1. Introduction
The brain is composed of cells called neurons, which are responsible for processing information. These neurons are surrounded by “feeder” cells, known as glial cells, which account at least 50% to 90% of the brain’s composition[
1]. Among these glial cells, the star-shaped astrocytes play a crucial role in supplying the nutrients required for neuron function, managing interneuronal connections, and regulating processes such as memory, movement, or odor processing[
2].
A glioma is a tumor that develops from these glial cells in the brain or spinal cord. Most brain tumors develop in the cerebral hemispheres, specifically in the white matter, which contains a large number of axons (projections from neurons) extensively surrounded by glial cells. However, gliomas can also be found throughout the central nervous system. Gliomas are the most common brain tumors in children, adolescents, and adults. Glioma is also the most common form of primary (i.e., non-metastatic) brain tumor in adults. The most severe form of glial tumor is glioblastoma. It’s also the most common central nervous tumor in US population (14.2% of all tumors and 50.1% of all malignant tumors) with 13272 new case in 2022, primarily affecting adults, and more frequently men than women.[
3]
Progressively, World Health Organization (WHO) classifications have incorporated correlations between the tumor’s genetic status and its phenotype, culminating in the 2021 classification, which clearly separates childhood and adult glial tumors[
4]. Diffuse adult gliomas are now divided into three classes:
IDH1- and
IDH2-mutated oligodendrogliomas, with a 1p/19q codeletion (loss of the p arm of chromosome 1 and the q arm of chromosome 19) and longer survival (low-grade gliomas);
IDH1- and
IDH2-mutated diffuse astrocytomas, with variable prognosis (ranging from low-grade to high-grade gliomas); and finally, glioblastomas without
IDH1 or
IDH2 mutations, but with an activating mutation in the
TERT promoter (a telomere-building protein), EGFR amplification, and specific karyotypic features, making them the tumors with the worst prognosis (high-grade gliomas). Methylation of the
MGMT gene (encoding the DNA repair enzyme O-6-methylguanine-DNA methyltransferase) is a factor linked to the efficacy of temozolomide chemotherapy; if this gene is demethylated, the cancer cell can repair the DNA damage caused by temozolomide [
3,
4].
Magnetic resonance imaging (MRI) of the brain seem to be the gold standard for the detection of brain tumors, whether discovered incidentally or as a result of neurological symptoms. MRI can determine the extent of the disease and indicate whether surgical resection of the tumor is feasible. It also helps identify the area to be biopsied via stereotactic biopsy, the only method for performing histological and molecular analyses to confirm the diagnosis and characterize the tumor. This histo-molecular characterization is essential for optimal, patient-specific therapeutic management. However, this biopsy is not feasible for all patients, and it is a gamble to sample the most severe area of the tumor, which is not always representative of the tumor as a whole and its prognosis. Additionally, biopsy is an invasive procedure and carries risks for the patient.
Since 2012, the classification of non-medical images has been dominated by Deep Learning (DL) algorithms utilizing convolutional neural networks[
7]. The potential application and use of these technologies in the genetic characterization of adult glial tumors are therefore compelling.
The aim of our scoping review is to provide a comprehensive state-of-the-art overview of the use of DL algorithms in the genetic characterization of adult glial tumors, encompassing their reported performance, potential limitations, the data used and available in open access, as well as future directions to advance this field towards a reliable and reproducible capacity for virtual biopsy.
2. Materials and Methods
We used for this scoping review was developed with reference for systematic reviews to the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) [
8].
2.1. PICOs (Inclusion Criteria)
We define PICOs as: Population: adults with gliomas or glioblastomas; Intervention: the development and use of AI models to predict somatic genetics from MRI; Comparison: not applicable; and for Outcomes: the capability to predict, evaluated by ROC curves.
We focused on Artificial Intelligence model used DL technology and not just Machine Learning (ML).
