Submitted:
01 August 2024
Posted:
02 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
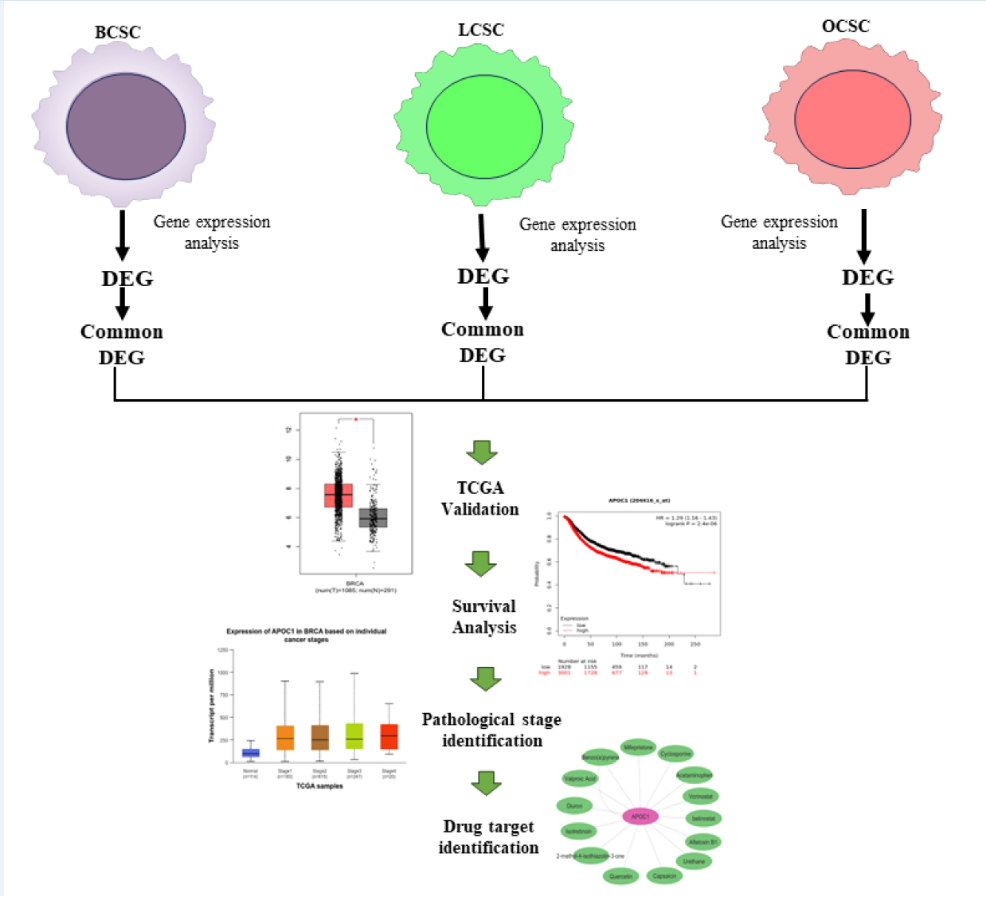
2.1. Collection of Datasets
2.2. Identification of DEGs
2.3. Validation of the Genes from TCGA Database
2.4. Survival Analysis
2.5. Identification of Pathological Stages
2.6. Identification of Protein Expression Level of the Genes by Using Human Protein Atlas (HPA)
2.7. Immune Infiltration Analysis
2.8. Drug Target Identification
3. Results
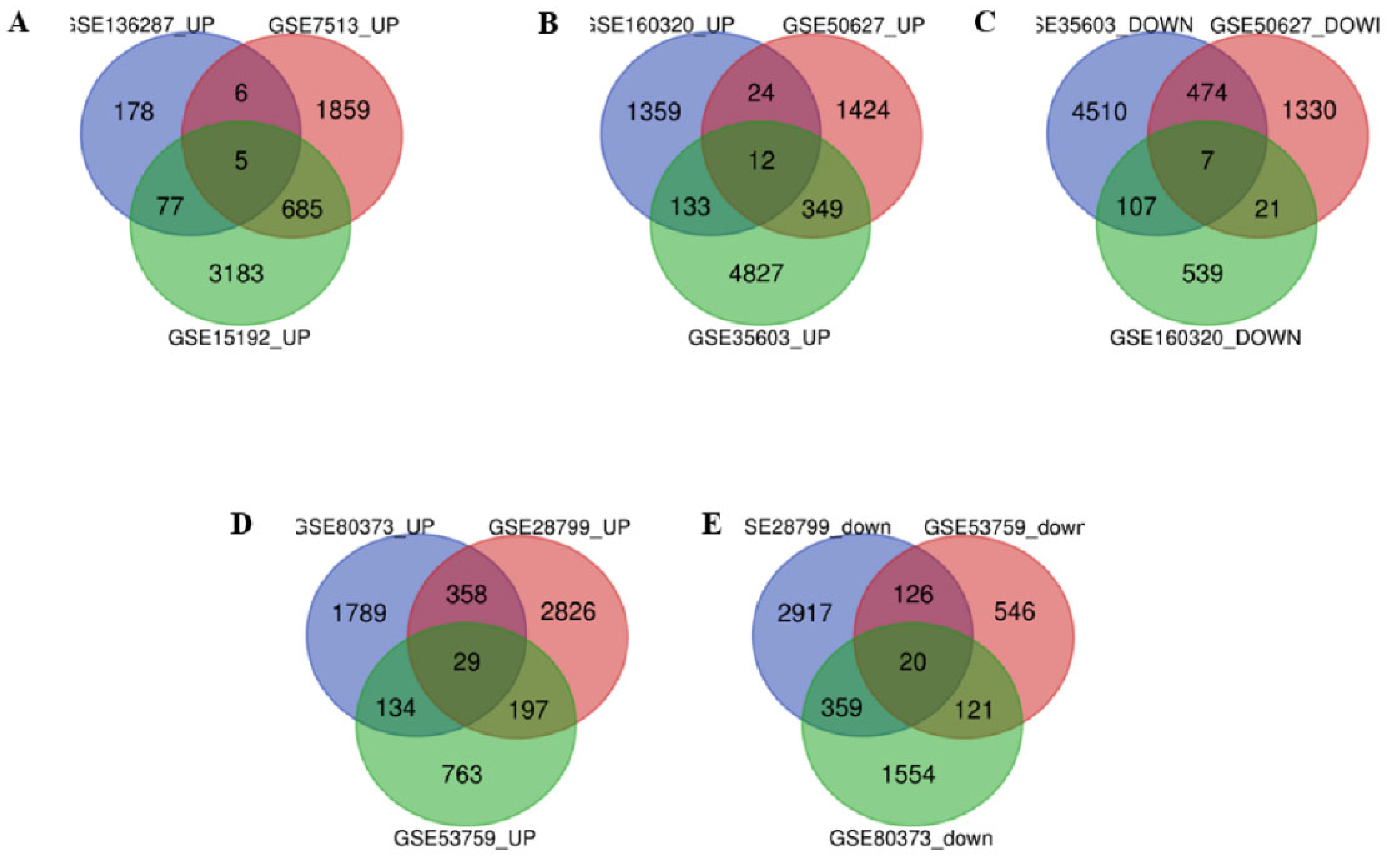
3.1. Identification of DEGs
