the Classification of Fibromyalgia. Arthritis Rheumatism 1990, 33, 160–172. [Google Scholar] [CrossRef]
- Wolfe, F.; Clauw, D.J.; Fitzcharles, M.; Goldenberg, D.L.; Katz, R.S.; Mease, P.; Russell, A.S.; Russell, I.J.; Winfield, J.B.; Yunus, M.B. The American College of Rheumatology Preliminary Diagnostic Criteria for Fibromyalgia and Measurement of Symptom Severity. Arthrit Care Res 2010, 62, 600–610. [Google Scholar] [CrossRef]
- Light, A.R.; Bateman, L.; Jo, D.; Hughen, R.W.; VanHaitsma, T.A.; White, A.T.; Light, K.C. Gene Expression Alterations at Baseline and Following Moderate Exercise in Patients with Chronic Fatigue Syndrome and Fibromyalgia Syndrome. J. Intern. Med. 2012, 271, 64–81. [Google Scholar] [CrossRef]
- Groven, N.; Reitan, S.K.; Fors, E.A.; Guzey, I.C. Kynurenine Metabolites and Ratios Differ between Chronic Fatigue Syndrome, Fibromyalgia, and Healthy Controls. Psychoneuroendocrinology 2021, 131, 105287. [Google Scholar] [CrossRef]
- Nepotchatykh, E.; Caraus, I.; Elremaly, W.; Leveau, C.; Elbakry, M.; Godbout, C.; Rostami-Afshari, B.; Petre, D.; Khatami, N.; Franco, A.; et al. Circulating MicroRNA Expression Signatures Accurately Discriminate Myalgic Encephalomyelitis from Fibromyalgia and Comorbid Conditions. Sci Rep-uk 2023, 13, 1896. [Google Scholar] [CrossRef]
- Giménez-Orenga, K.; Martín-Martínez, E.; Nathanson, L.; Oltra, E. HERV Activation Segregates ME/CFS from Fibromyalgia and Defines a Novel Nosological Entity for Patients Fulfilling Both Clinical Criteria. [CrossRef]
- Furness, P.J.; Vogt, K.; Ashe, S.; Taylor, S.; Haywood-Small, S.; Lawson, K. What Causes Fibromyalgia? An Online Survey of Patient Perspectives. Heal. Psychol. Open 2018, 5, 2055102918802683. [Google Scholar] [CrossRef]
- Chu, L.; Valencia, I.J.; Garvert, D.W.; Montoya, J.G. Onset Patterns and Course of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Front. Pediatr. 2019, 7, 12. [Google Scholar] [CrossRef]
- Tschopp, R.; König, R.S.; Rejmer, P.; Paris, D.H. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS): A Preliminary Survey among Patients in Switzerland. Heliyon 2023, 9, e15595. [Google Scholar] [CrossRef] [PubMed]
- Chapenko, S.; Krumina, A.; Logina, I.; Rasa, S.; Chistjakovs, M.; Sultanova, A.; Viksna, L.; Murovska, M. Association of Active Human Herpesvirus-6, -7 and Parvovirus B19 Infection with Clinical Outcomes in Patients with Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Adv Virology 2012, 2012, 205085. [Google Scholar] [CrossRef] [PubMed]
- Rasa, S.; Nora-Krukle, Z.; Henning, N.; Eliassen, E.; Shikova, E.; Harrer, T.; Scheibenbogen, C.; Murovska, M.; Prusty, B.K.; (EUROMENE), E.N. on M. Chronic Viral Infections in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS). J Transl Med 2018, 16, 268. [Google Scholar] [CrossRef] [PubMed]
- Saez-Cirion, A.; Manel, N. Immune Responses to Retroviruses. Annu. Rev. Immunol. 2016, 36, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Nie, Y.; Wang, Y.-Y. Innate Immune Responses to DNA Viruses. Protein Cell 2013, 4, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Smatti, M.K.; Cyprian, F.S.; Nasrallah, G.K.; Thani, A.A.A.; Almishal, R.O.; Yassine, H.M. Viruses and Autoimmunity: A Review on the Potential Interaction and Molecular Mechanisms. Viruses 2019, 11, 762. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hong, T.; Parameswaran, S.; Ernst, K.; Marazzi, I.; Weirauch, M.T.; Bass, J.I.F. Human Virus Transcriptional Regulators. Cell 2020, 182, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, P.; Harrer, T.; Scheibenbogen, C.; Lamer, S.; Schlosser, A.; Naviaux, R.K.; Prusty, B.K. Human Herpesvirus-6 Reactivation, Mitochondrial Fragmentation, and the Coordination of Antiviral and Metabolic Phenotypes in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome. Immunohorizons 2020, 4, 201–215. [Google Scholar] [CrossRef] [PubMed]