2.2. Search Strategy
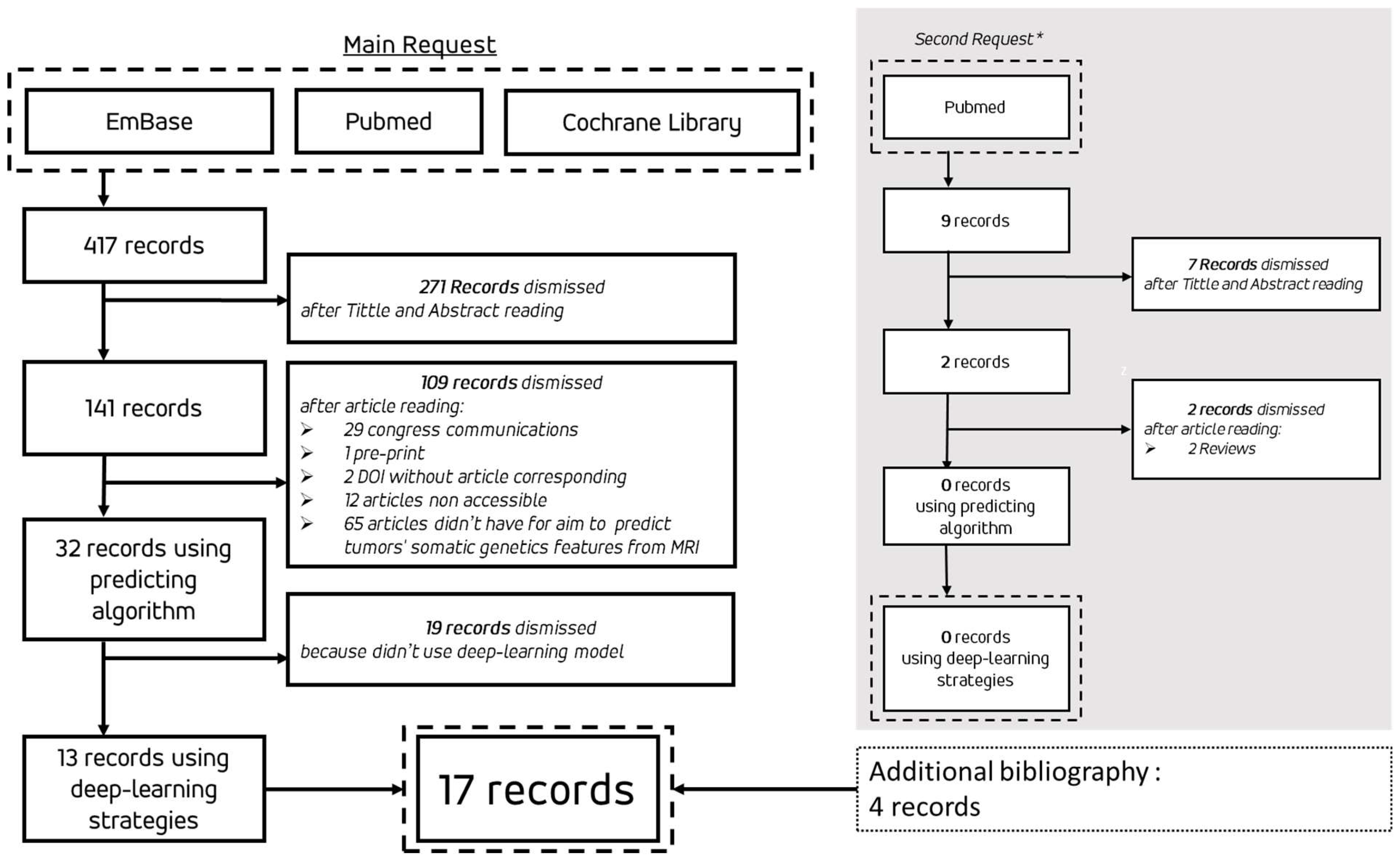
For research strategy we selected three databases: PubMed, EmBase and Cochrane Library. 3 equivalent requests were design (supplemental data S1). Double reading of title and abstract selected a first group of record follow by a double reading of article to selected record for this scoping review.
