1. Introduction
Lactic acid bacteria (LAB) are among the most important and beneficial groups of microorganisms found in various environments, including plants, terrestrial and marine animals, fermented foods, and the mucosal surfaces of humans. Traditionally, they have been associated with fermented foods for their health benefits and close ties to human culture. Therefore, LAB is the primary group of microbes responsible for many fermentation processes [
1,
2,
3]. In recent years, there has been a growing interest in studying plants as a source for LAB screening. However, LAB derived from flowers have been less isolated and studied, yet they hold great potential for creating new starter cultures for plant-based fermentation [
4]. LAB is utilized in fermentation to enhance the stability of raw materials, sensory qualities, and nutritional value by producing lactic acid and other metabolites. Due to limitations in developing fermented plant-based products using traditional starter cultures, isolating and culturing specific LAB strains from natural sources could be solution. Therefore, it is intriguing to isolate and characterize strains from flowers to develop innovative plant-based starter cultures [
5,
6,
7].
It is well-known that LAB can synthesize a variety of polysaccharides, attracting attention due to their safe use and potential probiotic properties [
8]. Polysaccharides are high molecular weight polymeric carbohydrate structures formed of monosaccharide units linked together by glycosidic bonds. They exhibit a variety of structures, functional properties, and biological activities. Microbial polysaccharides can be soluble or insoluble, found in the form of capsules tightly attached to the cell wall, mucus loosely attached to the cell, or completely released into the environment [
9,
10].
Polysaccharides produced by bacteria are classified based on their biological functions. These include intracellular cytoplasmic storage polysaccharides like glycogen, and cell surface associated polysaccharides. In lactic acid bacteria (LAB), the exocellular polysaccharides can be classified into two categories: exopolysaccharides (EPS) and capsular polysaccharides (CPS), both are important for texture development in the food industry [
11]. CPS are closely linked to the cell surface, form the outermost layer of the bacterial cell and provide a mechanism to protect the cell, mediating direct interactions with the environment, whereas EPS are polysaccharides that are loosely associated with the cell surface or released into the extracellular medium, They can either be produced extracellularly by enzymes secreted by the bacterium, or synthesized intracellularly and secreted outside the cells and often forming a slime layer [
1,
9,
10].
Polysaccharides are one of the main components involved in the formation of the extracellular biofilm matrix. Exo-cellular polysaccharides are among the main techno-functional metabolites of LAB species. They play an important role in protecting bacteria from adverse environmental factors, such as protection against abiotic or biotic stress, competition, pH, and temperature. Exo-cellular polysaccharides have physicochemical properties that have potential for the food and pharmaceutical industries. like xanthan, alginate, and cellulose, that are important for biofilm formation and pathogenicity [
10,
12].
Exo-cellular polysaccharides can be classified as homo-polysaccharides when composed of a single type of monosaccharide, or hetero-polysaccharides if composed of two or more different sugars in their repeating unit. Moreover, the sugars may be modified by non-carbohydrate moieties such as acetate, pyruvate, sulfate, and succinate. Bacterial exo-cellular polysaccharides vary according to their source. LAB can produce exo-cellular polysaccharides in both hetero-polysaccharides and homo-polysaccharides structure. Exo-cellular polysaccharides in hetero-polysaccharides structure are produced at a much lower rate because they have a more complex biosynthesis mechanism and require more enzymes for production. The homo-polysaccharides produced by LAB classified as glucans, fructans, and galactans, which consist of D-glucose, D-fructose, or D-galactose, respectively. For the production of exo-cellular polysaccharides in homo-polysaccharides structure, a simpler metabolic pathway is followed and the presence of sucrose as a carbon source and glucan sucrase or fructan sucrase as an enzyme is sufficient. In some studies, galactan is also detected as a major residue of EPS produced by LAB, however, the genes and enzymes behind this are not very clear yet [
13]. In contrast, hetero-polysaccharides are composed of several repeating units of sugars, such as pentose (D-ribose, D-arabinose, D-xylose), hexose (D-glucose, D-galactose, D-mannose), N-acetylated monosaccharides (N-acetylglucosamine and N-acetyl-galactosamine), or uronic acids (D-glucuronic acid, D-galacturonic acid) [
8,
9,
10].