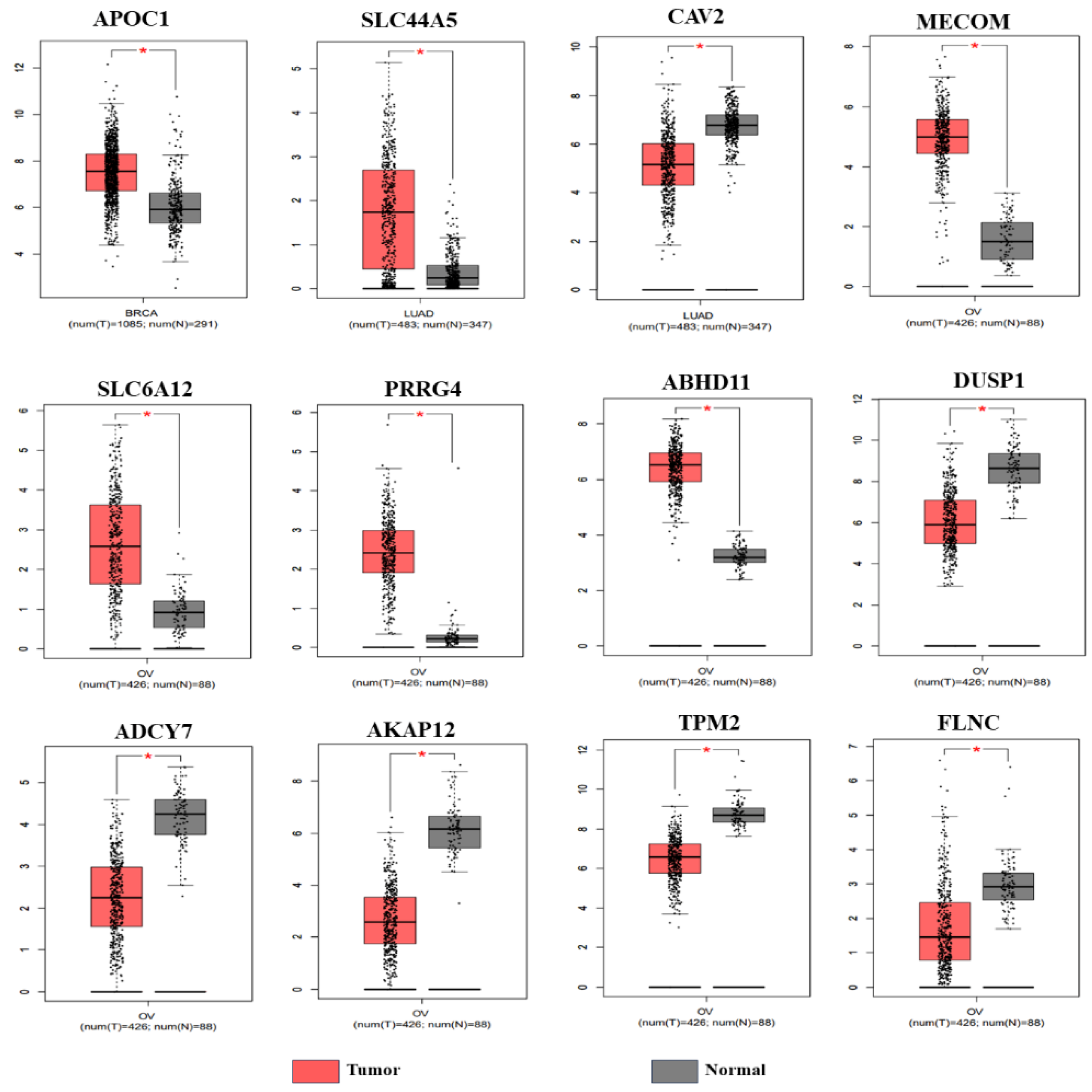
3.2. Validation of the Genes from TCGA Database
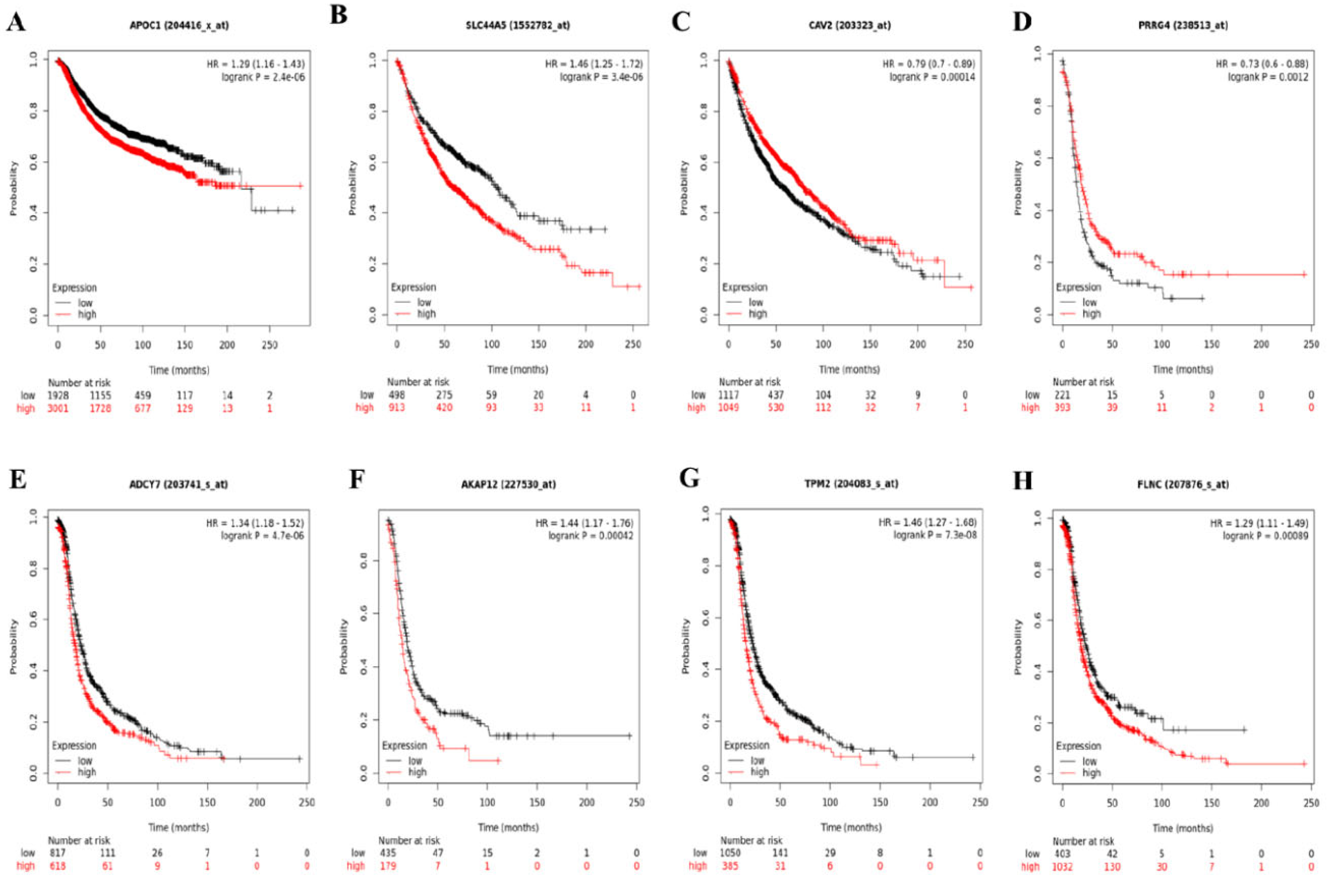
3.3. Survival Analysis
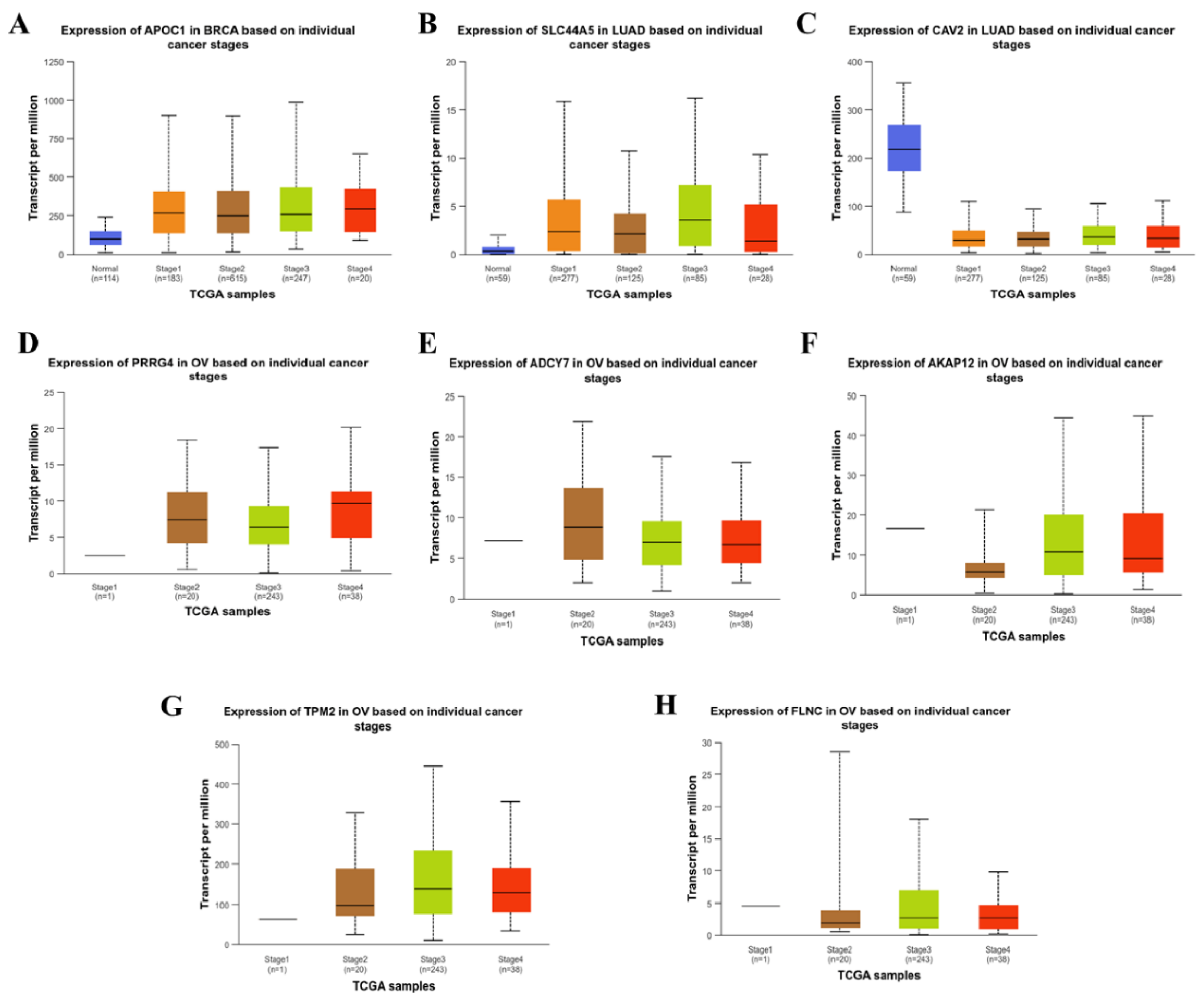
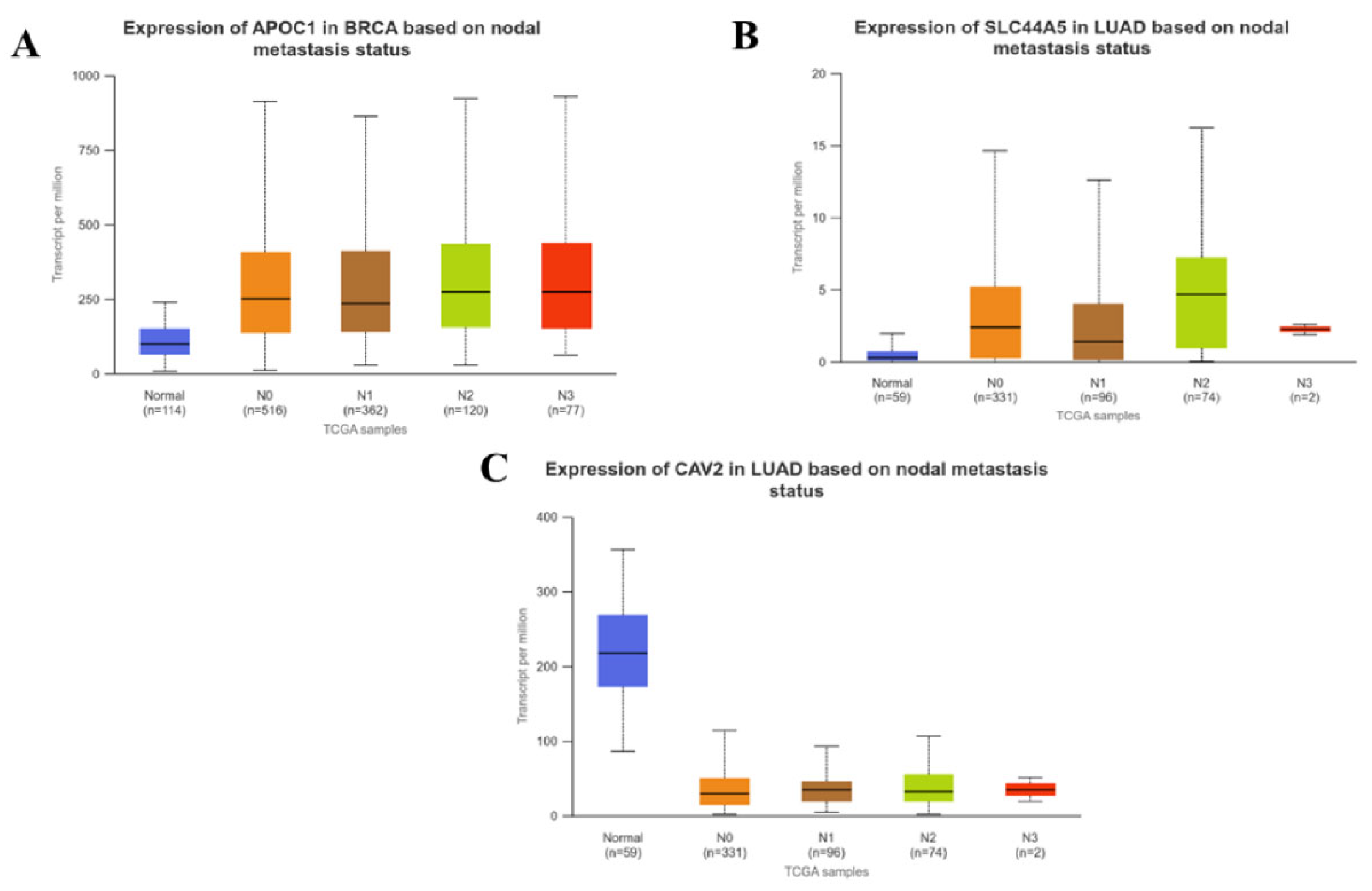
3.4. Correlation between the Expression of CSC Associated Genes and Pathological Stages of Carcinomas
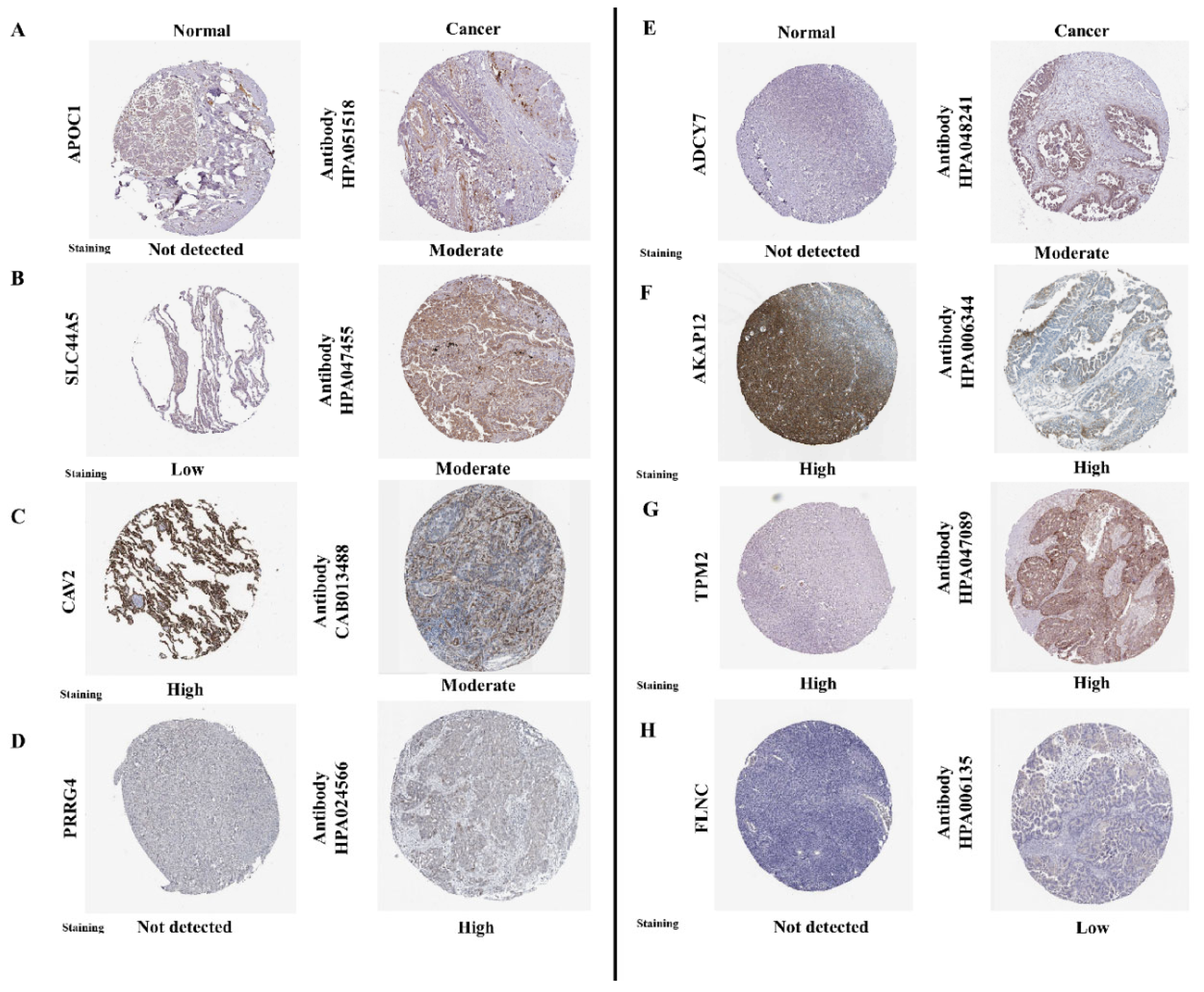
3.5. Immunohistochemical Analysis
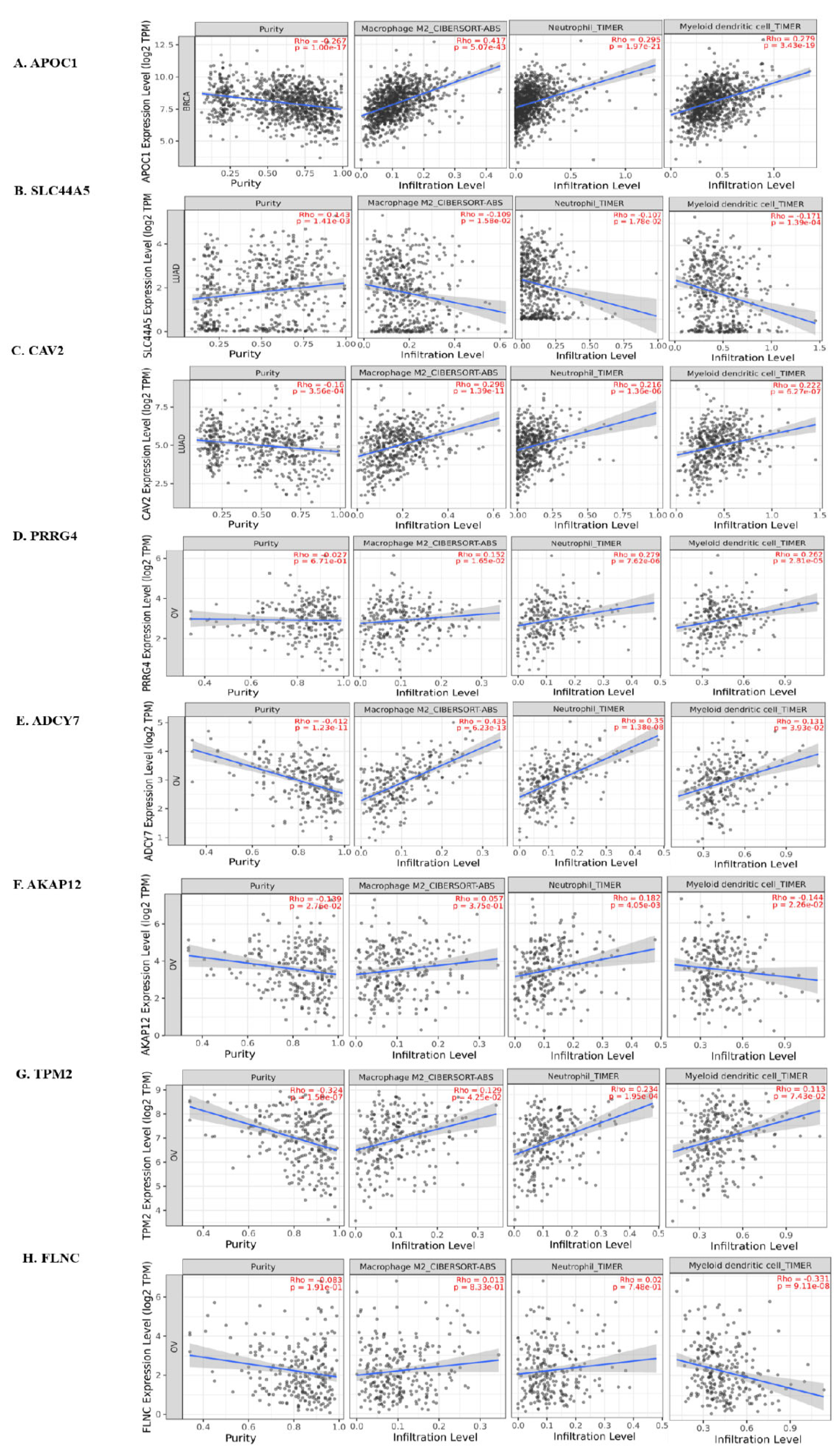
3.6. Immune Infiltration Analysis
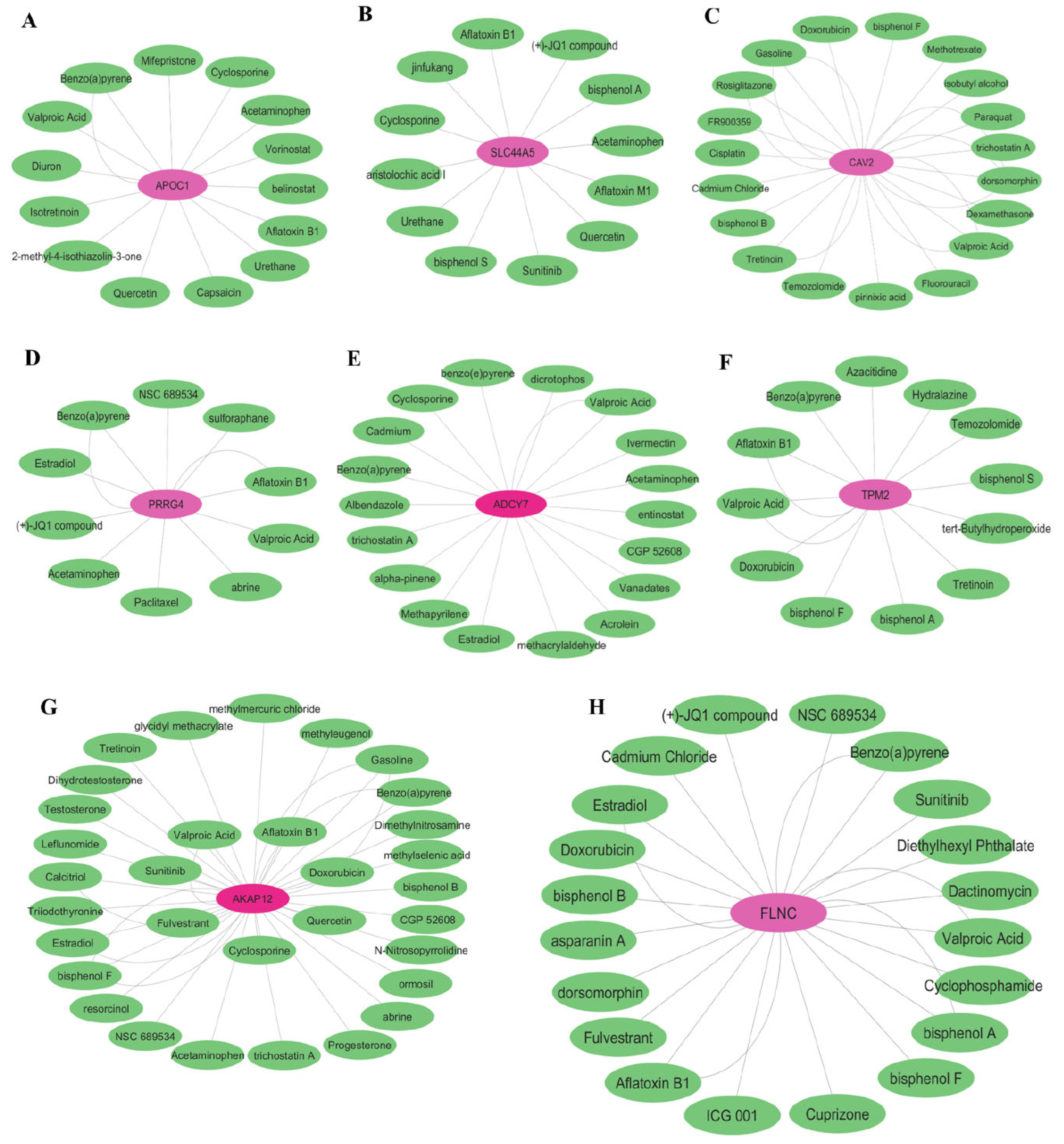