We have defined a rule on access to OpenAcces as follows, if the article cannot be read by OpenAcces or Shiboleth library access by our institutions. We exclude preprint and article wrote in other language than English. For PubMed Request, we exclude all references without full text associated. We planned to request one bases again during the writing of the reviews. For simplicity to used we have chosen PubMed for this second request.
As matter, if an article known to ours but not include in the results of our request, we take the liberty to submitting it in our reviews. Similarly, if an article was cited by one of the query results and corresponding to PICOs but was not included in the query results, we were allowed to add it to our review.
2.3. Data Extraction
Requests results were compiled on Zotero (open access bibliographic tools), duplicates and retired research were automatically excluded. For data extraction we used tabulate file at format “.csv”.
3. Results
Flowchart of review was on
Figure 1. We did a first request on 3 databases the 30th December 2023 and an additional request on PubMed for 2024 research was realized the 1st June 2024 during reviews article writing. The results of the 17 articles retained for this review have been grouped in
Table 1. Among them, three presented an algorithmic methodology using DL strategies for the segmentation and extraction of radiomics features which are then pipelined into a ML algorithm (Support Vector Machine (SVM), k-Nears Neighbors (k-NN) or Random Forest) S. RATHORE et al. [
9], S. KIHIRA et al. [
10]. and S. QURESHI et al.[
11].
3.1. Bibliographical and Descriptive Data on Publications
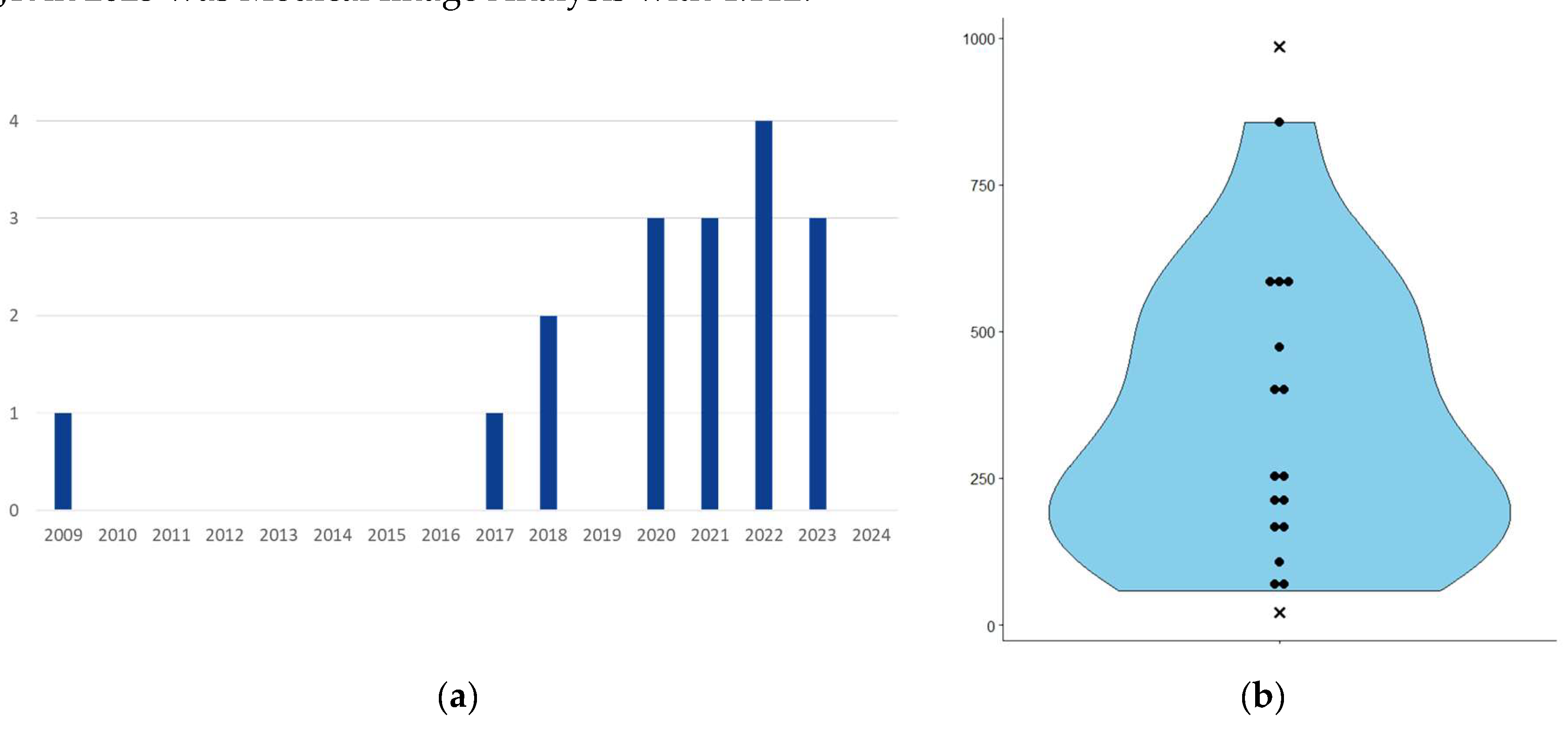
The 17 publications range from 2009 to 2023 (
Figure 2.a). For each of these publications, we collected the Scimago Journal Ranking (SJR) score for the year of publication; median 1.312, min 0.297, max 4.112. For comparison, the SJR scores for 2023 for Nature, Cell and the New England Journal of Medicine were 18.509, 24.342 and 20.544 respectively. The medical imaging journal with the highest SJR in 2023 was Medical Image Analysis with 4.112.
