1. Introduction
Unconventional computing is a field that investigates different ways of processing information and performing computations, going beyond the use of classic silicon-based technologies [
1]. Bio-inspired computing systems have attracted considerable attention in this field because of their promise for energy economy, versatility, and parallel processing capabilities [
2]. Recent progress in the field has resulted in the examination of several biological and chemical materials for use in computation. These include DNA-based computing [
3], reaction-diffusion systems [
4], and neuromorphic computing [
5]. The latter, which draws inspiration from the form and function of biological neural networks, has demonstrated significant potential in tasks such as pattern recognition and adaptive learning [
6]. Our research specifically examines the relationship between omeprazole-proteinoid complexes and Izhikevich neurone models. Omeprazole, a widely used proton pump inhibitor for gastric disorders [
7], is combined with proteinoids, which are thermal proteins capable of forming microspheres and have been extensively explored in the field of artificial cells [
8]. The Izhikevich neurone model, known for its computational efficacy and capacity to replicate a diverse array of neuronal firing patterns [
9], serves as our framework for exploring the computational characteristics of these biochemical complexes. We analyse five specific neuronal behaviours: accommodation, chattering, triggered spiking, phasic spiking, and tonic spiking. These behaviours indicate diverse ways in which biological neural networks do computations [
10]. Our objective is to discover new methods for information processing and computing by studying the effects of omeprazole-proteinoid complexes on neuronal dynamics. This technique connects the fields of pharmacology, proteinoid chemistry, and neuromorphic computing, potentially creating opportunities for the advancement of adaptive, bio-inspired computational systems [
11,
12,
13]. Our research adds to the expanding field of neuromorphic substrates and could impact the development of future hybrid bio-synthetic computational architectures. Moreover, it offers valuable information about the possible neuromodulatory impacts of pharmacological drugs when paired with proteinoids, which could have significant implications for the fields of computing and neuropharmacology. To fully understand the spiking behaviour and signal processing abilities of the omeprazole-proteinoid complex, it is essential to have a solid foundation of the molecular structures of the proteinoid (L-Glu:L-Asp:L-Phe) and omeprazole.
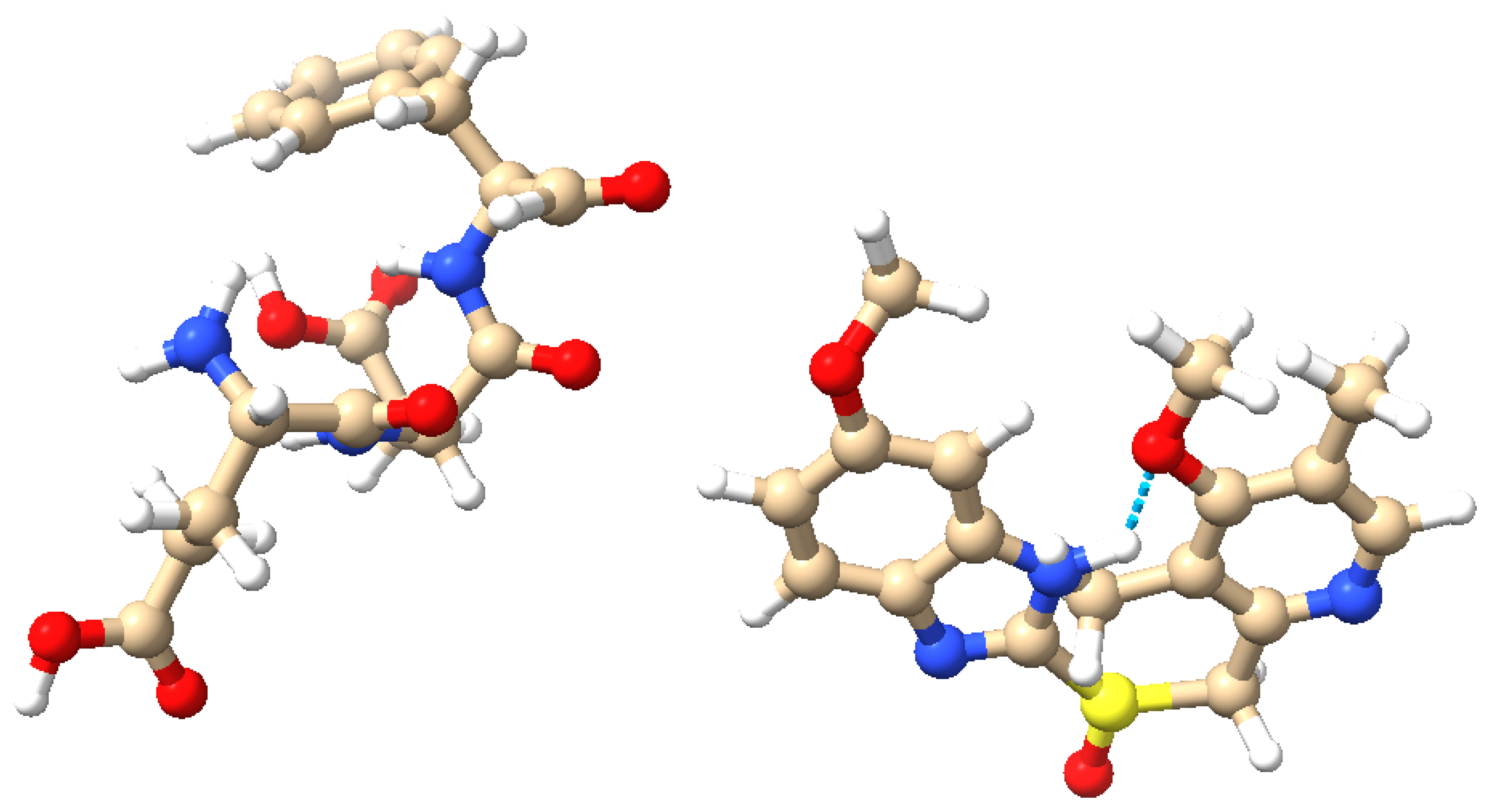
Figure 1 depicts these structures and their possible interactions. The amino acid composition of the proteinoid offers a range of functional groups that can engage in hydrogen bonding and other non-covalent interactions with omeprazole. These interactions are highly likely to have a crucial impact on the voltage-sensitive conformational changes and charge redistribution pathways that are described in our mechanistic model (Figure 10). The precise configuration of atoms and chemical bonds in omeprazole, specifically its sulfoxide group and benzimidazole ring, could potentially impact the electrical characteristics of the proteinoid. This influence may arise from the modulation of local pH gradients or the modification of conductive pathways within the complex.
The Izhikevich neuron model is described by a system of two differential equations:
with the auxiliary after-spike resetting:
Here, v represents the membrane potential of the neuron, u is a recovery variable, and I is the input current. The parameters a, b, c, and d are dimensionless parameters that can be adjusted to produce various types of neuronal behavior:
a: the time scale of the recovery variable u
b: the sensitivity of the recovery variable u to the subthreshold fluctuations of the membrane potential v
c: the after-spike reset value of the membrane potential v
d: after-spike reset of the recovery variable u
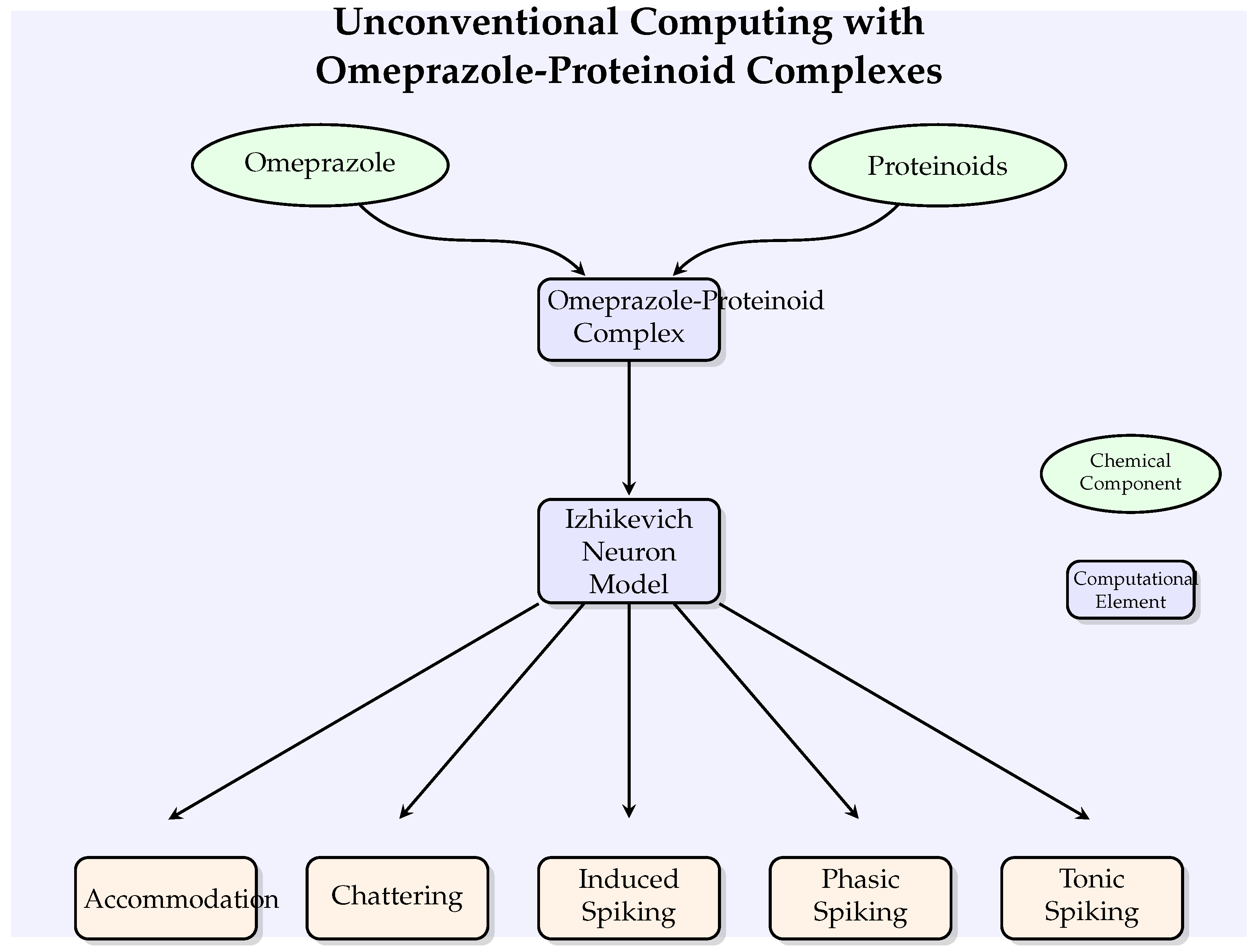
Figure 2 illustrates the conceptual framework of our unconventional computing approach, showing how omeprazole-proteinoid complexes interact with the Izhikevich neuron model to potentially modulate various neuronal behaviours.