Recently research is focused on the application of exo-cellular polysaccharides in the food industry, due to their structural properties, such as texturization, emulsification, gelling, sweetening, water-binding capacity, and bioactive properties. Also studies have demonstrated the health-promoting potential of exo-cellular polysaccharides, including immunomodulatory, prebiotic, anti-inflammatory, anti-biofilm, and antioxidant activities [
2,
6]. In recent years, interest in vegetarian and vegan diets has increased for many reasons. The challenge for manufacturers of plant-based milk alternatives is to produce products with acceptable taste and texture for customers. The application of exo-cellular polysaccharide producing strains to improve the sensory and organoleptic analysis of these products, can positively influence texture, mouthfeel, and decrease syneresis in plant-based milk alternatives [
14]. In the production of functional and fermented products, LAB that produce EPS can lead to a reduction in the use of food additives by enhancing texture, as a natural thickener [
8,
15,
16].
The aim of our study was to identify novel LAB strains, from various Danish flowers that produce exo-cellular polysaccharides. We focused on strains that produce more exo-cellular polysaccharides on MLS agar medium, as these have the best performance in fermentation. To achieve this, we applied a high-throughput texture screening method based on plant drink fermentations in microtiter plates (MTP) combined with Total Aspirate Dispense Monitoring (TADM) pressure measurements. Additionally, we investigated the structural characteristics of the exo-cellular polysaccharides produced by all strains.
2. Materials and Methods
2.1. Preparation of Flowers
As the source of LAB species, 47 flower samples were collected from different locations in Denmark on summer season and have remained in 3.5% NaCl solution (saline solution) in 15 ml. After 5 days incubation at 22 ℃, glycerol was added to each flower sample and store at -80 ℃ until use.
2.2. Screening and Isolation of LAB Strain from Flowers
The frozen flower samples were inoculated into MLS-agar supplemented with 1% glucose and 0.5% fructose for LAB screening. The composition of MLS medium is 5 g/L Meat extract, 2.5 g/L Meat peptone, 5 g/L KH
2PO
4, 8 g/L Soy peptone, 10 g/L tryptone, 4 g/L Yeast extract, 5 g/L Sodium acetate, 2 g/L Ammonium citrate, 0.1 g/L ascorbic acid, 0.3 g/L MgSO
4, 0.1 g/L MnSO
4, 0.034 g/L FeSO
4, 1 ml Tween 80 and 15 g/L Agar. After pH was adjusted to 6.2, 0.4 g/L Cycloheximide was also added to agar-plates for inhibiting yeast and fungi growth [
5]. For isolation, the plates were made by directly streaked of the flower samples supernatant after brief vortex. After incubating the plates at 30 °C for 48 h, plates were transferred to fridge for 24 h for showing more distinctive morphology. Then 10 to 15 single colonies were picked mainly based on their colony morphology, followed by re-streak, and sub cultured on the MLS-agar at least one time for colony purification [
5,
17].