The number of patients used to develop the algorithms ranges from 21 to 985 (
Figure 2.b). However, I. HRAPȘA et al. [
21] presented replication work on 21 patients without re-training, based on the work of Y.S. CHOI et al.[
20] The article of B-H KIM et al.[
23] used up to 985 patients were merged from the Seoul National University Hospital (SNUH) dataset (400 patients) and the MICCAI BrATS 2021 dataset (585 patients). The algorithm proposed in this was first successfully trained on the 400 patients of the SNUH set.
Among the 17 articles, seven used data from patients with glioblastomas, 5 used data from patients with glioma and five used data from patients with both glioblastoma and glioma.
The MRI sequences used in the various articles were primarily T2-weighted (T2w) and T2 Fluid Attenuated Inversion Recovery (T2-FLAIR) (
Table 2).
3.2. Deep-Learning Strategy
All the articles used algorithms corresponding to the definition of deep-learning. Of these, 3 used DL algorithms but the task of classifying tumors according to their genetic characteristics was subjected to a ML algorithm[
2,
3,
4].
Among the algorithmic strategies, 10 of the 17 papers used convolutional neural networks (CNN) with fully convolutional architecture. These strategies were used either directly on the images or after automatic segmentation by a deep-learning algorithm with a specific architecture or after human segmentation.
Algorithms with specific architectures included DenseNet, ResNet and U-Net. One of this article have worked on transfer learning from non-medical image classification into medical image classification (H SAKLY et al. 2023 [
24]).
3.3. Tumor Genetics’ Explored
The articles have mainly focused on the prediction of genetic or epigenetic disorders: presence of the IDH1/2 mutation, methylation of the MGMT promoter, over-expression of EGFR and the presence of the 1p/19q co-deletion (
Table 3). One of the papers also worked on predicting the presence of tumors variations affecting
PTEN,
ATRX,
TERT,
CDKN2A/
B,
TP53 and chromosomic rearrangement Trisomy7-Monosomy10 [
9].