Proton pump inhibitors (PPIs) are a type of drug that decreases the production of acid in the gut by permanently blocking the hydrogen/potassium adenosine triphosphatase enzyme system in the cells of the stomach lining [
15]. Omeprazole, the subject of this investigation, is among a number of Proton Pump Inhibitors (PPIs) presently being used in clinical practice.
Table 1 displays a comparison of omeprazole and other prevalent PPIs, emphasising their chemical formulae, half-lives, and pKa values. Although these medications have a similar way of working, they have slight variations in their pharmacokinetic and pharmacodynamic characteristics [
16,
17,
18,
19]. We selected omeprazole for this work due to its extensive usage and well-established characteristics, which make it an excellent candidate for investigating potential neuromodulatory effects in conjunction with proteinoid structures.
4. Discussion
The detailed examination of omeprazole-proteinoid complexes using several spiking modes demonstrates a complex and adaptable signal processing system. The results of our study indicate that these complexes have unique responses to various input patterns, indicating the possibility of processing many types of information at the molecular level.
4.1. Comparative Analysis of Spiking Modes
We detected considerable signal attenuation and change in all spiking modes, including accommodation, chattering, induced, phasic, and tonic. Nevertheless, the extent and characteristics of this change differed significantly among different modes:
Amplitude Modulation: All modes showed a significant decrease in signal amplitude, with output ranges constantly falling within a range of ±4 mV, whereas input ranges often exceeded ±60 mV. This implies the presence of a strong buffering mechanism that could protect molecular fluctuations downstream from extreme fluctuations in voltage.
Temporal Dynamics: The temporal delay between the input and output signals exhibited significant variation across different modes, ranging from ms in the chattering mode to 1590 milliseconds in the induced mode. The negative lag found in accommodation ( ms), phasic ( ms), and tonic ( ms) modes is particularly remarkable. This suggests the presence of anticipatory behaviour, which could have important consequences for information processing and response preparation in biological systems.
Signal Correlation: The correlation between the input and output signals varied from moderate (0.4503 in phasic mode) to strong (0.7937 in chattering mode). This suggests that the complexes effectively modify the input signal while retaining different levels of the original signal properties.
Distribution Transformation: The Kolmogorov-Smirnov tests consistently revealed significant differences between the distributions of the input and output data in all modes. The KS statistics ranged from 0.9276 (tonic) to 0.9945 (phasic). This implies the use of non-linear processing techniques that have the potential to amplify specific signal characteristics while simultaneously reducing the prominence of others.
4.2. Implications for Molecular Computing
The behaviours shown by omeprazole-proteinoid complexes when subjected to various spiking regimes have significant implications for molecular computing and bio-inspired signal processing.
Multi-modal Processing: The diverse reactions to various spiking patterns indicate that these complexes have the ability to function as versatile molecular processors, adjusting their behaviour according to input parameters.
Non-linear Transformation: The persistent non-linear alteration of input signals, as indicated by the results of the KS test and Q-Q plots, suggests that these complexes perform complex signal processing procedures that go beyond mere filtering or amplification.
Anticipatory Behaviour: The presence of negative time delays in several modes indicates the occurrence of predictive processing at the molecular level. These findings could have important consequences for the development of molecular systems that can anticipate events or for understanding biological reactions that occur before an event.
Robust Signal Normalization: The consistent output range observed in all input modes indicates that these complexes have the potential to function as reliable signal normalisers, which could be valuable in molecular-scale sensor systems [
26] or signal processing units [
27].
4.3. Potential Mechanisms and Future Directions
The observed behaviours are most likely a result of complex interactions among the omeprazole molecules, the proteinoid structure, and the electrical fluctuations that were applied. Possible mechanisms encompass voltage-dependent alterations in the proteinoid structure, emergence and disintegration of transient conductive pathways within the complex, accumulation and redistribution of charges with distinct time constants, and interactions between omeprazole’s inhibition of proton pumps and local pH gradients. Future research should prioritise conducting molecular dynamics simulations to uncover the underlying structural mechanisms of the observed behaviours. In addition, it should investigate the frequency-dependent responses of these complexes, explore potential applications in molecular-scale signal processing and computing, and examine how these properties can be adjusted or changed through chemical modifications. Our analysis concludes that omeprazole-proteinoid complexes possess diverse signal processing capacities that vary depending on the mode. These findings not only improve our understanding of molecular-scale information processing but also create new opportunities for the advancement of bio-inspired computing systems and smart drug delivery mechanisms.
The complex behaviours observed in various spiking modes are presumably the result of a combination of molecular-level mechanisms within the omeprazole-proteinoid complexes.
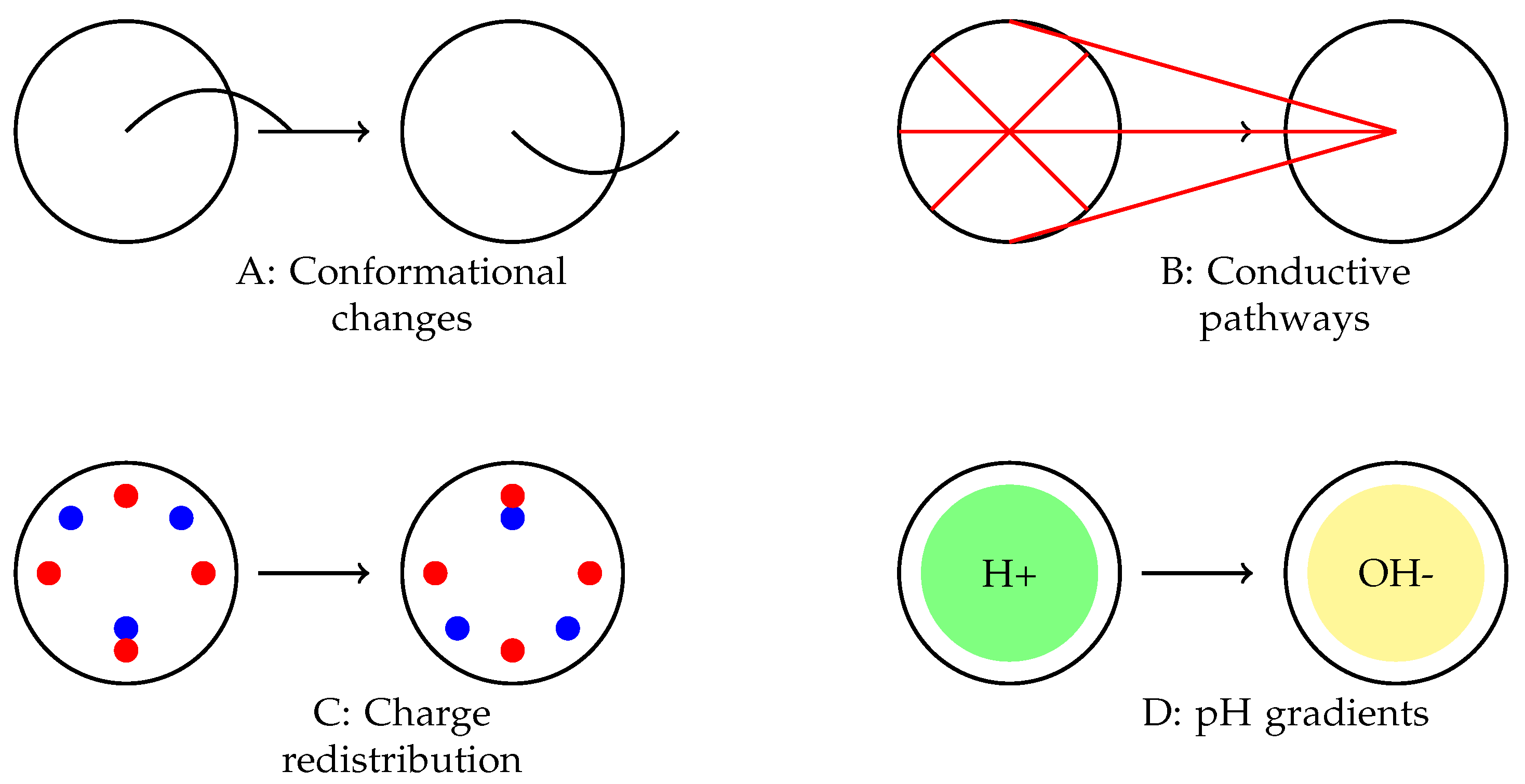
Figure 10 depicts many suggested mechanisms that could potentially contribute to the observed signal processing capabilities. These factors include changes in the proteinoid structure that are sensitive to voltage (
Fig. 10A), which could explain the different responses depending on the mode; the creation and breakdown of temporary pathways for conducting signals (
Figure 10B), which may account for the non-linear transformation of the signal; processes of accumulating and redistributing charges (
Figure 10C), which could explain the observed delays and anticipatory behaviours; and the interaction between omeprazole’s inhibition of proton pumps and local differences in pH (
Figure 10D), which may contribute to the consistent normalisation of the signal across all modes. The interaction of these mechanisms could elucidate the diverse and flexible behaviour of the omeprazole-proteinoid system under different patterns of stimulation. Additional research into these processes at the molecular level will be essential for gaining a complete understanding and perhaps utilising these capabilities in molecular computing [
28] and smart drug delivery systems [
29].