2.3. Identification of Isolated LAB Strains from Flowers Using MALDI-TOF, PCR, and De-Replication
The species identification of the isolated LABs was performed by protein extraction from 24 h grown cultures on MLS-agar plates from purified single colony using MALDI-TOF Biotype (Bruker Daltonics, Bremen, Germany), which can identify species based on the protein mass to charge (m/z) spectra. The sample preparation process for MALDI-TOF identification is described as follows: Fresh pure cultures from an agar plate are picked using inoculation loops and spread out on spots on a target MALDI-TOF plate. The plate is then treated with 1 µl 75% ethanol, mixed well and left to dry. Then 1µl of 70% formic acid is added to each spot and the plate is left to dry again. Finally, 1µl of a saturated matrix is then added to each spot. Matrix is prepared by mixing α-cyano-4-hydroxycinnamic acid with 475µl of water, 500µl of 100% Acetonitrile, and 25µl of Trifluoroacetic acid. The plate is left to dry completely before being scanned using a MALDI-TOF Biotype for protein mass spectra detection and identification at the species level by matching with its integrated spectral database library. The identification process will provide a species ID along with a score indicating the similarity between the protein mass spectrum and the database. The instrument gives a score of 1-3–score values from 1 to 1.69 are considered not reliable, and thus the genus/species cannot be determined; scores between 1.70 and 1.99 refer to identifications that are only reliable on a genus level; and scores above 2 correspond to identifications that are reliable on a species and genus level [
5]. The not reliable strains (14 strains) with low score were identified by 16S rRNA sequencing. The fragment (300 bp) was amplified by using a universal primer: following forward (5’ TGGCTCAGGACGAACGCTGGCGGC 3’) and reverse (5’ CCTACTGCTGCCTCCCGTAGGAGT 3’). The PCR procedure is as follows: using PCR master mix (2X) 25 µl, forward primer 1 µM, reverse primer 1 µM, template DNA 10 pg-1µg, nuclease-free water to 50 µl. Gently vortex the samples. and perform PCR using thermal cycling conditions. The PCR program was carried out in a thermal cycler as 5 min of initial denaturation at 95°C, followed by 35 amplification cycles of denaturation at 95°C for 30 s, annealing at 58°C for 30s and extension at 72°C for 40 s. The final elongation was set as 72°C for 7 min. The PCR products were analyzed via 1% agarose gel electrophoresis, stained with DNA Gold Viewer Dye, and visualized under UV light using mini gel documentation device (VMR,). Then the samples (11 strains) were sent for sequencing. To remove duplicate strains from the isolates, a strict de-replication procedure was then employed. If two strains originating from the same sample received the same MALDI-TOF ID with similar scores, only one of the strains will be saved and characterized for further studies [
5]. After dereplication, strains were saved to DTU National Food Institute Culture Collection (NFICC) with a designated NFICC number.
2.4. Screening for Exo-Cellular Polysaccharides Producers
A modified MLS-agar medium supplemented with 1% glucose, 0.5% fructose as control, and 2% or 6% of sucrose was prepared to observe the exo-cellular polysaccharide production abilities of 61 selected strains and compare the exo-cellular polysaccharide production in 2% and 6% sucrose by control. Overnight cultures were inoculated to modified MLS agar plates and the slime formation was monitored following the incubation period of 24, 48, and 72 h at 30 °C, described previously by [
3], with slight modifications. Morphologically slimy colonies were further selected for exo-cellular polysaccharides production on modified MLS-agar medium supplemented with 2% sucrose to observe the exocellular polysaccharides production abilities of 40 selected strains and compare the Exo-cellular polysaccharide production. Exo-cellular polysaccharide production was assessed through visual inspection of colonies.
2.5. Isolation and Purification of Exo-Cellular Polysaccharides
The 40 selected LAB strains were grown in modified MLS-broth medium supplemented with 2% sucrose, at 30 °C for 48 h. Isolation of exo-cellular polysaccharide was conducted by following the method as depicted previously [
8]. Briefly, following the incubation period, 2 vol of chilled ethanol was added to the culture supernatants obtained following the centrifugation of the bacterial cultures, and supernatants were left at 4 °C overnight to precipitate the exocellular polysaccharides. The exocellular polysaccharides pellet was then recovered by centrifugation at 10,000×
g for 20 min at 4 °C and subjected to resuspension process with distilled water. This process was repeated twice with the use of less distilled water at each time for the resuspension process.