3.3.1. IDH Mutation Prediction
Based on the AUC (Area Under the ROC (Receiver Operating Characteristic) Curve), the algorithm proposed by Y. S. CHOI et al.[
20] showed the best results at 0.96 (CI 95%: 0.93-0.99) (
Table 4). However, the same algorithm replicated on external data by I. HRAPȘA et al. found an AUC of 0.74 (CI 95%: 0.53-0.91) with a sensitivity of 78% and a specificity of 75%[
21]. The first of these articles worked on patients labelled as having gliomas and glioblastomas, while the second submitted only patients labelled as having glioblastomas. One of the highest performance was proposed by P. EICHINGER et al.[
13] with an AUC of 0.952 and archived this result with 79 patients in comparison of 856 patients. of the article by Y. S. CHOI et al.[
20]
3.3.2. MGMT Promoter Methylation Prediction
The best performance was achieved by algorithm of S. A. QURESHI et al. with an AUC of 0.96 (CI95%: 0.94-0.98)[
11] (
Table 5). However, this performance was obtained via an algorithm that combined deep-learning for segmentation and radiomics extraction but then pipped a classifier based on a Radom Forest algorithm (which is a machine-learning strategy).
The second best performance was obtained by P. CHANG et al. with an AUC of 0.81 (CI 95%: 0.76–0.84)[
14]. Among the results, two have an AUC with a confidence interval of 0.5, and are therefore potentially no better than chance[
19]. Finally, the work of I. LEVNER, who in 2009 proposed an innovative work using an extremely small neural network, based on MRI textural studies to predict MGMT promoter methylation with an accuracy of 87.7%, trained on a very small dataset. This work is more of a proof of concept. H. SAKLY et al.[
24] also proposes to test transfer learning on image classification algorithms.
Finally, we should mention the paper by SAEED et al. who did not present the best results (Table 5), but did present results comparing several deep-learning algorithm architectures. An important part of their work was not to create and train the most robust algorithm, but to understand where disparities in results from the same algorithm might come from.
3.3.3. EGFR Amplification Prediction
In our review, only three articles sought to predict the status of EGFR amplification (overexpression) (
Table 6). Among them, M. HEDYEHZADEH et al. did not present any results we could compare, but a deep-learning strategy was able to statistically detect EGFR amplification in glioblastomas [
16].
In the remaining two articles, the results presented by S. RATHORE et al. were slightly better, but the final classifier was not a deep-learning algorithm but a random forest classifier [
9]. In the article of E. CALABRESE et al. thy also presented better results for their Random Forest classifier than for their CNN classifier.[
22]
3.3.4. Chromosome 1p19q Co-Deletion Prediction
The algorithm with the best performance for predicting 1p19q co-deletion status was proposed by P. CHANG et al.[
14] (
Table 7). We noted among these four articles, the paper of B. KOCAK et al. [
18] who tested a CNN and ML algorithms (k-NN, Random Forest, SVM, etc.) on the same dataset with better results for the CNN.
4. Discussion
4.1. Evolution of Publications
A chronological analysis of publications shows a notable gap between the first study in 2009 and subsequent studies from 2017 onwards. This hiatus could be explained by several factors. In 2009, the classification of gliomas and glioblastomas was based mainly on histological criteria, with limited involvement of genetic characteristics in therapeutic decisions. Methylation of the
MGMT promoter influenced the choice of chemotherapy. Thus, I. LEVNER et al.[
12] proposed a proof-of-concept using a two-layer CNN in 2009 to predict this methylation status with 87% accuracy on a training dataset. It wasn’t until 2017 that more sophisticated work integrating CNN architectures, and then more advanced models such as ResNet and DenseNet, emerged, reflecting advances in DL and the growing importance of genetics in glioma classification and treatment.