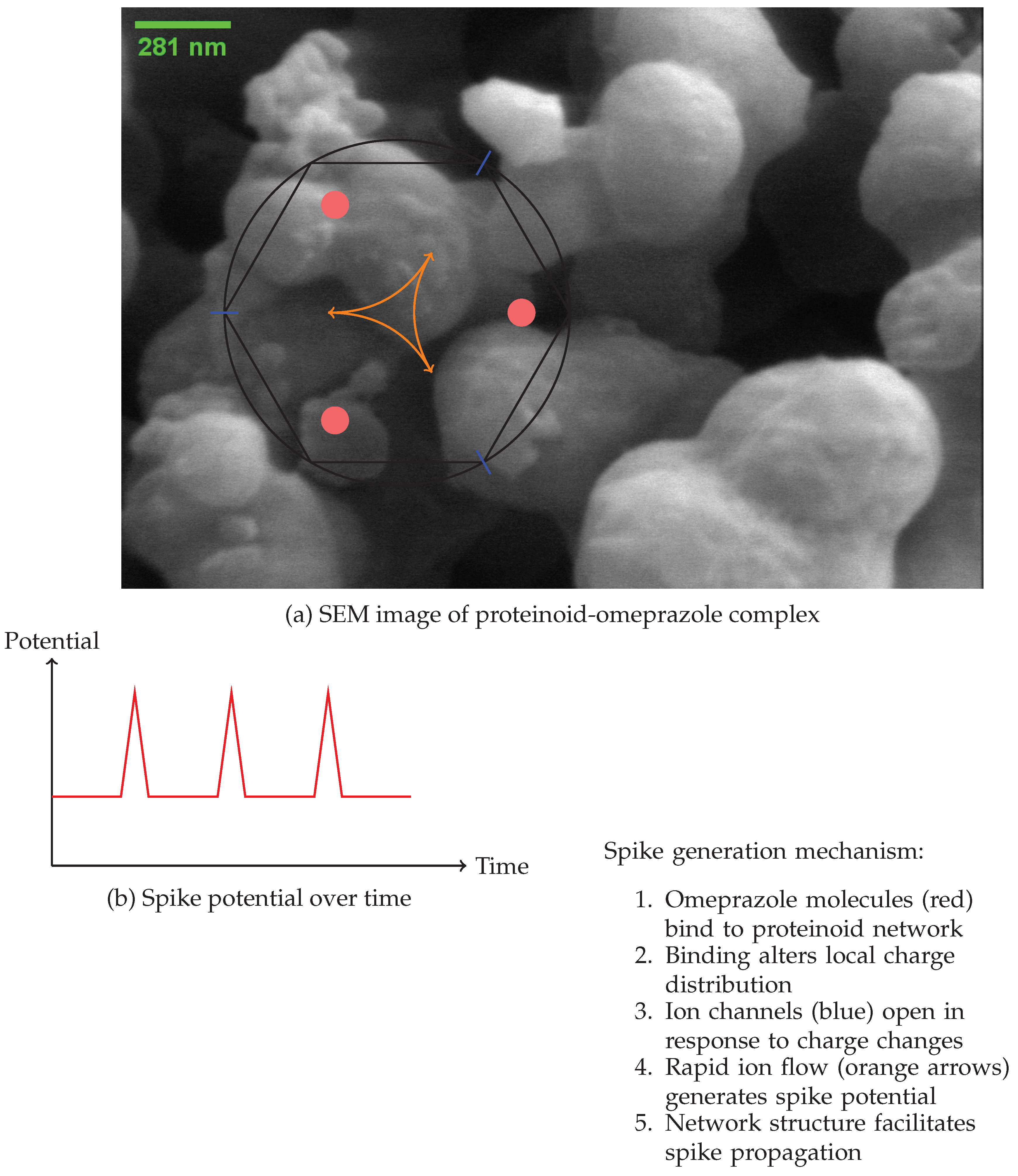
The study of the proteinoid-omeprazole complex unveils a complex mechanism [
30] for spike emergence, as depicted in
Figure 11. The scanning electron microscope (SEM) image (
Figure 11a) depicts a complex network structure that serves as the foundation for the observed electrical behaviour. Our hypothesis suggests that the process of spike formation consists of a sequence of stages, starting with the attachment of omeprazole molecules to the proteinoid network (
Figure 11a). The occurrence of this binding event can be mathematically represented by the equation [
15]:
where P represents the proteinoid binding site, O represents omeprazole, and PO is the bound complex. Omeprazole binding modifies the local distribution of electric charge inside the complex. This alteration can be represented as a disturbance to the nearby electric field:
where
is the initial electric field and
is the change induced by the omeprazole binding. The modified electric field stimulates the activation of ion channels [
31] located near the binding site. The probability of an ion channel opening can be described by a Boltzmann distribution:
where z is the gating charge, V is the membrane potential, is the half-activation voltage, k is the Boltzmann constant, and T is the temperature.
The activation of these channels results in a fast movement of ions, which in turn generates a spike potential. The membrane potential during a spike can be represented using the Hodgkin-Huxley equations [
22], which have been simplified in this context for the sake of simplicity.
where is the membrane capacitance, represents the sum of ionic currents, and is any external current.
The resulting spike potential over time is depicted in
Figure 11c, showing the characteristic rapid rise and fall of the membrane potential.
The network structure of the proteinoid-omeprazole complex, as observed in the SEM image, plays a crucial role in facilitating the propagation of these spikes. The interconnected nature of the complex allows for the spread of the electrical signal, which can be modeled as a reaction-diffusion process [
32]:
where D is the diffusion coefficient and represents the non-linear reaction terms that account for the spike generation and propagation dynamics.
This proposed process establishes a connection between the structural characteristics exhibited in the SEM image and the functional electrical properties of the proteinoid-omeprazole mixture. The observed spiking behaviour is a result of the interaction between omeprazole binding, ion channel kinetics, and the network topology of the complex. This suggests a new method for bio-inspired signal processing [
33] and possible uses in neuromorphic computing [
34].
Figure 1.
Molecular structures of the proteinoid and omeprazole. Left: The proteinoid composed of L-Glutamic acid (L-Glu), L-Aspartic acid (L-Asp), and L-Phenylalanine (L-Phe). Right: The omeprazole molecule. Atoms are color-coded: blue (nitrogen), red (oxygen), brown (carbon), yellow (sulfur), and white (hydrogen). Potential hydrogen bonding is highlighted, illustrating the possible interactions between the proteinoid and omeprazole. These molecular structures and their interactions are key to understanding the complex signal processing behaviour observed in the omeprazole-proteinoid system. (Visualization created using UCSF Chimera [
14].)
Figure 1.
Molecular structures of the proteinoid and omeprazole. Left: The proteinoid composed of L-Glutamic acid (L-Glu), L-Aspartic acid (L-Asp), and L-Phenylalanine (L-Phe). Right: The omeprazole molecule. Atoms are color-coded: blue (nitrogen), red (oxygen), brown (carbon), yellow (sulfur), and white (hydrogen). Potential hydrogen bonding is highlighted, illustrating the possible interactions between the proteinoid and omeprazole. These molecular structures and their interactions are key to understanding the complex signal processing behaviour observed in the omeprazole-proteinoid system. (Visualization created using UCSF Chimera [
14].)
Figure 2.
A conceptual diagram illustrating the unconventional computing strategy involving omeprazole-proteinoid complexes and Izhikevich neurone models. The graphic depicts the combination of chemical components (shown by ellipses) with computational components (represented by rectangles) to regulate different neural behaviours.
Figure 2.
A conceptual diagram illustrating the unconventional computing strategy involving omeprazole-proteinoid complexes and Izhikevich neurone models. The graphic depicts the combination of chemical components (shown by ellipses) with computational components (represented by rectangles) to regulate different neural behaviours.
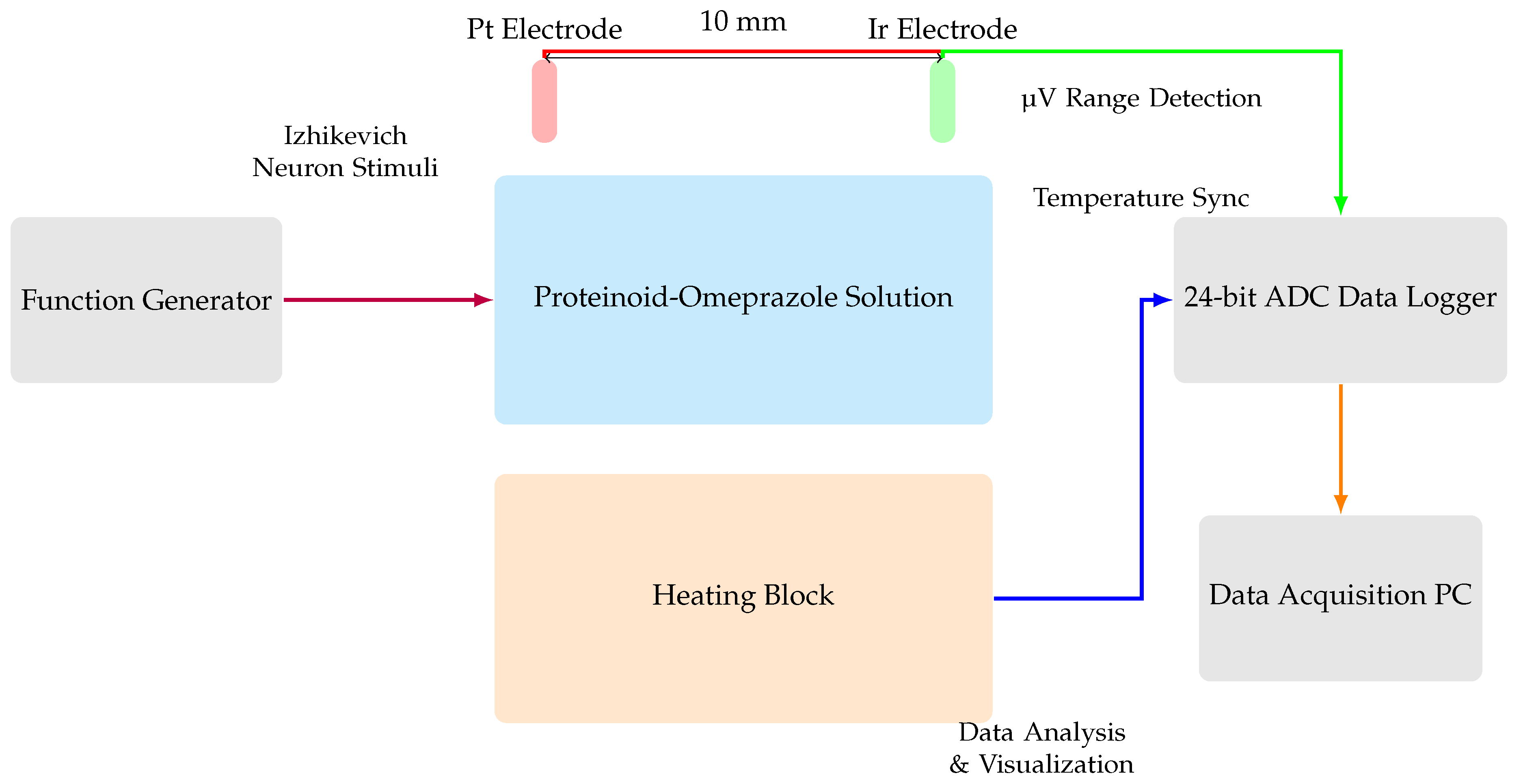
Figure 3.