2.6. Determination of Monosaccharides by High Performance Liquid Chromatography (HPLC) Analysis
The monosaccharide composition is determined by HPLC after treatment for the purified polysaccharides as previously described [
8]. HPLC (ThermoFisher, Boston, US) analysis is set up in a system equipped with an Aminex HPX-87H (Bio-Rad, Hercules, CA) and a Shodex RI-101 refractive index detector (Showa Denko K.K., Tokyo, Japan). The mobile phase was 5 mM sulfuric acid with flow rate 0.5 mL/min. The column oven temperature was maintained at 60°C. Glucose, galactose, and fructose were used as standard sugars for determining the composition of purified polysaccharides. Chromatograms for samples and standards were analyzed using Chromeleon 2.0 software (ThermoFisher, Boston, US).
2.7. High-Throughput Screening for Texturing Strains in Plant-Base Drink
The ability of strains to acidify milk was investigated using color of pH method and their texturing abilities were investigated using TADM as described in [
18]. Here, three types of commercial plant-based milk were used, and their nutritional content is listed in
Table 1. TADM results (pressure versus time curves) were converted into single descriptors (TADM area) by accumulating all the measured pressure points below zero. The pressure was measured every 0.01 s for 3 s. Strains resulting in fermented samples with high texture were represented by large TADM areas, whereas non texturing strains, by small TADM areas. Strains were considered texturing when giving rise to elevated texture in fermented milk samples (TADM area ≥ 800,000 Pa × ms).
3. Results and Discussion
3.1. Diversity of plant-based LAB strains according to MALDI-TOF, PCR, and de-replication
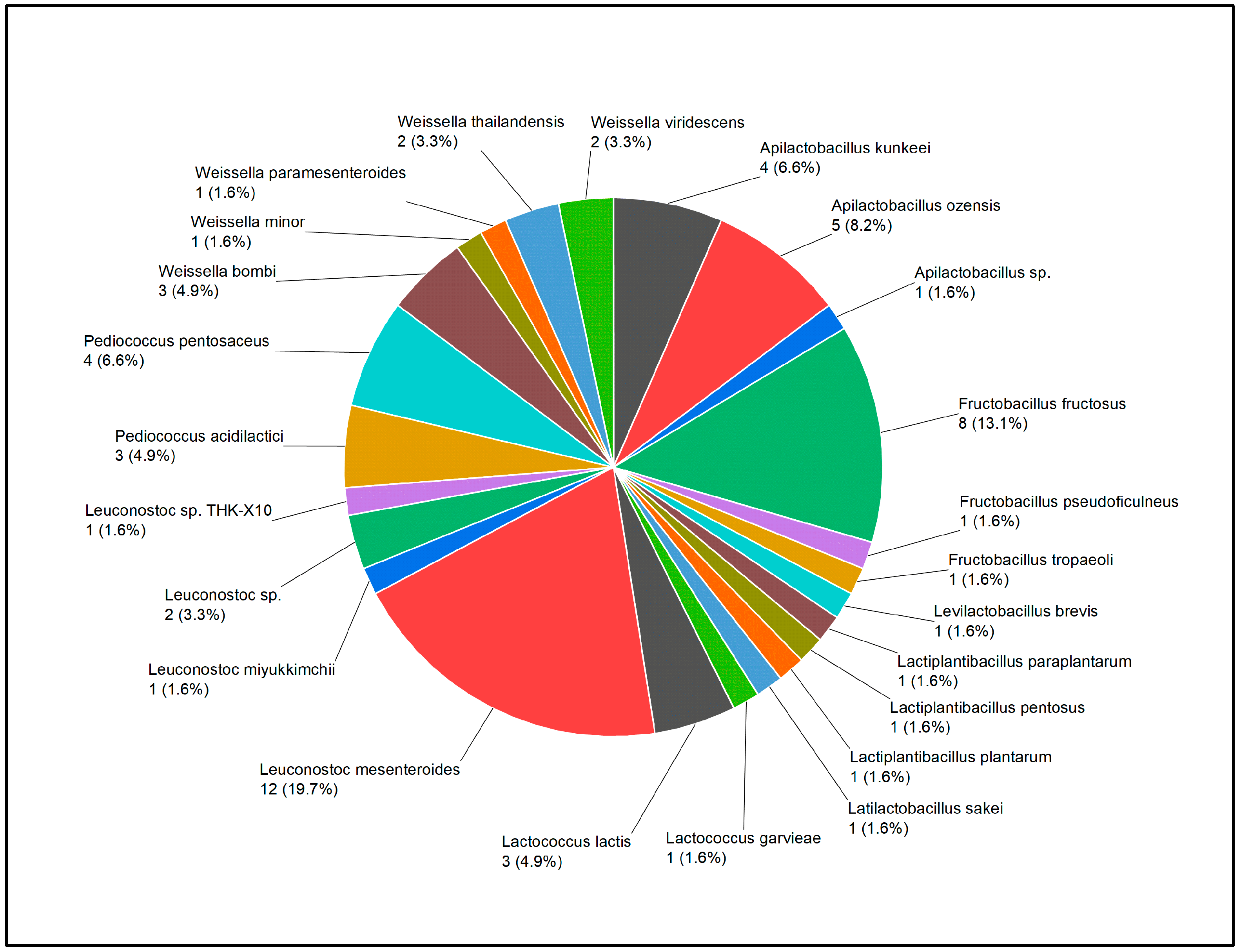
To isolate flower-derived LAB, we collected 46 flower samples representing 34 genera from various locations in Denmark during the summer season. By employing MALDI-TOF and 16SrRNA identification, a total of 61 LAB strains belonging to 24 species were isolated after a strict de-replication process (
Table 2,
Figure 1). The strains belong to 9 genera under
Apilactobacillus,
Fructobacillus, Levilactobacillus, Lactiplantibacillus, Latilactobacillus, Lactococcus,