3.7. Drug Target Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cooper GM. The Cell: A Molecular Approach. 2nd edition. Sunderland (MA): Sinauer Associates; 2000. The Development and Causes of Cancer. Available from: https://www.ncbi.nlm.nih.gov/books/NBK9963/.
- Nagai, H., & Kim, Y. H. (2017). Cancer prevention from the perspective of global cancer burden patterns. Journal of thoracic disease, 9(3), 448–451. [CrossRef]
- Walcher, L., Kistenmacher, A. K., Suo, H., Kitte, R., Dluczek, S., Strauß, A., Blaudszun, A. R., Yevsa, T., Fricke, S., & Kossatz-Boehlert, U. (2020). Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Frontiers in immunology, 11, 1280. [CrossRef]
- Najafi, M., Mortezaee, K., & Majidpoor, J. (2019). Cancer stem cell (CSC) resistance drivers. Life sciences, 234, 116781. [CrossRef]
- Biserova, K., Jakovlevs, A., Uljanovs, R., & Strumfa, I. (2021). Cancer Stem Cells: Significance in Origin, Pathogenesis and Treatment of Glioblastoma. Cells, 10(3), 621. [CrossRef]
- Ordaz-Ramos, A., Tellez-Jimenez, O., & Vazquez-Santillan, K. (2023). Signaling pathways governing the maintenance of breast cancer stem cells and their therapeutic implications. Frontiers in cell and developmental biology, 11, 1221175. [CrossRef]
- Bisht, S., Nigam, M., Kunjwal, S. S., Sergey, P., Mishra, A. P., & Sharifi-Rad, J. (2022). Cancer Stem Cells: From an Insight into the Basics to Recent Advances and Therapeutic Targeting. Stem cells international, 2022, 9653244. [CrossRef]
- Yang, L., Shi, P., Zhao, G., Xu, J., Peng, W., Zhang, J., Zhang, G., Wang, X., Dong, Z., Chen, F., & Cui, H. (2020). Targeting cancer stem cell pathways for cancer therapy. Signal transduction and targeted therapy, 5(1), 8. [CrossRef]
- Łukasiewicz, S., Czeczelewski, M., Forma, A., Baj, J., Sitarz, R., & Stanisławek, A. (2021). Breast Cancer-Epidemiology, Risk Factors, Classification, Prognostic Markers, and Current Treatment Strategies-An Updated Review. Cancers, 13(17), 4287. [CrossRef]
- Menon G, Alkabban FM, Ferguson T. Breast Cancer. [Updated 2024 Feb 25]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK482286/.