Diagram illustrating the setup used for electrochemical characterisation of the proteinoid-omeprazole complex. The proteinoid-omeprazole solution is stored within the central container, which is equipped with two needle electrodes coated with platinum (Pt) and iridium (Ir) respectively. These electrodes are positioned 10 mm apart. A high-precision 24-bit ADC data recorder captures voltage responses, while a heating block regulates temperature. A waveform generator produces stimuli that resemble those experienced by neurones, while an oscilloscope displays the response of the system. This sophisticated setup allows for the identification of very small changes in voltage (in the range of microvolts) and a thorough analysis of the spatial and temporal patterns of voltage responses in the proteinoid-omeprazole system.
Figure 3.
Diagram illustrating the setup used for electrochemical characterisation of the proteinoid-omeprazole complex. The proteinoid-omeprazole solution is stored within the central container, which is equipped with two needle electrodes coated with platinum (Pt) and iridium (Ir) respectively. These electrodes are positioned 10 mm apart. A high-precision 24-bit ADC data recorder captures voltage responses, while a heating block regulates temperature. A waveform generator produces stimuli that resemble those experienced by neurones, while an oscilloscope displays the response of the system. This sophisticated setup allows for the identification of very small changes in voltage (in the range of microvolts) and a thorough analysis of the spatial and temporal patterns of voltage responses in the proteinoid-omeprazole system.
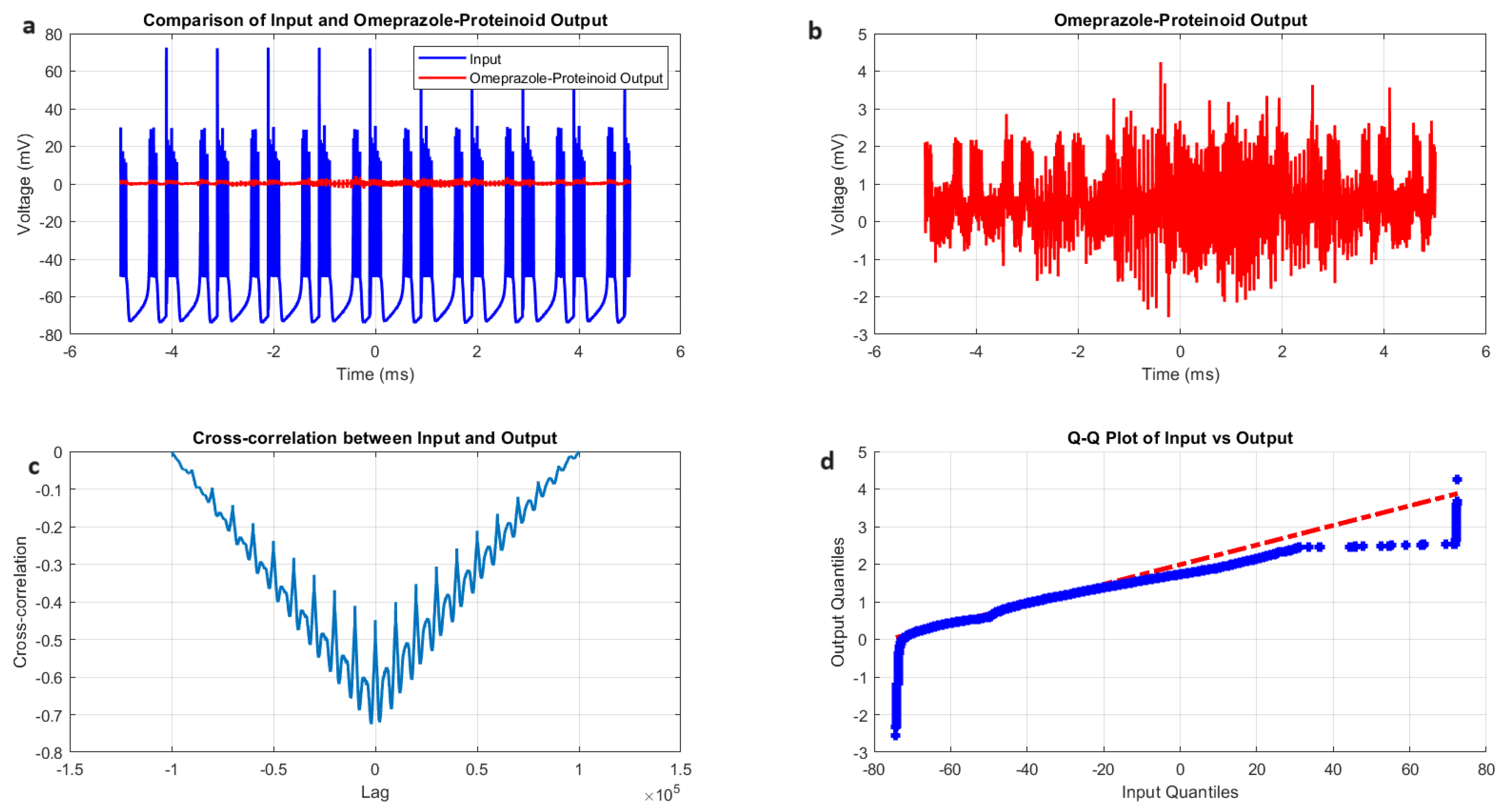
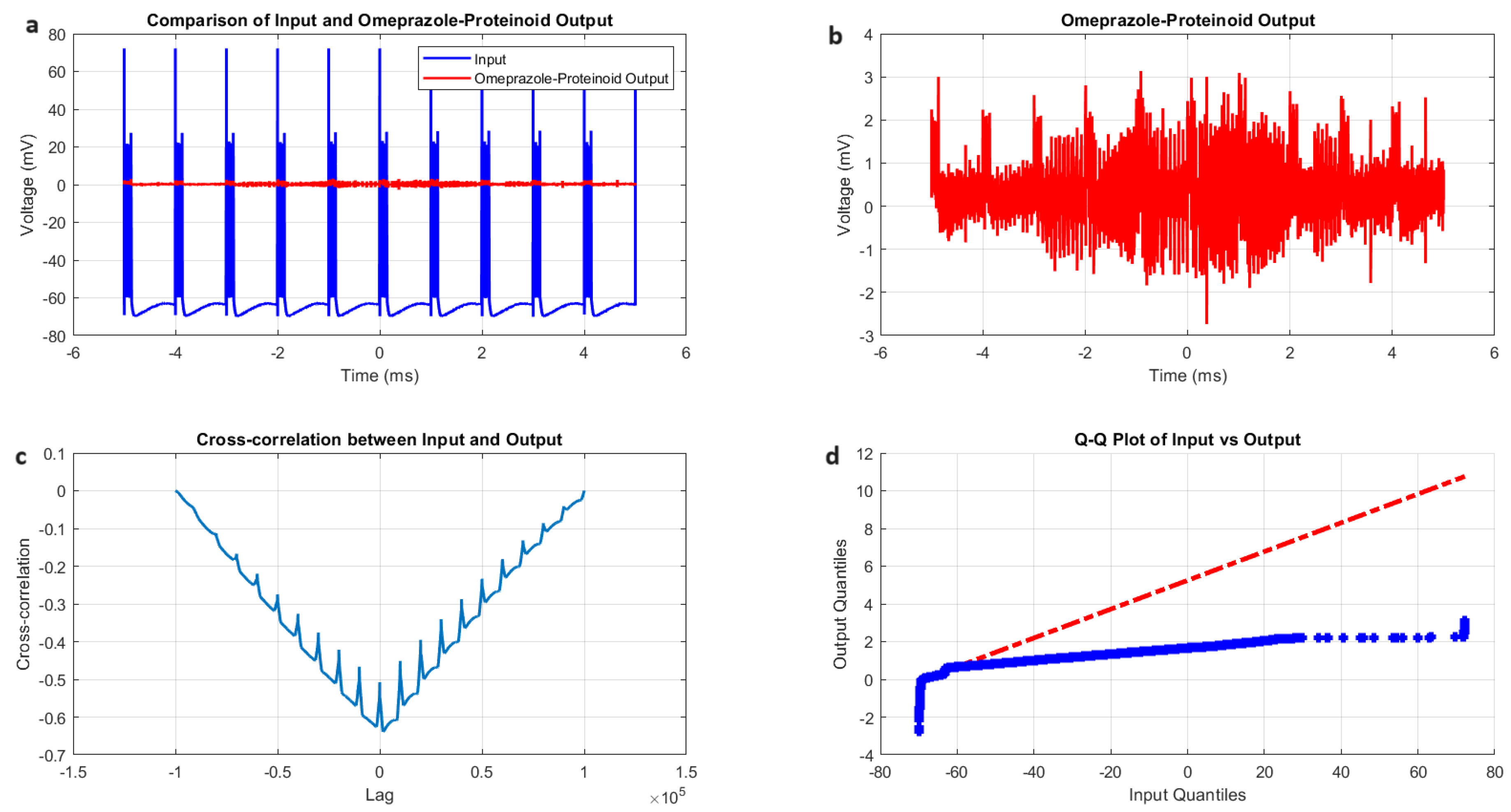
Figure 4.