Leuconostoc,
Pediococcus, and
Weissella. At the species level, the most prevalent LAB species found among all flowers were
Leuconostoc mesenteroides, with 12 isolated strains. Followed by other frequently occurring species such as
Fructobacillus fructosus with 8 isolates,
Apilactobacillus ozensis with 5 isolates and
Apilactobacillus Kunkeei with 4 isolates. Our results indicate a wide spread of LAB species on flowers.
In other studies focusing on LAB isolation from flowers, similar occurrences at the species level have been described, which aligns well with the findings of our study [
19,
20]. In contrast, LAB isolated from fermented vegetables or dairy sources is typically abundant in other species such as
Lactiplantibacillus plantarum, Lacticaseibacillus casei, Lacticaseibacillus paracase, Latilactobacillus curvatus, Latilactobacillus sakei, and
Lactobacillus delbrueckii [
5,
21,
22]. This indicates flowers harbor a niche for distinct LAB species, with high abundance of fructophilic LAB. Interestingly, such species are also frequently found inside of honeybee gut [
23], suggesting a potential microbiota exchange between flowers and pollinators. Unlike LAB derived from dairy and fermented vegetables, flower-derived LAB is less studied. Considering their distinctiveness, it could be a promising resource for investigating novel LAB in food applications.
Although LAB is ubiquitous in nature, isolating and identifying plant-derived LAB strains can be challenging due to their low abundance on plant surfaces and meticulous cultivation conditions [
24]. We attempted to directly use flower-washed water for plating but only yielded poor results. To get better isolation from flower samples, an enrichment procedure for LAB in each sample needs to be employed before isolation. In this study, inspired by the preparation of fermented vegetables, we introduced a simple method that preferment the flower samples in 3.5% NaCl solution at room temperature for 5 days. The pre-fermentation resulted in a low pH around 4 for most of the samples, indicating an enrichment of anaerobes. Additionally, the successful isolation of LAB from different flower samples confirmed the robustness and efficiency of this method.