4.2. Deep Learning Algorithms
The neural network architectures used in the studies reviewed have evolved over time. Initially, Fully Convolutional Neural Networks (FCNN) were used. Later, more complex architectures such as ResNet, DenseNet, and U-Net were explored. One team even carried out a proof-of-concept with transfer learning using models such as AlexNet, GoogleNet, or InceptionV2[
24]. However, there were significant variations in the validation methods used between studies. Some studies used a classic split train-test, while others opted for k-fold cross-validation with k-values of 3, 4, or 5, or LOOCV (Leave-One-Out Cross-Validation) for small samples. Validation on an external dataset was clearly not the norm, which could be a major shortcoming. However, we should note the article by Y. S. CHOI et al. who proposed an external validation on the TCIA and the SNUH set, with significantly poorer results on the TCIA and comparable results on the SNUH set[
20]. This difference in results between SNUH and TCIA could be due to the fact that the training data comes from patients managed in Seoul, whose management and imaging modalities (as well as the histological study of the tumor and the oncobiological study of molecular characteristics) are probably carried out in much the same way as the data aggregated in SNUH.
4.3. Performance and Reproducibility
Algorithms with very high AUCs (close to 1) raise questions about their generalizability. A publication by N. SAEED et al.[
25] explored this issue, highlighting that the limited size of datasets and inadequate validation methods can artificially inflate performance. Most studies are based on a single dataset, thus limiting the reproducibility of results.
Combining Genetic Traits Although some attempts have been made to predict multiple genetic traits in a single model, results are often presented individually for each trait. This suggests a juxtaposition of classifiers rather than an integrated model capable of distributed prediction in a multidimensional space.
4.4. Limitations and Challenges
The main limitations identified by the authors include the limited size of the datasets and their homogeneity. With fewer than 1,000 patients, the datasets cannot capture the necessary variability, especially as a single DICOM MRI can provide up to 1,000 radiomics parameters. Another crucial point seems to be the recurrent absence of clearly identified external validation datasets, serving only to assess the reliability and reproducibility of the algorithms. This is essential to guarantee the generalizability of the models in the future. On the other hand, it might be appropriate to integrate an explanatory dimension into the development of algorithms to better understand their limits and biases. This could also be necessary to ensure medical liability.
Another challenge to overcome would be to understand the genetic heterogeneity of a tumor. However, the datasets currently available will not allow this. It will therefore be necessary to develop datasets incorporating three-dimensional information on the biopsied area of the tumor (which could take the form of a three-dimensional biopsy probability field).
Finally, a point not clearly identified by the researchers is the disparity between one dataset and another (which we can suggest between TCIA and SNUH, given the difference in results from Y. S. CHOI et al.[
20]). Indeed, there are no guidelines on the initial MRI assessment of a brain tumor (type of sequences to be used, field to be proposed 3T versus 1.5T), or that the acquisition of genetic data from a tumor depends on many factors including the neurosurgeon’s expertise, the histological study, and the choice of fragment to be studied, to which molecular biology methods and their interpretation will be applied, which again are not set by guidelines.
Of course, to understand this theme, it’s important to bring together all the specialists involved, including mathematicians, radiologists, pathologists, geneticists, and neurosurgeons, to best respond to clinical demand.
5. Conclusion
Our literature review of 17 articles on the use of DL algorithms in the radiogenomic characterization of gliomas and glioblastomas highlights the advances but above all the persistent challenges in this emerging field. Since 2009 and especially since 2017, there are still many obstacles to overcome to guarantee the effectiveness and generalization of the models developed with a view to a hypothetical virtual biopsy.
The trend in publications shows that interest in this theme has increased since 2017, largely thanks to technological advances and growing recognition of the importance of genetic markers in the characterization, prognosis, and treatment of gliomas and glioblastomas (WHO 2021 classification). However, model validation methods and the limited size of the datasets used remain major weaknesses. The apparently high performance of some algorithms needs to be taken with caution, due to the recurrent lack of validation on external datasets, which raises questions about their generalizability.