Analysis of Izhikevich neuron accommodation spiking and omeprazole-proteinoid complex interaction. (A) Comparison of input Izhikevich neuron signal (blue, range: to 72.50 mV) and omeprazole-proteinoid output (red, range: to 3.99 mV), showing significant amplitude attenuation. (B) Isolated omeprazole-proteinoid output, revealing subtle voltage fluctuations (mean: 0.60 mV, SD: 0.43 mV) in response to input. (C) Cross-correlation between input and output, indicating a time lag of msec and a moderate positive correlation (r = 0.6841). (D) Q-Q plot demonstrating substantial deviation from the identity line, confirming significantly different distributions (Kolmogorov-Smirnov test: p < 0.0001, KS statistic: 0.9709). The omeprazole-proteinoid complex exhibits a marked filtering effect, reducing signal amplitude while preserving some temporal characteristics of the input. The RMSE of 50.4592 mV and maximum difference of 71.92 mV (at 1.39 ms) further quantify the substantial transformation of the signal. This analysis suggests complex neuromodulatory effects of the omeprazole-proteinoid interaction on accommodation spiking patterns.
Figure 4.
Analysis of Izhikevich neuron accommodation spiking and omeprazole-proteinoid complex interaction. (A) Comparison of input Izhikevich neuron signal (blue, range: to 72.50 mV) and omeprazole-proteinoid output (red, range: to 3.99 mV), showing significant amplitude attenuation. (B) Isolated omeprazole-proteinoid output, revealing subtle voltage fluctuations (mean: 0.60 mV, SD: 0.43 mV) in response to input. (C) Cross-correlation between input and output, indicating a time lag of msec and a moderate positive correlation (r = 0.6841). (D) Q-Q plot demonstrating substantial deviation from the identity line, confirming significantly different distributions (Kolmogorov-Smirnov test: p < 0.0001, KS statistic: 0.9709). The omeprazole-proteinoid complex exhibits a marked filtering effect, reducing signal amplitude while preserving some temporal characteristics of the input. The RMSE of 50.4592 mV and maximum difference of 71.92 mV (at 1.39 ms) further quantify the substantial transformation of the signal. This analysis suggests complex neuromodulatory effects of the omeprazole-proteinoid interaction on accommodation spiking patterns.
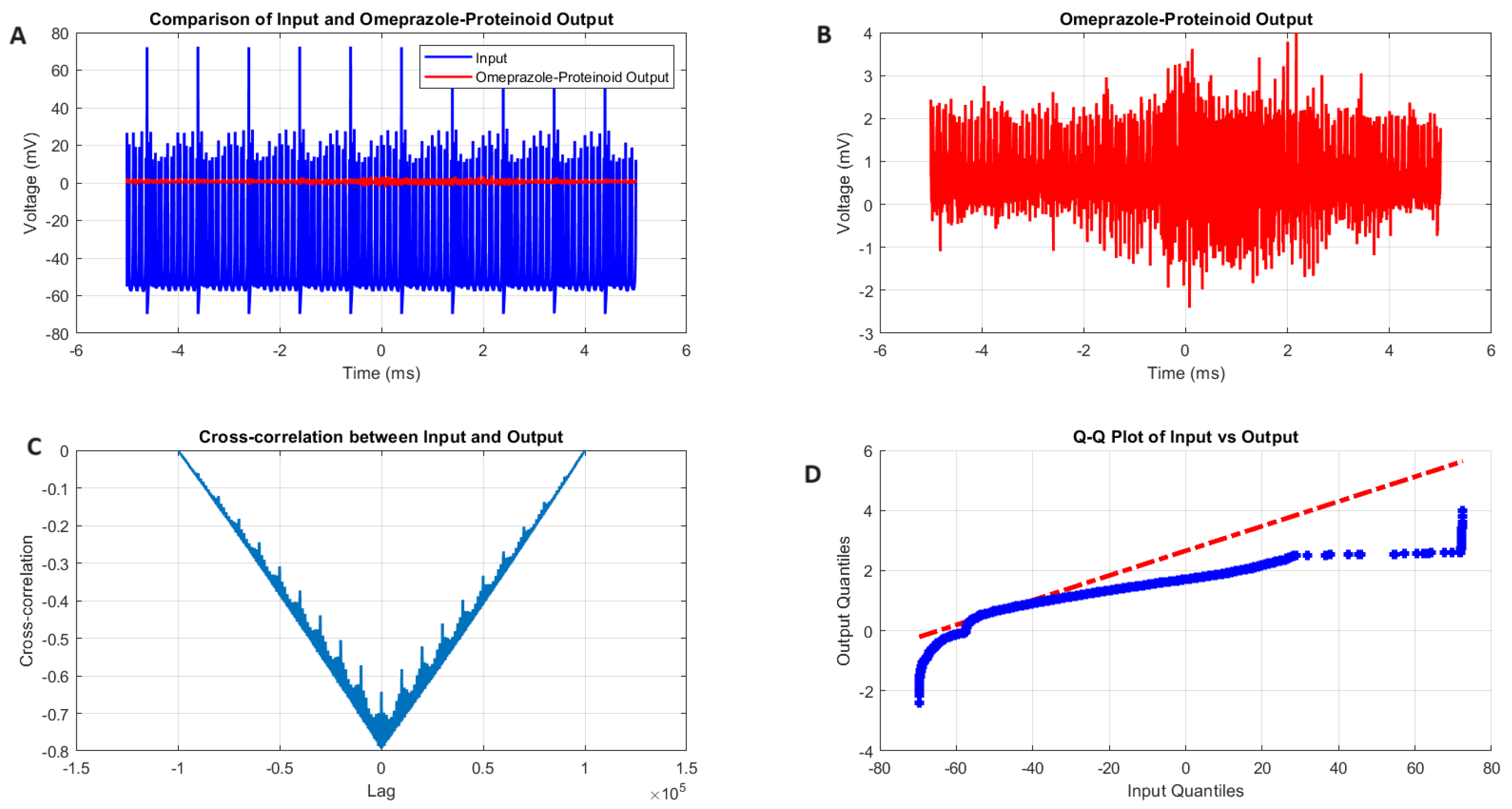
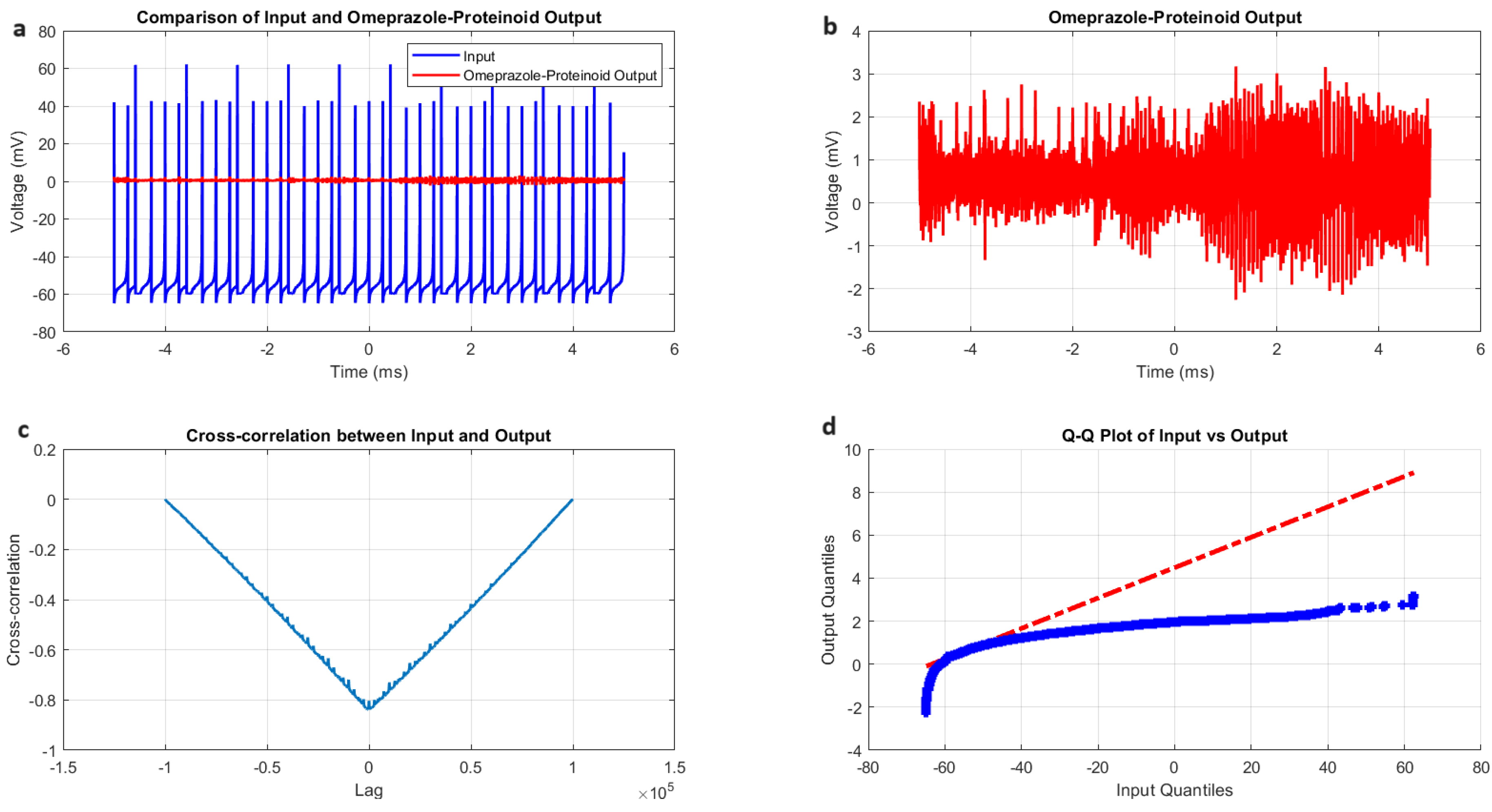
Figure 5.