Regarding the isolation and dereplication process, approximately 10 strains per sample were selected based on the appearance of the colonies on agar plates. Therefore, the isolates from each sample only represented the prevalent strains, not the entire LAB community. Dereplication is crucial during isolation as it enables high-quality outputs during strain isolation, furthermore, it allows people to identify new strains without wasting time and resources on strains that have already been discovered.
Overall, the study highlights the potential of flowers as a promising resource for LAB isolation. In comparison to other sources, flowers have received less attention, however, our findings emphasize the distinctness of LAB strains found in flower samples. To develop new starter cultures for plant-based fermentation, it’s important to study plant-derived LAB, as they are likely equipped with the capability to metabolize plant-derived sugars and protein, and detoxify phenolic compounds present in plant-based materials [
25].
3.2. Screening for Polysaccharides Producing LAB Strains on Different Sucrose-Supplemented Media
Exo-cellular polysaccharides, including exopolysaccharides (EPS) and capsular polysaccharides (CPS) are particularly important in the food industry. An efficient way to assess exo-cellular polysaccharides production is to visually observe the phenotypic characteristics of the colonies such as sliminess or ropiness [
8]. The slimy phenotype is recognized by mucilaginous colonies, while the ropy phenotype is identified by the formation of long filaments when an inoculation loop is lifted from the colony surface or cell pellet [