Moreover, efforts to combine the prediction of several genetic traits in a single integrated model still seem to be in their infancy. Most studies focus on individual predictions, suggesting a juxtaposition of classifiers rather than a true distributed multidimensional approach. To improve this situation, it will be crucial to develop larger and more varied datasets (ethnic origin, tumor size, histological type, molecular and cytogenetic features). In addition, to understand tumor genetic heterogeneity, it would be desirable to include three-dimensional information on the biopsied area(s). In the long term, this could help to determine the tumor characteristics of tissue at the edge of tumor resection, for example.
The current limitations of datasets and the lack of robust validation methods underline the need for interdisciplinary collaboration. Integrating mathematicians, radiologists, pathologists, geneticists, and neurosurgeons into the algorithm development process will create models that are more reliable and better aligned with clinical practice. Moreover, incorporating an explanatory dimension into the algorithms could help to understand and then mitigate their biases and limitations.
Finally, although DL offers promising prospects for the radiogenomic characterization of gliomas and glioblastomas, it is essential to continue improving datasets, validation methods, and interdisciplinary collaborations to ensure that these tools can be effectively used in the clinic and unlock virtual biopsy capability. This new technology in healthcare would enable objective early therapeutic decision-making, fitting perfectly into a 5P medicine (personalized, preventive, predictive, participative, and pertinent).
Supplementary Materials
The following supporting information can be downloaded at:
www.mdpi.com/xxx/s1, S1: Requests of 3 bases;
Author Contributions
Conceptualization, Xavier LE GUILLOU HORN and François LECELLIER; methodology Mathieu NAUDIN and Clement GIRAUD; software, Pierre FAYOLLE and Céline THOMARAT; formal analysis, Xavier LE GUILLOU HORN and François LECELLIER; writing—original draft preparation, Xavier LE GUILLOU HORN; writing—review and editing, Christine FERNANDEZ MALOIGNE and Rémy GUILLEVIN.; visualization, Clement GIRAUD; supervision, Rémy GUILLEVIN. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article
Conflicts of Interest
The authors declare no conflicts of interest.
References
- S. Herculano-Houzel, « The human brain in numbers: a linearly scaled-up primate brain », Front. Hum. Neurosci., vol. 3, nov. 2009. [CrossRef]
- V. Parpura et al., « Glial cells in (patho)physiology », J Neurochem, vol. 121, no 1, p. 4-27, avr. 2012. [CrossRef]
- Q. T. Ostrom et al., « CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2015–2019 », Neuro Oncol, vol. 24, no Suppl 5, p. v1-v95, oct. 2022. [CrossRef]
- D. N. Louis et al., « The 2021 WHO Classification of Tumors of the Central Nervous System: a summary », Neuro Oncol, vol. 23, no 8, p. 1231-1251, août 2021. [CrossRef]
- M. E. Hegi et al., « MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma », The New England Journal of Medicine, vol. 352, no 10, p. 997-1003, mars 2005. [CrossRef]
- R. Stupp et al., « Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma », N Engl J Med, vol. 352, no 10, p. 987-996, mars 2005. [CrossRef]
- A. Krizhevsky, I. Sutskever, et G. E. Hinton, « ImageNet classification with deep convolutional neural networks », Commun. ACM, vol. 60, no 6, p. 84-90, mai 2017. [CrossRef]
- D. Moher, A. Liberati, J. Tetzlaff, D. G. Altman, et PRISMA Group, « Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement », J Clin Epidemiol, vol. 62, no 10, p. 1006-1012, oct. 2009. [CrossRef]