Chattering spiking stimulation analysis of omeprazole-proteinoid sample. (a) Input-output comparison showing the input signal (mean: mV, SD: 19.72 mV) and the omeprazole-proteinoid output (mean: 0.48 mV, SD: 0.51 mV). Correlation coefficient: 0.7937, RMSE: 59.2964 mV. (b) Output plot highlighting the response characteristics of the omeprazole-proteinoid sample. (c) Cross-correlation lag plot demonstrating a time difference of msec between input and output signals. (d) Q-Q plot comparing input and output distributions (Kolmogorov-Smirnov test: H=1, p<0.0001, KS statistic=0.9717).
Figure 5.
Chattering spiking stimulation analysis of omeprazole-proteinoid sample. (a) Input-output comparison showing the input signal (mean: mV, SD: 19.72 mV) and the omeprazole-proteinoid output (mean: 0.48 mV, SD: 0.51 mV). Correlation coefficient: 0.7937, RMSE: 59.2964 mV. (b) Output plot highlighting the response characteristics of the omeprazole-proteinoid sample. (c) Cross-correlation lag plot demonstrating a time difference of msec between input and output signals. (d) Q-Q plot comparing input and output distributions (Kolmogorov-Smirnov test: H=1, p<0.0001, KS statistic=0.9717).
Figure 6.
Characterization of induced-mode spiking in omeprazole-proteinoid samples. (a) Input-output comparison: Input signal (mean: mV, SD: 14.20 mV) and omeprazole-proteinoid output (mean: 0.34 mV, SD: 0.40 mV). Correlation coefficient: 0.6644, RMSE: 62.8671 mV. (b) Detailed output plot showing the response characteristics of the omeprazole-proteinoid sample (range: mV to 3.14 mV). (c) Cross-correlation analysis revealing a time lag of 1590 msec between input and output signals. Maximum difference: 71.91 mV at 2.00 ms. (d) Q-Q plot comparing input and output distributions (Kolmogorov-Smirnov test: KS statistic = 0.9844, p < 0.0001), indicating significantly different signal distributions.
Figure 6.
Characterization of induced-mode spiking in omeprazole-proteinoid samples. (a) Input-output comparison: Input signal (mean: mV, SD: 14.20 mV) and omeprazole-proteinoid output (mean: 0.34 mV, SD: 0.40 mV). Correlation coefficient: 0.6644, RMSE: 62.8671 mV. (b) Detailed output plot showing the response characteristics of the omeprazole-proteinoid sample (range: mV to 3.14 mV). (c) Cross-correlation analysis revealing a time lag of 1590 msec between input and output signals. Maximum difference: 71.91 mV at 2.00 ms. (d) Q-Q plot comparing input and output distributions (Kolmogorov-Smirnov test: KS statistic = 0.9844, p < 0.0001), indicating significantly different signal distributions.
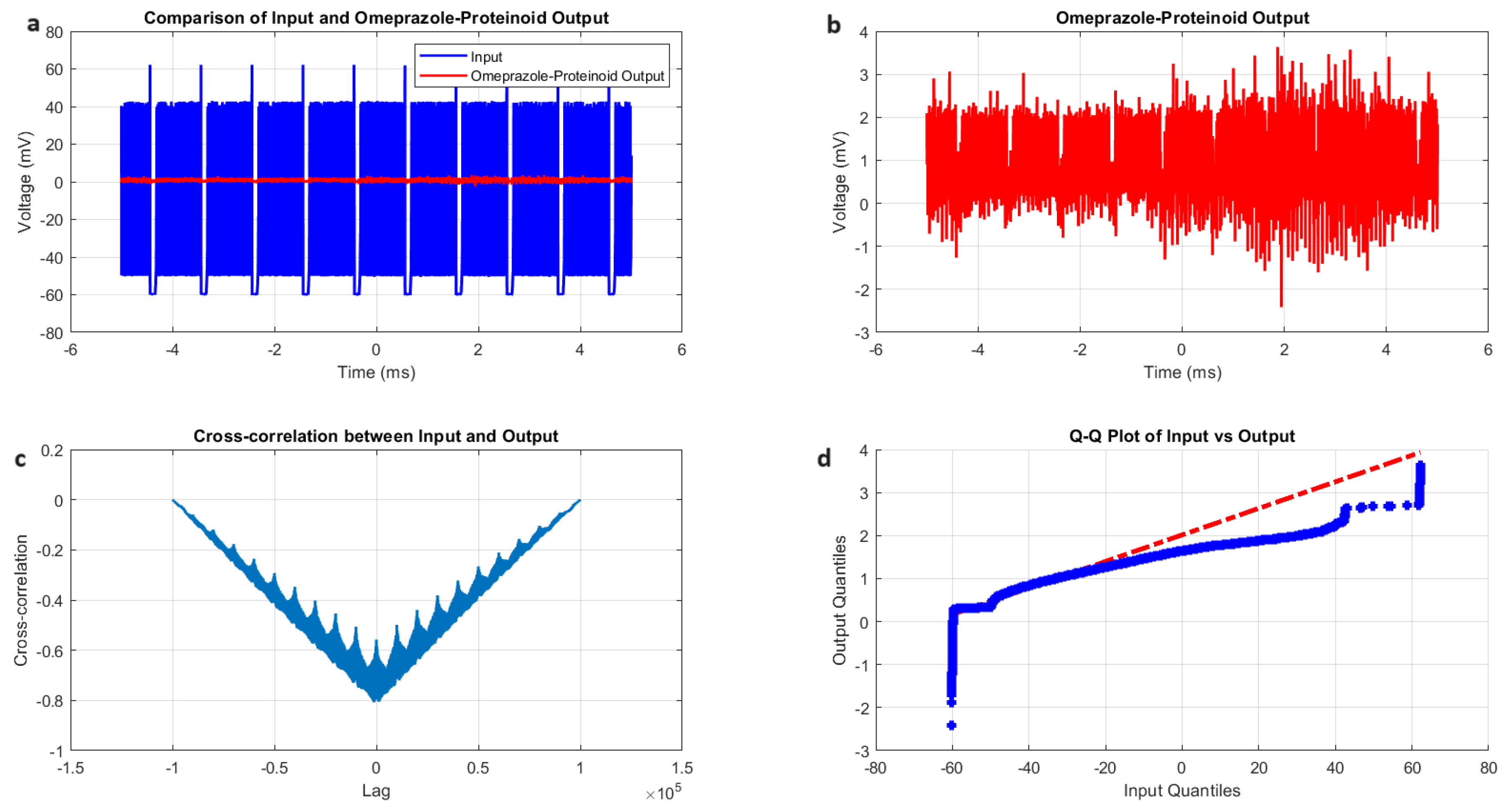
Figure 7.
Characterization of phasic spiking dynamics in proteinoid-omeprazole complexes. (a) Input-output comparison: Input signal (mean: mV, SD: 8.23 mV) and proteinoid-omeprazole output (mean: 0.54 mV, SD: 0.33 mV). Correlation coefficient: 0.4503, RMSE: 55.9366 mV. (b) Detailed output plot illustrating the response characteristics of the proteinoid-omeprazole complex (range: mV to 3.17 mV). (c) Cross-correlation analysis revealing a time lag of msec between input and output signals. Maximum difference: 67.00 mV at 2.00 ms. (d) Q-Q plot comparing input and output distributions (Kolmogorov-Smirnov test: KS statistic = 0.9945, p < 0.0001), demonstrating significantly different signal distributions and non-linear response properties.
Figure 7.
Characterization of phasic spiking dynamics in proteinoid-omeprazole complexes. (a) Input-output comparison: Input signal (mean: mV, SD: 8.23 mV) and proteinoid-omeprazole output (mean: 0.54 mV, SD: 0.33 mV). Correlation coefficient: 0.4503, RMSE: 55.9366 mV. (b) Detailed output plot illustrating the response characteristics of the proteinoid-omeprazole complex (range: mV to 3.17 mV). (c) Cross-correlation analysis revealing a time lag of msec between input and output signals. Maximum difference: 67.00 mV at 2.00 ms. (d) Q-Q plot comparing input and output distributions (Kolmogorov-Smirnov test: KS statistic = 0.9945, p < 0.0001), demonstrating significantly different signal distributions and non-linear response properties.
Figure 8.
Characterization of tonic spiking dynamics in omeprazole-proteinoid complexes. (a) Input-output comparison: Input signal (mean: mV, SD: 20.55 mV) and omeprazole-proteinoid output (mean: 0.80 mV, SD: 0.48 mV). Correlation coefficient: 0.6823, RMSE: 43.2821 mV. (b) Detailed output plot illustrating the sustained response characteristics of the omeprazole-proteinoid complex (range: mV to 3.63 mV). (c) Cross-correlation analysis revealing a time lag of -1231 samples between input and output signals. Maximum difference: 62.11 mV at 0.56 ms. (d) Q-Q plot comparing input and output distributions (Kolmogorov-Smirnov test: KS statistic = 0.9276, p < 0.0001), demonstrating significantly different signal distributions and non-linear response properties in tonic spiking mode.
Figure 8.
Characterization of tonic spiking dynamics in omeprazole-proteinoid complexes. (a) Input-output comparison: Input signal (mean: mV, SD: 20.55 mV) and omeprazole-proteinoid output (mean: 0.80 mV, SD: 0.48 mV). Correlation coefficient: 0.6823, RMSE: 43.2821 mV. (b) Detailed output plot illustrating the sustained response characteristics of the omeprazole-proteinoid complex (range: mV to 3.63 mV). (c) Cross-correlation analysis revealing a time lag of -1231 samples between input and output signals. Maximum difference: 62.11 mV at 0.56 ms. (d) Q-Q plot comparing input and output distributions (Kolmogorov-Smirnov test: KS statistic = 0.9276, p < 0.0001), demonstrating significantly different signal distributions and non-linear response properties in tonic spiking mode.