- Apostolou, E.; Rizwan, M.; Moustardas, P.; Sjögren, P.; Bertilson, B.C.; Bragée, B.; Polo, O.; Rosén, A. Saliva Antibody-Fingerprint of Reactivated Latent Viruses after Mild/Asymptomatic COVID-19 Is Unique in Patients with Myalgic-Encephalomyelitis/Chronic Fatigue Syndrome. Front. Immunol. 2022, 13, 949787. [Google Scholar] [CrossRef]
- Shikova, E.; Reshkova, V.; Kumanova, А.; Raleva, S.; Alexandrova, D.; Capo, N.; Murovska, M.; (EUROMENE), on behalf of the E.N. on M. Cytomegalovirus, Epstein-Barr Virus, and Human Herpesvirus-6 Infections in Patients with Myalgic Еncephalomyelitis/Chronic Fatigue Syndrome. J Med Virol 2020, 92, 3682–3688. [Google Scholar] [CrossRef]
- Macchietto, M.G.; Langlois, R.A.; Shen, S.S. Virus-Induced Transposable Element Expression up-Regulation in Human and Mouse Host Cells. Life Sci. Alliance 2020, 3, e201900536. [Google Scholar] [CrossRef] [PubMed]
- Sotzny, F.; Blanco, J.; Capelli, E.; Castro-Marrero, J.; Steiner, S.; Murovska, M.; Scheibenbogen, C.; (EUROMENE), on behalf of the E.N. on M. Myalgic Encephalomyelitis/Chronic Fatigue Syndrome – Evidence for an Autoimmune Disease. Autoimmun Rev 2018, 17, 601–609. [Google Scholar] [CrossRef]
- Becker, J.; Pérot, P.; Cheynet, V.; Oriol, G.; Mugnier, N.; Mommert, M.; Tabone, O.; Textoris, J.; Veyrieras, J.-B.; Mallet, F. A Comprehensive Hybridization Model Allows Whole HERV Transcriptome Profiling Using High Density Microarray. Bmc Genomics 2017, 18, 286. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, B.S.; Irizarry, R.A. A Framework for Oligonucleotide Microarray Preprocessing. Bioinformatics 2010, 26, 2363–2367. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47–e47. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2, Elegant Graphics for Data Analysis. R 2016. [Google Scholar] [CrossRef]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2019, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Steen, C.B.; Liu, C.L.; Gentles, A.J.; Chaudhuri, A.A.; Scherer, F.; Khodadoust, M.S.; Esfahani, M.S.; Luca, B.A.; Steiner, D.; et al. Determining Cell Type Abundance and Expression from Bulk Tissues with Digital Cytometry. Nat. Biotechnol. 2019, 37, 773–782. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust Enumeration of Cell Subsets from Tissue Expression Profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Robin, X.; Turck, N.; Hainard, A.; Tiberti, N.; Lisacek, F.; Sanchez, J.-C.; Müller, M. PROC: An Open-Source Package for R and S+ to Analyze and Compare ROC Curves. BMC Bioinform. 2011, 12, 77. [Google Scholar] [CrossRef]
- Burckhardt, C.S.; Clark, S.R.; Bennett, R.M. The Fibromyalgia Impact Questionnaire: Development and Validation. J Rheumatology 1991, 18, 728–733. [Google Scholar]
- Smets, E.M.A.; Garssen, B.; Bonke, B.; Haes, J.C.J.M.D. The Multidimensional Fatigue Inventory (MFI) Psychometric Qualities of an Instrument to Assess Fatigue. J Psychosom Res 1995, 39, 315–325. [Google Scholar] [CrossRef] [PubMed]
- MCHORNEY, C.A.; WARE, J.; RACZEK, A. The MOS 36-Item Short-Form Health Survey (SF-36). Med Care 1993, 31, 247–263. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-H.; Lee, J.-S.; Oh, H.-M.; Lee, E.-J.; Lim, E.-J.; Son, C.-G. Evaluation of Viral Infection as an Etiology of ME/CFS: A Systematic Review and Meta-Analysis. J. Transl. Med. 2023, 21, 763. [Google Scholar] [CrossRef] [PubMed]
- Varsani, A.; Opriessnig, T.; Celer, V.; Maggi, F.; Okamoto, H.; Blomström, A.-L.; Cadar, D.; Harrach, B.; Biagini, P.; Kraberger, S. Taxonomic Update for Mammalian Anelloviruses (Family Anelloviridae). Arch. Virol. 2021, 166, 2943–2953. [Google Scholar] [CrossRef] [PubMed]