- S. Rathore et al., « Multi-institutional noninvasive in vivo characterization of IDH, 1p/19q, and EGFRvIII in glioma using neuro-Cancer Imaging Phenomics Toolkit (neuro-CaPTk). », Neurooncol Adv, vol. 2, no Suppl 4, p. iv22-iv34, déc. 2020,. [CrossRef]
- S. Kihira et al., « U-Net Based Segmentation and Characterization of Gliomas », Cancers, vol. 14, no 18, 2022. [CrossRef]
- S. A. Qureshi et al., « Radiogenomic classification for MGMT promoter methylation status using multi-omics fused feature space for least invasive diagnosis through mpMRI scans », Sci Rep, vol. 13, no 1, p. 3291, 2023. [CrossRef]
- I. Levner, S. Drabycz, G. Roldan, P. De Robles, J. G. Cairncross, et R. Mitchell, « Predicting MGMT methylation status of glioblastomas from MRI texture », Med Image Comput Comput Assist Interv, vol. 12, no Pt 2, p. 522-530, 2009. [CrossRef]
- P. Eichinger et al., « Diffusion tensor image features predict IDH genotype in newly diagnosed WHO grade II/III gliomas », Sci Rep, vol. 7, no 1, p. 13396, 2017. [CrossRef]
- P. Chang et al., « Deep-Learning Convolutional Neural Networks Accurately Classify Genetic Mutations in Gliomas », AJNR Am J Neuroradiol, vol. 39, no 7, p. 1201-1207, juill. 2018. [CrossRef]
- S. Liang et al., « Multimodal 3D densenet for IDH genotype prediction in gliomas », Genes, vol. 9, no 8, 2018. [CrossRef]
- M. Hedyehzadeh, K. Maghooli, M. MomenGharibvand, et S. Pistorius, « A Comparison of the Efficiency of Using a Deep CNN Approach with Other Common Regression Methods for the Prediction of EGFR Expression in Glioblastoma Patients », J Digit Imaging, vol. 33, no 2, p. 391-398, 2020. [CrossRef]
- Y. Matsui et al., « Prediction of lower-grade glioma molecular subtypes using deep learning », J. Neuro-Oncol., vol. 146, no 2, p. 321-327, 2020. [CrossRef]
- B. Kocak et al., « Radiogenomics of lower-grade gliomas: machine learning–based MRI texture analysis for predicting 1p/19q codeletion status », Eur. Radiol., vol. 30, no 2, p. 877-886, 2020. [CrossRef]
- C. G. B. Yogananda et al., « MRI-Based Deep-Learning Method for Determining Glioma MGMT Promoter Methylation Status », AJNR Am J Neuroradiol, vol. 42, no 5, p. 845-852, mai 2021. [CrossRef]
- Y. S. Choi et al., « Fully automated hybrid approach to predict the IDH mutation status of gliomas via deep learning and radiomics », Neuro-Oncology, vol. 23, no 2, p. 304-313, févr. 2021. [CrossRef]
- I. Hrapșa, I. A. Florian, S. Șușman, M. Farcaș, L. Beni, et I. S. Florian, « External Validation of a Convolutional Neural Network for IDH Mutation Prediction », Medicina (Kaunas), vol. 58, no 4, 2022. [CrossRef]
- [22] E. Calabrese et al., « Combining radiomics and deep convolutional neural network features from preoperative MRI for predicting clinically relevant genetic biomarkers in glioblastoma », NeuroOncol. Adv., vol. 4, no 1, 2022. [CrossRef]
- B.-H. Kim et al., « Validation of MRI-Based Models to Predict MGMT Promoter Methylation in Gliomas: BraTS 2021 Radiogenomics Challenge », Cancers, vol. 14, no 19, 2022. [CrossRef]
- H. Sakly, M. Said, J. Seekins, R. Guetari, N. Kraiem, et M. Marzougui, « Brain Tumor Radiogenomic Classification of O6-Methylguanine-DNA Methyltransferase Promoter Methylation in Malignant Gliomas-Based Transfer Learning », Cancer Control, vol. 30, no (Sakly H., houneida.sakly@esiee.fr; Kraiem N.) RIADI Laboratory, ENSI, Manouba University, Campus Universitaire de La Manouba, La Manouba, Tunisia, 2023. [CrossRef]
- N. Saeed, M. Ridzuan, H. Alasmawi, I. Sobirov, et M. Yaqub, « MGMT promoter methylation status prediction using MRI scans? An extensive experimental evaluation of deep learning models. », Med Image Anal, vol. 90, p. 102989, déc. 2023. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).