- Bade, B. C., & Dela Cruz, C. S. (2020). Lung Cancer 2020: Epidemiology, Etiology, and Prevention. Clinics in chest medicine, 41(1), 1–24. [CrossRef]
- Siddiqui F, Vaqar S, Siddiqui AH. Lung Cancer. [Updated 2023 May 8]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK482357/.
- Webb, P. M., & Jordan, S. J. (2017). Epidemiology of epithelial ovarian cancer. Best practice & research. Clinical obstetrics & gynaecology, 41, 3–14. [CrossRef]
- Roett, M. A., & Evans, P. (2009). Ovarian cancer: an overview. American family physician, 80(6), 609–616.
- Wooller, S. K., Benstead-Hume, G., Chen, X., Ali, Y., & Pearl, F. M. G. (2017). Bioinformatics in translational drug discovery. Bioscience reports, 37(4), BSR20160180. [CrossRef]
- Gauthier, J., Vincent, A. T., Charette, S. J., & Derome, N. (2019). A brief history of bioinformatics. Briefings in bioinformatics, 20(6), 1981–1996. [CrossRef]
- Clough, E., & Barrett, T. (2016). The Gene Expression Omnibus Database. Methods in molecular biology (Clifton, N.J.), 1418, 93–110. [CrossRef]
- Diboun, I., Wernisch, L., Orengo, C. A., & Koltzenburg, M. (2006). Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC genomics, 7, 252. [CrossRef]
- Love, M. I., Huber, W., & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology, 15(12), 550. [CrossRef]
- Tomczak, K., Czerwińska, P., & Wiznerowicz, M. (2015). The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemporary oncology (Poznan, Poland), 19(1A), A68–A77. [CrossRef]
- Tang, Z., Li, C., Kang, B., Gao, G., Li, C., & Zhang, Z. (2017). GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic acids research, 45(W1), W98–W102. [CrossRef]
- Győrffy B. (2024). Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation (Cambridge (Mass.)), 5(3), 100625. [CrossRef]
- Chandrashekar, D. S., Bashel, B., Balasubramanya, S. A. H., Creighton, C. J., Ponce-Rodriguez, I., Chakravarthi, B. V. S. K., & Varambally, S. (2017). UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (New York, N.Y.), 19(8), 649–658. [CrossRef]
- Asplund, A., Edqvist, P. H., Schwenk, J. M., & Pontén, F. (2012). Antibodies for profiling the human proteome-The Human Protein Atlas as a resource for cancer research. Proteomics, 12(13), 2067–2077. [CrossRef]
- Li, T., Fu, J., Zeng, Z., Cohen, D., Li, J., Chen, Q., Li, B., & Liu, X. S. (2020). TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic acids research, 48(W1), W509–W514. [CrossRef]
- Davis, A. P., Wiegers, T. C., Johnson, R. J., Sciaky, D., Wiegers, J., & Mattingly, C. J. (2023). Comparative Toxicogenomics Database (CTD): update 2023. Nucleic acids research, 51(D1), D1257–D1262. [CrossRef]
- Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B., & Ideker, T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research, 13(11), 2498–2504. [CrossRef]
- Lin, Y., Xu, J., & Lan, H. (2019). Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications. Journal of hematology & oncology, 12(1), 76. [CrossRef]
- Giese, M. A., Hind, L. E., & Huttenlocher, A. (2019). Neutrophil plasticity in the tumor microenvironment. Blood, 133(20), 2159–2167. [CrossRef]
- Tran Janco, J. M., Lamichhane, P., Karyampudi, L., & Knutson, K. L. (2015). Tumor-infiltrating dendritic cells in cancer pathogenesis. Journal of immunology (Baltimore, Md. : 1950), 194(7), 2985–2991. [CrossRef]
- Borlongan, M. C., & Wang, H. (2023). Profiling and targeting cancer stem cell signaling pathways for cancer therapeutics. Frontiers in cell and developmental biology, 11, 1125174. [CrossRef]
- Pal, A. K., Sharma, P., Zia, A., Siwan, D., Nandave, D., Nandave, M., & Gautam, R. K. (2022). Metabolomics and EMT Markers of Breast Cancer: A Crosstalk and Future Perspective. Pathophysiology : the official journal of the International Society for Pathophysiology, 29(2), 200–222. [CrossRef]
- Xiao, D., & He, J. (2010). Epithelial mesenchymal transition and lung cancer. Journal of thoracic disease, 2(3), 154–159. [CrossRef]
- Loret, N., Denys, H., Tummers, P., & Berx, G. (2019). The Role of Epithelial-to-Mesenchymal Plasticity in Ovarian Cancer Progression and Therapy Resistance. Cancers, 11(6), 838. [CrossRef]
- Tang, W., Liu, H., Li, X., Ooi, T. C., Rajab, N. F., Cao, H., & Sharif, R. (2023). Upregulation of APOC1 Promotes Colorectal Cancer Progression and Serves as a Potential Therapeutic Target Based on Bioinformatics Analysis. Journal of oncology, 2023, 2611105. [CrossRef]
- Shi, X., Wang, J., Dai, S., Qin, L., Zhou, J., & Chen, Y. (2020). Apolipoprotein C1 (APOC1): A Novel Diagnostic and Prognostic Biomarker for Cervical Cancer. OncoTargets and therapy, 13, 12881–12891. [CrossRef]
- Zhang, H., Wang, Y., Liu, C., Li, W., Zhou, F., Wang, X., & Zheng, J. (2022). The Apolipoprotein C1 is involved in breast cancer progression via EMT and MAPK/JNK pathway. Pathology, research and practice, 229, 153746. [CrossRef]