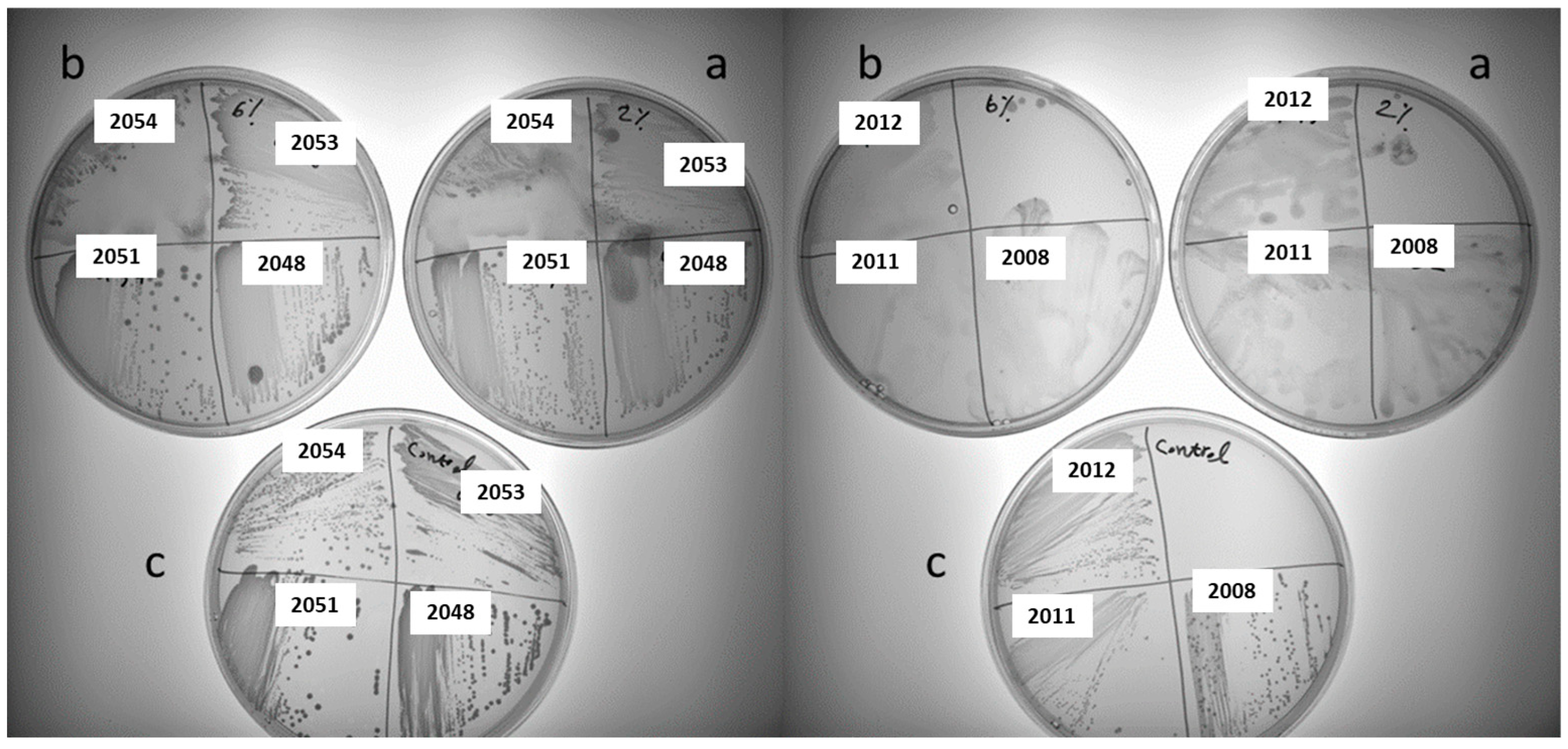
26]. As sucrose is usually used for stimulating homo-EPS production in LAB, the screening of homo-EPS production for 61 LAB strains was characterized on the MLS plates containing 2% or 6% sucrose. In total 40 of the tested strains showed varying degree of sliminess and ropiness, indicating the production of homo-EPS at various levels. Additionally, more slime was observed by using 2% sucrose compared to that with 6% sucrose (
Figure 2). No sliminess or ropiness observed in the control plates with no sucrose added for all tested strains, only the growth of colonies. Hence, we focus on these 40 slime-producing strains for further studies.
Sugar metabolism significantly influences exo-cellular polysaccharides production in LAB. Studies have demonstrated that exo-cellular polysaccharides can be varied in both amount and composition when LAB grows on different sugars [
27,
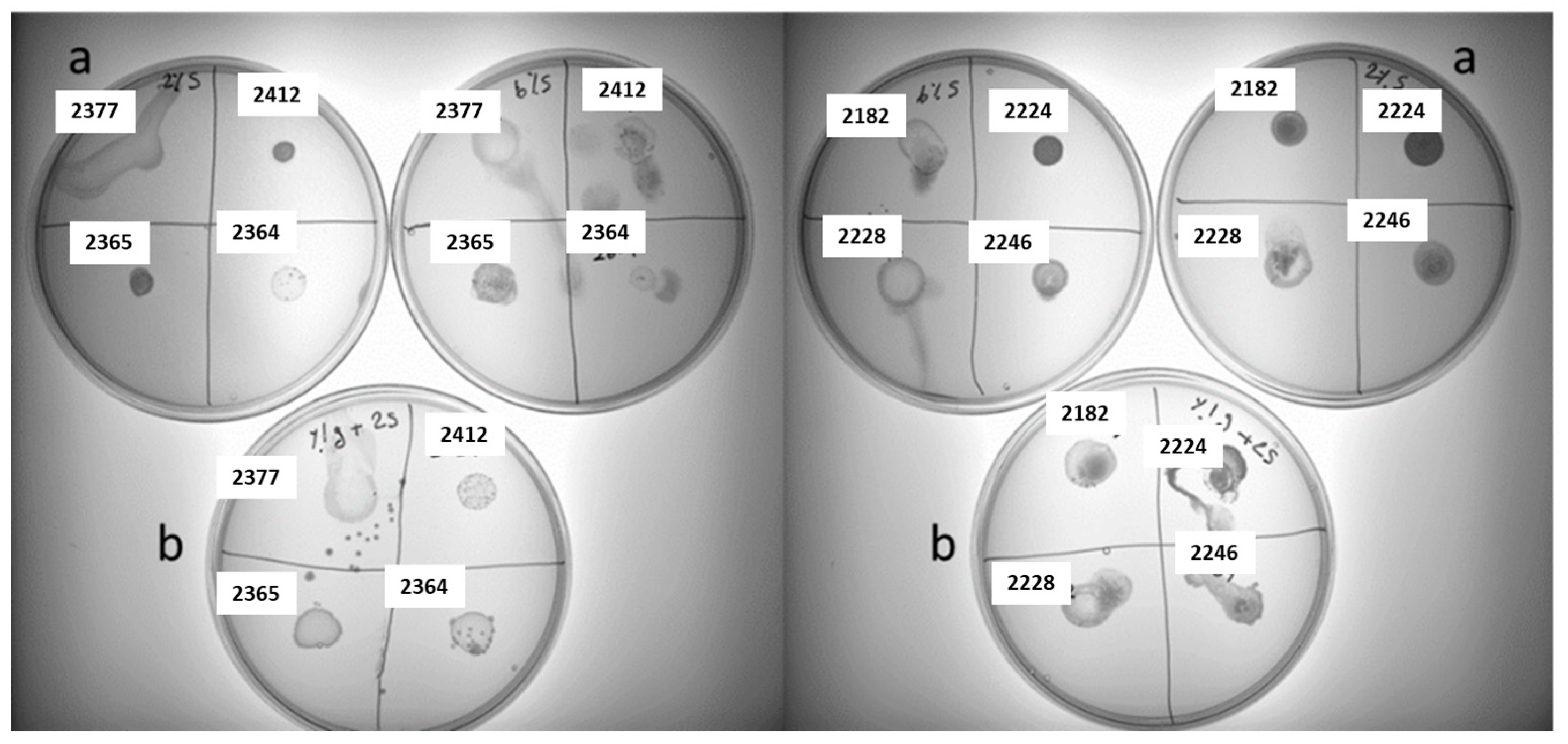
28]. Unlike dairy products, which mainly contain lactose as fermentable sugar, the sugar composition in plant-based materials is more complex. Hence, we attempted to investigate exo-cellular polysaccharides production in LAB under mixed sugar conditions. To simplify the model, exo-cellular polysaccharides production was evaluated in 2% sucrose plate supplemented with 1% glucose (exemplified in
Figure 3). As summarized in
Table 3, The results revealed a highly species-dependent pattern in sliminess production. For example, most
Apilactobacillus ozensis,
Pediococcus pentosaceus, Weissella viridescens,