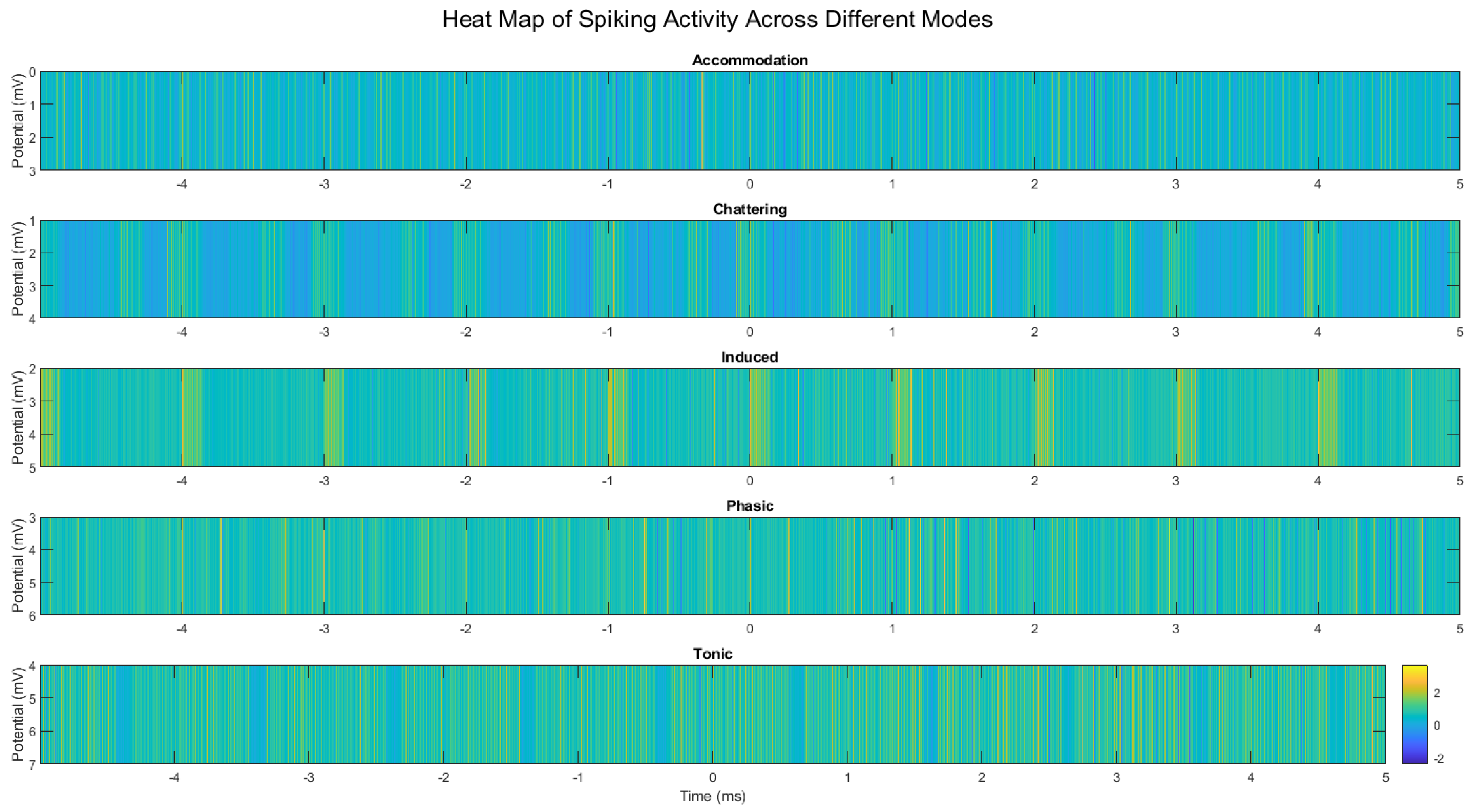
Figure 9.
Heatmap visualization of spike patterns across different spiking modes in omeprazole-proteinoid complexes. (a) Accommodation, (b) Chattering, (c) Induced, (d) Phasic, and (e) Tonic spiking modes. Color intensity represents the membrane potential, with warmer colors indicating higher potentials. The heatmap reveals distinct temporal patterns and intensity distributions characteristic of each spiking mode, highlighting the complex and mode-specific signal processing capabilities of the omeprazole-proteinoid system.
Figure 9.
Heatmap visualization of spike patterns across different spiking modes in omeprazole-proteinoid complexes. (a) Accommodation, (b) Chattering, (c) Induced, (d) Phasic, and (e) Tonic spiking modes. Color intensity represents the membrane potential, with warmer colors indicating higher potentials. The heatmap reveals distinct temporal patterns and intensity distributions characteristic of each spiking mode, highlighting the complex and mode-specific signal processing capabilities of the omeprazole-proteinoid system.
Figure 10.
Proposed mechanisms underlying the observed spiking behaviour in omeprazole-proteinoid complexes. (A) Voltage-sensitive conformational changes in the proteinoid structure. (B) Formation and breakdown of temporary conductive pathways. (C) Charge accumulation and redistribution processes. (D) Interactions between omeprazole’s proton pump inhibition and local pH gradients. The interplay of these mechanisms likely contributes to the complex, mode-dependent signal processing observed across different spiking regimes.
Figure 10.
Proposed mechanisms underlying the observed spiking behaviour in omeprazole-proteinoid complexes. (A) Voltage-sensitive conformational changes in the proteinoid structure. (B) Formation and breakdown of temporary conductive pathways. (C) Charge accumulation and redistribution processes. (D) Interactions between omeprazole’s proton pump inhibition and local pH gradients. The interplay of these mechanisms likely contributes to the complex, mode-dependent signal processing observed across different spiking regimes.
Figure 11.
Proposed mechanism for the generation of spikes in complexes formed by proteinoids and omeprazole. (a) SEM image showing the complex network structure. Schematic representation of the spike generation mechanism, illustrating omeprazole binding sites (red), ion channels (blue), and charge movement (orange arrows). (b) Graph illustrating the temporal evolution of the spike potential.
Figure 11.
Proposed mechanism for the generation of spikes in complexes formed by proteinoids and omeprazole. (a) SEM image showing the complex network structure. Schematic representation of the spike generation mechanism, illustrating omeprazole binding sites (red), ion channels (blue), and charge movement (orange arrows). (b) Graph illustrating the temporal evolution of the spike potential.
Table 1.
Comparison of Proton Pump Inhibitors
Table 1.
Comparison of Proton Pump Inhibitors
| PPI |
Chemical Formula |
Half-life (h) |
pKa |
Reference |
| Omeprazole |
C17H19N3O3S |
1.0 |
4.0 |
[15] |
| Esomeprazole |
C17H19N3O3S |
1.5 |
4.0 |
[16,20] |
| Lansoprazole |
C16H14F3N3O2S |
1.5 |
4.0 |
[17] |
| Pantoprazole |
C16H15F2N3O4S |
1.0 |
3.9 |
[18] |
| Rabeprazole |
C18H21N3O3S |
1.0 |
4.9 |
[19] |
Table 2.
Summary of Izhikevich Neuron Input and Omeprazole-Proteinoid Output Characteristics for Accommodation Spiking Model. This table presents key metrics comparing the input signal generated by the Izhikevich neuron model configured for accommodation spiking and the corresponding output from the omeprazole-proteinoid complex. Accommodation spiking is characterized by a gradual increase in interspike intervals in response to sustained stimulation. The significant differences in mean, standard deviation, and range between input and output highlight the complex signal processing occurring within the omeprazole-proteinoid system. Comparative metrics, including correlation coefficient and time lag, provide insights into the temporal relationship between input and output signals. The Kolmogorov-Smirnov test results confirm the statistically significant difference in signal distributions, underscoring the non-linear transformation induced by the omeprazole-proteinoid complex on the accommodation spiking pattern.
Table 2.
Summary of Izhikevich Neuron Input and Omeprazole-Proteinoid Output Characteristics for Accommodation Spiking Model. This table presents key metrics comparing the input signal generated by the Izhikevich neuron model configured for accommodation spiking and the corresponding output from the omeprazole-proteinoid complex. Accommodation spiking is characterized by a gradual increase in interspike intervals in response to sustained stimulation. The significant differences in mean, standard deviation, and range between input and output highlight the complex signal processing occurring within the omeprazole-proteinoid system. Comparative metrics, including correlation coefficient and time lag, provide insights into the temporal relationship between input and output signals. The Kolmogorov-Smirnov test results confirm the statistically significant difference in signal distributions, underscoring the non-linear transformation induced by the omeprazole-proteinoid complex on the accommodation spiking pattern.
| Metric |
Input Signal |
Omeprazole-Proteinoid Output |
| Mean (mV) |
|
0.60 |
| Standard Deviation (mV) |
15.30 |
0.43 |
| Maximum (mV) |
72.50 |
3.99 |
| Minimum (mV) |
|
|
| Comparative Metrics |
| Correlation Coefficient |
0.6841 |
| Root Mean Square Error (mV) |
50.4592 |
| Maximum Difference (mV) |
71.92 at 1.39 ms |
| Time Lag (msec) |
|
| Kolmogorov-Smirnov Test |
| H-value |
1 (distributions are different) |
| p-value |
< 0.0001 |
| KS statistic |
0.9709 |
Table 3.
Comparative analysis of input signal and omeprazole-proteinoid output characteristics under chattering spiking stimulation. The table displays important statistical parameters for the omeprazole-proteinoid sample’s input signal and output response. The results showed that the correlation coefficient between the input and output was 0.79, the time lag was msec, the greatest variance was 75.71 mV (at time ms), and the root mean square error (RMSE) was 59.3 mV. A Kolmogorov-Smirnov test revealed that the input and output signals had significantly different distributions (KS statistic = 0.9717, p < 0.0001). These findings show that the omeprazole-proteinoid sample significantly changed the signal while retaining a moderate correlation with the input patterns.
Table 3.
Comparative analysis of input signal and omeprazole-proteinoid output characteristics under chattering spiking stimulation. The table displays important statistical parameters for the omeprazole-proteinoid sample’s input signal and output response. The results showed that the correlation coefficient between the input and output was 0.79, the time lag was msec, the greatest variance was 75.71 mV (at time ms), and the root mean square error (RMSE) was 59.3 mV. A Kolmogorov-Smirnov test revealed that the input and output signals had significantly different distributions (KS statistic = 0.9717, p < 0.0001). These findings show that the omeprazole-proteinoid sample significantly changed the signal while retaining a moderate correlation with the input patterns.
| Metric |
Input Signal |
Omeprazole-Proteinoid Output |
| Mean (mV) |
|
0.48 |
| Standard Deviation (mV) |
19.72 |
0.51 |
| Maximum (mV) |
72.50 |
4.24 |
| Minimum (mV) |
|
|
Table 4.