- Spandole, S.; Cimponeriu, D.; Berca, L.M.; Mihăescu, G. Human Anelloviruses: An Update of Molecular, Epidemiological and Clinical Aspects. Arch. Virol. 2015, 160, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Kaczorowska, J.; Hoek, L. van der Human Anelloviruses: Diverse, Omnipresent and Commensal Members of the Virome. FEMS Microbiol. Rev. 2020, 44, 305–313. [Google Scholar] [CrossRef] [PubMed]
- Rezahosseini, O.; Drabe, C.H.; Sørensen, S.S.; Rasmussen, A.; Perch, M.; Ostrowski, S.R.; Nielsen, S.D. Torque-Teno Virus Viral Load as a Potential Endogenous Marker of Immune Function in Solid Organ Transplantation. Transplant. Rev. 2019, 33, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Sabbaghian, M.; Gheitasi, H.; Shekarchi, A.A.; Tavakoli, A.; Poortahmasebi, V. The Mysterious Anelloviruses: Investigating Its Role in Human Diseases. BMC Microbiol. 2024, 24, 40. [Google Scholar] [CrossRef] [PubMed]
- Taylo, L.J.; Keeler, E.L.; Bushman, F.D.; Collman, R.G. The Enigmatic Roles of Anelloviridae and Redondoviridae in Humans. Curr. Opin. Virol. 2022, 55, 101248. [Google Scholar] [CrossRef]
- Liou, S.; Cohen, N.; Zhang, Y.; Acharekar, N.M.; Rodgers, H.; Islam, S.; Zeheb, L.; Pitts, J.; Arze, C.; Swaminathan, H.; et al. Anellovirus Structure Reveals a Mechanism for Immune Evasion. bioRxiv 2022, 2022.07.01.498313. [Google Scholar] [CrossRef]
- Gergely, P.; Pullmann, R.; Stancato, C.; Otvos, L.; Koncz, A.; Blazsek, A.; Poor, G.; Brown, K.E.; Phillips, P.E.; Perl, A. Increased Prevalence of Transfusion-Transmitted Virus and Cross-Reactivity with Immunodominant Epitopes of the HRES-1/P28 Endogenous Retroviral Autoantigen in Patients with Systemic Lupus Erythematosus. Clin. Immunol. 2005, 116, 124–134. [Google Scholar] [CrossRef]
- Perl, A.; Nagy, G.; Koncz, A.; Gergely, P.; Fernandez, D.; Doherty, E.; Telarico, T.; Bonilla, E.; Phillips, P.E. Molecular Mimicry and Immunomodulation by the HRES-1 Endogenous Retrovirus in SLE. Autoimmunity 2008, 41, 287–297. [Google Scholar] [CrossRef]
- Pioli, P.D. Plasma Cells, the Next Generation: Beyond Antibody Secretion. Front. Immunol. 2019, 10, 2768. [Google Scholar] [CrossRef]
- Kang, Y.-J.; Zhou, M.-F.; Huang, W.; Deng, C.; Yan, G.; Lu, Z.-H. Identification of a Novel Torque Teno Mini Virus in Cerebrospinal Fluid from a Child with Encephalitis. Virol. Sin. 2017, 32, 541–544. [Google Scholar] [CrossRef]
- Hagkarim, N.C.; Ryan, E.L.; Byrd, P.J.; Hollingworth, R.; Shimwell, N.J.; Agathanggelou, A.; Vavasseur, M.; Kolbe, V.; Speiseder, T.; Dobner, T.; et al. Degradation of a Novel DNA Damage Response Protein, Tankyrase 1 Binding Protein 1, Following Adenovirus Infection. J. Virol. 2018, 92, e02034-17. [Google Scholar] [CrossRef]
- Peppenelli, M.A.; Miller, M.J.; Altman, A.M.; Cojohari, O.; Chan, G.C. Aberrant Regulation of the Akt Signaling Network by Human Cytomegalovirus Allows for Targeting of Infected Monocytes. Antivir. Res. 2018, 158, 13–24. [Google Scholar] [CrossRef]
- Daimary, U.D.; Girisa, S.; Parama, D.; Verma, E.; Kumar, A.; Kunnumakkara, A.B. Embelin: A Novel XIAP Inhibitor for the Prevention and Treatment of Chronic Diseases. J. Biochem. Mol. Toxicol. 2022, 36, e22950. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, D.; Zheng, H.; He, Z.; Qian, F.; Wu, X.; Yin, Z.; Bao, P.T.; Jin, M. A Novel LncRNA-miRNA-mRNA Competing Endogenous RNA Regulatory Network in Lung Adenocarcinoma and Kidney Renal Papillary Cell Carcinoma. Thorac. Cancer 2021, 12, 2526–2536. [Google Scholar] [CrossRef]
- Kish, S.J.; Mastrogiacomo, F.; Guttman, M.; Furukawa, Y.; Taanman, J.; Dozic, S.; Pandolfo, M.; Lamarche, J.; DiStefano, L.; Chang, L. Decreased Brain Protein Levels of Cytochrome Oxidase Subunits in Alzheimer’s Disease and in Hereditary Spinocerebella Ataxia Disorders. J. Neurochem. 1999, 72, 700–707. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).