- Peng, G. Z., Ye, Q. F., Wang, R., Li, M. X., & Yang, Z. X. (2016). Knockdown by shRNA identifies SLC44A5 as a potential therapeutic target in hepatocellular carcinoma. Molecular medicine reports, 13(6), 4845–4852. [CrossRef]
- Xie, L., Vo-Ransdell, C., Abel, B., Willoughby, C., Jang, S., & Sowa, G. (2011). Caveolin-2 is a negative regulator of anti-proliferative function and signaling of transforming growth factor-β in endothelial cells. American journal of physiology. Cell physiology, 301(5), C1161–C1174. [CrossRef]
- Zhang, L., Qin, Y., Wu, G., Wang, J., Cao, J., Wang, Y., Wu, D., Yang, K., Zhao, Z., He, L., Lyu, J., Li, H., & Gu, H. (2020). PRRG4 promotes breast cancer metastasis through the recruitment of NEDD4 and downregulation of Robo1. Oncogene, 39(49), 7196–7208. [CrossRef]
- Wang, Y., Wang, J., Chen, L., Chen, Z., Wang, T., Xiong, S., Zhou, T., Wu, G., He, L., Cao, J., Liu, M., Li, H., & Gu, H. (2023). PRRG4 regulates mitochondrial function and promotes migratory behaviors of breast cancer cells through the Src-STAT3-POLG axis. Cancer cell international, 23(1), 323. [CrossRef]
- Chen, S. L., Hu, F., Wang, D. W., Qin, Z. Y., Liang, Y., & Dai, Y. J. (2020). Prognosis and regulation of an adenylyl cyclase network in acute myeloid leukemia. Aging, 12(12), 11864–11877. [CrossRef]
- Li H. (2022). Physiologic and pathophysiologic roles of AKAP12. Science progress, 105(3), 368504221109212. [CrossRef]
- Cui, J., Cai, Y., Hu, Y., Huang, Z., Luo, Y., Kaz, A. M., Yang, Z., Chen, D., Fan, X., Grady, W. M., & Wang, J. (2016). Epigenetic silencing of TPM2 contributes to colorectal cancer progression upon RhoA activation. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine, 37(9), 12477–12483. [CrossRef]
- Zhang, J., Zhang, J., Xu, S., Zhang, X., Wang, P., Wu, H., Xia, B., Zhang, G., Lei, B., Wan, L., Zhang, D., & Pang, D. (2018). Hypoxia-Induced TPM2 Methylation is Associated with Chemoresistance and Poor Prognosis in Breast Cancer. Cellular physiology and biochemistry : international journal of experimental cellular physiology, biochemistry, and pharmacology, 45(2), 692–705. [CrossRef]
- Qiao, J., Cui, S. J., Xu, L. L., Chen, S. J., Yao, J., Jiang, Y. H., Peng, G., Fang, C. Y., Yang, P. Y., & Liu, F. (2015). Filamin C, a dysregulated protein in cancer revealed by label-free quantitative proteomic analyses of human gastric cancer cells. Oncotarget, 6(2), 1171–1189. [CrossRef]
- Yang, D., Wang, T., Long, M., & Li, P. (2020). Quercetin: Its Main Pharmacological Activity and Potential Application in Clinical Medicine. Oxidative medicine and cellular longevity, 2020, 8825387. [CrossRef]
| Serial No. | Accession no. | Cancer Type | Sample groups used in this study | Platform |
|---|---|---|---|---|
| 1. | GSE7513 | Breast Cancer | Cancer Stem Cell (n=14) vs. Non-Cancer Stem Cell (n=15) | GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array |
| 2. | GSE15192 | Breast Cancer | Cancer Stem Cell (n=4) vs. Non-Cancer Stem Cell (n=4) | GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array |
| 3. | GSE136287 | Breast Cancer | Cancer Stem Cell (n=9) vs. Non-Cancer Stem Cell (n=9) | GPL6244 [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version] |
| 4. | GSE35603 | Lung Cancer | Cancer Stem Cell (n=3) vs. Parental Tumor cell (n=3) | GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array |
| 5. | GSE50627 | Lung Cancer | Normal Stem Cell (n=6) vs. Cancer Stem Cell (n=9) | GPL6244 [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version] |
| 6. | GSE160320 | Lung Cancer | Cancer Stem Cell (n=3) vs. Non-Cancer Stem Cell (n=3) | GPL26963 Agilent-085982 Arraystar human IncRNA V5 microarray |
| 7. | GSE28799 | Ovarian Cancer | Cancer Stem Cell (n=3) vs. Non-Cancer Stem Cell (n=3) | GPL570 [HG-U133_Plus_2] Affymetrix Human Genome U133 Plus 2.0 Array |
| 8. | GSE53759 | Ovarian Cancer | Cancer Stem Cell (n=3) vs. Non-Cancer Stem Cell (n=3) | GPL6244 [HuGene-1_0-st] Affymetrix Human Gene 1.0 ST Array [transcript (gene) version] |
| 9. | GSE80373 | Ovarian Cancer | Cancer Stem Cell (n=4) vs. Non-Cancer Stem Cell (n=4) | GPL13667 [HG-U219] Affymetrix Human Genome U219 Array |
| Cancer Type | Up-regulated common genes | Down-regulated common genes |
|---|---|---|
| Breast Cancer | VWA5A, LXN, CLIC4, APOC1, MCFD2. | ------- |
| Lung Cancer | ABCB7, SLC44A5, AIF1L, SYNE1, ID2, ID4, RPS6KA, PPM1D, TP53BP2, ANGPT1, RHOBT, SLCO4C1. | TRAM2, CAV2, CAP1, GLIPR1, TFPI2, PLAUR, MAN2A1 |
| Ovarian Cancer | ABCA3, DUSP4, EPHA4, ASAH1, FOXO1, MECOM, SLC6A12, PRRG4, ZDHHC14, AKR1C3, FRAS1, TSC22D1, MBD5, SLC5A3, PRDM1, ZFX, CTNS, AKR1B1, TGFBR3, AKR1C1, CAT, PKD2, C7ORF26, TMEM222, ARID5B, CCNG2, ABHD11, CDK19, PXK. | ITGA3, ANXA3, NFE2L3, GJC1, CYR61, DARS2, UGCG, DEPDC1B, DUSP1(MKP-1), BUB1, TBC1D1, ADCY7, TPX2, AURKA, AKAP12, TPM2, FLNC, PGM2, COTL1, HJURP. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