Summary of Izhikevich Neuron Input and Omeprazole-Proteinoid Output Characteristics for Induced Spiking Mode. This table presents a comparison between the input signal generated by the Izhikevich neuron model configured for induced spiking and the corresponding output from the omeprazole-proteinoid complex. The input signal’s wide voltage range ( mV to 72.21 mV) is dramatically compressed in the output ( mV to 3.14 mV), indicating a powerful attenuation effect. The positive mean of the output (0.34 mV) compared to the negative input mean ( mV) suggests a baseline shift in the signal processing. The moderate correlation coefficient (0.6644) implies that while the output preserves some characteristics of the input, substantial non-linear processing occurs. The large time lag of 1590 ms points to complex internal dynamics within the omeprazole-proteinoid complex, possibly involving slow chemical or conformational changes. The high RMSE (62.8671 mV) and maximum difference (71.91 mV) further quantify the extent of signal transformation. The Kolmogorov-Smirnov test results (KS statistic = 0.9844, p < 0.0001) confirm that the input and output signals follow significantly different distributions, underscoring the non-linear nature of the signal processing in the omeprazole-proteinoid system during induced spiking stimulation.
Table 4.
Summary of Izhikevich Neuron Input and Omeprazole-Proteinoid Output Characteristics for Induced Spiking Mode. This table presents a comparison between the input signal generated by the Izhikevich neuron model configured for induced spiking and the corresponding output from the omeprazole-proteinoid complex. The input signal’s wide voltage range ( mV to 72.21 mV) is dramatically compressed in the output ( mV to 3.14 mV), indicating a powerful attenuation effect. The positive mean of the output (0.34 mV) compared to the negative input mean ( mV) suggests a baseline shift in the signal processing. The moderate correlation coefficient (0.6644) implies that while the output preserves some characteristics of the input, substantial non-linear processing occurs. The large time lag of 1590 ms points to complex internal dynamics within the omeprazole-proteinoid complex, possibly involving slow chemical or conformational changes. The high RMSE (62.8671 mV) and maximum difference (71.91 mV) further quantify the extent of signal transformation. The Kolmogorov-Smirnov test results (KS statistic = 0.9844, p < 0.0001) confirm that the input and output signals follow significantly different distributions, underscoring the non-linear nature of the signal processing in the omeprazole-proteinoid system during induced spiking stimulation.
| Metric |
Input Signal |
Omeprazole-Proteinoid Output |
| Mean (mV) |
|
0.34 |
| Standard Deviation (mV) |
14.20 |
0.40 |
| Maximum (mV) |
72.21 |
3.14 |
| Minimum (mV) |
|
|
| Comparative Metrics |
| Correlation Coefficient |
0.6644 |
| Root Mean Square Error (mV) |
62.8671 |
| Maximum Difference (mV) |
71.91 at 2.00 ms |
| Time Lag (msec) |
1590 |
| Kolmogorov-Smirnov Test |
| H-value |
1 (distributions are different) |
| p-value |
< 0.0001 |
| KS statistic |
0.9844 |
Table 5.
Summary of Phasic Spiking Characteristics in Proteinoid-Omeprazole Complexes. This table presents a detailed comparison between the input signal from the Izhikevich neuron model configured for phasic spiking and the output from the proteinoid-omeprazole complex. The results reveal significant signal transformation by the proteinoid-omeprazole system. The input signal’s wide voltage range ( mV to 62.46 mV) is markedly compressed in the output ( mV to 3.17 mV), demonstrating strong attenuation. The shift from a negative input mean ( mV) to a positive output mean (0.54 mV) suggests a fundamental change in signal characteristics. The reduced standard deviation in the output (0.33 mV vs. 8.23 mV input) indicates a smoothing effect. The low correlation coefficient (0.4503) implies substantial non-linear processing, more pronounced than in other spiking modes. The negative time lag of ms is particularly noteworthy, suggesting anticipatory behaviour in the proteinoid-omeprazole complex. This contrasts with the positive lag observed in induced spiking, highlighting mode-specific processing. The high RMSE (55.9366 mV) and maximum difference (67.00 mV) further quantify the extent of signal transformation. The Kolmogorov-Smirnov test results (KS statistic = 0.9945, p < 0.0001) indicate the most significant distributional difference among all spiking modes studied, emphasizing the unique processing characteristics of the proteinoid-omeprazole system during phasic spiking stimulation.
Table 5.
Summary of Phasic Spiking Characteristics in Proteinoid-Omeprazole Complexes. This table presents a detailed comparison between the input signal from the Izhikevich neuron model configured for phasic spiking and the output from the proteinoid-omeprazole complex. The results reveal significant signal transformation by the proteinoid-omeprazole system. The input signal’s wide voltage range ( mV to 62.46 mV) is markedly compressed in the output ( mV to 3.17 mV), demonstrating strong attenuation. The shift from a negative input mean ( mV) to a positive output mean (0.54 mV) suggests a fundamental change in signal characteristics. The reduced standard deviation in the output (0.33 mV vs. 8.23 mV input) indicates a smoothing effect. The low correlation coefficient (0.4503) implies substantial non-linear processing, more pronounced than in other spiking modes. The negative time lag of ms is particularly noteworthy, suggesting anticipatory behaviour in the proteinoid-omeprazole complex. This contrasts with the positive lag observed in induced spiking, highlighting mode-specific processing. The high RMSE (55.9366 mV) and maximum difference (67.00 mV) further quantify the extent of signal transformation. The Kolmogorov-Smirnov test results (KS statistic = 0.9945, p < 0.0001) indicate the most significant distributional difference among all spiking modes studied, emphasizing the unique processing characteristics of the proteinoid-omeprazole system during phasic spiking stimulation.
| Metric |
Input Signal |
Proteinoid-Omeprazole Output |
| Mean (mV) |
|
0.54 |
| Standard Deviation (mV) |
8.23 |
0.33 |
| Maximum (mV) |
62.46 |
3.17 |
| Minimum (mV) |
|
|
| Comparative Metrics |
| Correlation Coefficient |
0.4503 |
| Root Mean Square Error (mV) |
55.9366 |
| Maximum Difference (mV) |
67.00 at 2.00 ms |
| Time Lag (msec) |
|
| Kolmogorov-Smirnov Test |
| H-value |
1 (distributions are different) |
| p-value |
< 0.0001 |
| KS statistic |
0.9945 |
Table 6.
Summary of Tonic Spiking Characteristics in Omeprazole-Proteinoid Complexes. This table presents an analysis of the input signal from the Izhikevich neuron model configured for tonic spiking and the corresponding output from the omeprazole-proteinoid complex. The results reveal significant and unique signal processing by the omeprazole-proteinoid system under tonic stimulation. The input signal’s broad voltage range ( mV to 62.17 mV) is substantially compressed in the output ( mV to 3.63 mV), demonstrating potent signal attenuation. Notably, the mean potential shifts from a negative input ( mV) to a positive output (0.80 mV), the highest positive shift observed among all spiking modes, suggesting a robust baseline alteration in signal characteristics. The dramatic reduction in standard deviation (from 20.55 mV to 0.48 mV) indicates a strong smoothing effect, potentially filtering out high-frequency components of the input signal. The correlation coefficient (0.6823) is higher than in phasic mode but comparable to induced mode, implying a more linear relationship between input and output while still preserving significant non-linear processing. The negative time lag of ms is the largest among all modes, suggesting a highly pronounced anticipatory behaviour in the omeprazole-proteinoid complex under tonic stimulation. This could indicate the development of a strong predictive response mechanism during sustained, regular input. The root mean square error (43.2821 mV) is the lowest among all modes, suggesting that tonic spiking may induce the most consistent and predictable response in the complex. The Kolmogorov-Smirnov test results (KS statistic = 0.9276, p < 0.0001), while still indicating significantly different distributions, show the lowest KS statistic among all modes. This suggests that the output distribution in tonic mode, while distinct, may be closer to the input distribution compared to other spiking patterns.
Table 6.
Summary of Tonic Spiking Characteristics in Omeprazole-Proteinoid Complexes. This table presents an analysis of the input signal from the Izhikevich neuron model configured for tonic spiking and the corresponding output from the omeprazole-proteinoid complex. The results reveal significant and unique signal processing by the omeprazole-proteinoid system under tonic stimulation. The input signal’s broad voltage range ( mV to 62.17 mV) is substantially compressed in the output ( mV to 3.63 mV), demonstrating potent signal attenuation. Notably, the mean potential shifts from a negative input ( mV) to a positive output (0.80 mV), the highest positive shift observed among all spiking modes, suggesting a robust baseline alteration in signal characteristics. The dramatic reduction in standard deviation (from 20.55 mV to 0.48 mV) indicates a strong smoothing effect, potentially filtering out high-frequency components of the input signal. The correlation coefficient (0.6823) is higher than in phasic mode but comparable to induced mode, implying a more linear relationship between input and output while still preserving significant non-linear processing. The negative time lag of ms is the largest among all modes, suggesting a highly pronounced anticipatory behaviour in the omeprazole-proteinoid complex under tonic stimulation. This could indicate the development of a strong predictive response mechanism during sustained, regular input. The root mean square error (43.2821 mV) is the lowest among all modes, suggesting that tonic spiking may induce the most consistent and predictable response in the complex. The Kolmogorov-Smirnov test results (KS statistic = 0.9276, p < 0.0001), while still indicating significantly different distributions, show the lowest KS statistic among all modes. This suggests that the output distribution in tonic mode, while distinct, may be closer to the input distribution compared to other spiking patterns.
| Metric |
Input Signal |
Omeprazole-Proteinoid Output |
| Mean (mV) |
|
0.80 |
| Standard Deviation (mV) |
20.55 |
0.48 |
| Maximum (mV) |
62.17 |
3.63 |
| Minimum (mV) |
|
|
| Comparative Metrics |
| Correlation Coefficient |
0.6823 |
| Root Mean Square Error (mV) |
43.2821 |
| Maximum Difference (mV) |
62.11 at 0.56 ms |
| Time Lag (msec) |
|
| Kolmogorov-Smirnov Test |
| H-value |
1 (distributions are different) |
| p-value |
< 0.0001 |
| KS statistic |
0.9276 |