Fructobacillus fructosus hampered in slime formation in the presence of glucose. Interestingly, some species behaved completely in an opposite way, like
Apilactobacillus kunkeei,
Leuconostoc miyukkimchii,
Lactococcus lactis,
Lactococcus garvieae, Leuconostoc mesenteroides
, Weissella bombi, and
Weissella minor, an enhanced homo-EPS production was detected in the presence of glucose.
The quantity of exo-cellular polysaccharides synthesized by LAB largely depends on several parameters, including pH, temperature, oxygen tension, incubation period, metabolic activity, and microbial growth conditions. Nevertheless, the most important factor is the composition of the culture medium and its sugar compounds [
26,
29]. Exo-cellular polysaccharides production usually can be induced by adding sucrose, which plays a key role in the EPS biosynthesis pathway [
30]. However, in this study, an inhibited slime formation production is observed in some strains when glucose is present. This inhibition may be attributed to carbon catabolite repression, which may inhibit sucrose uptake when glucose is present [
31]. Conversely, some strains showed enhanced homo-EPS production in the presence of glucose, which may benefit from accelerated sugar metabolism. This suggests a diverse regulation mechanism in EPS production among different LAB, which has been reviewed in other studies [
11]. Although the mechanism behind the inconsistent slime formation with and without glucose remains unclear, our findings provide valuable insights when selecting LAB strains for texturizing plant-based materials with different sugar compositions.
3.3. Determination of Monosaccharide Composition by HPLC Analysis
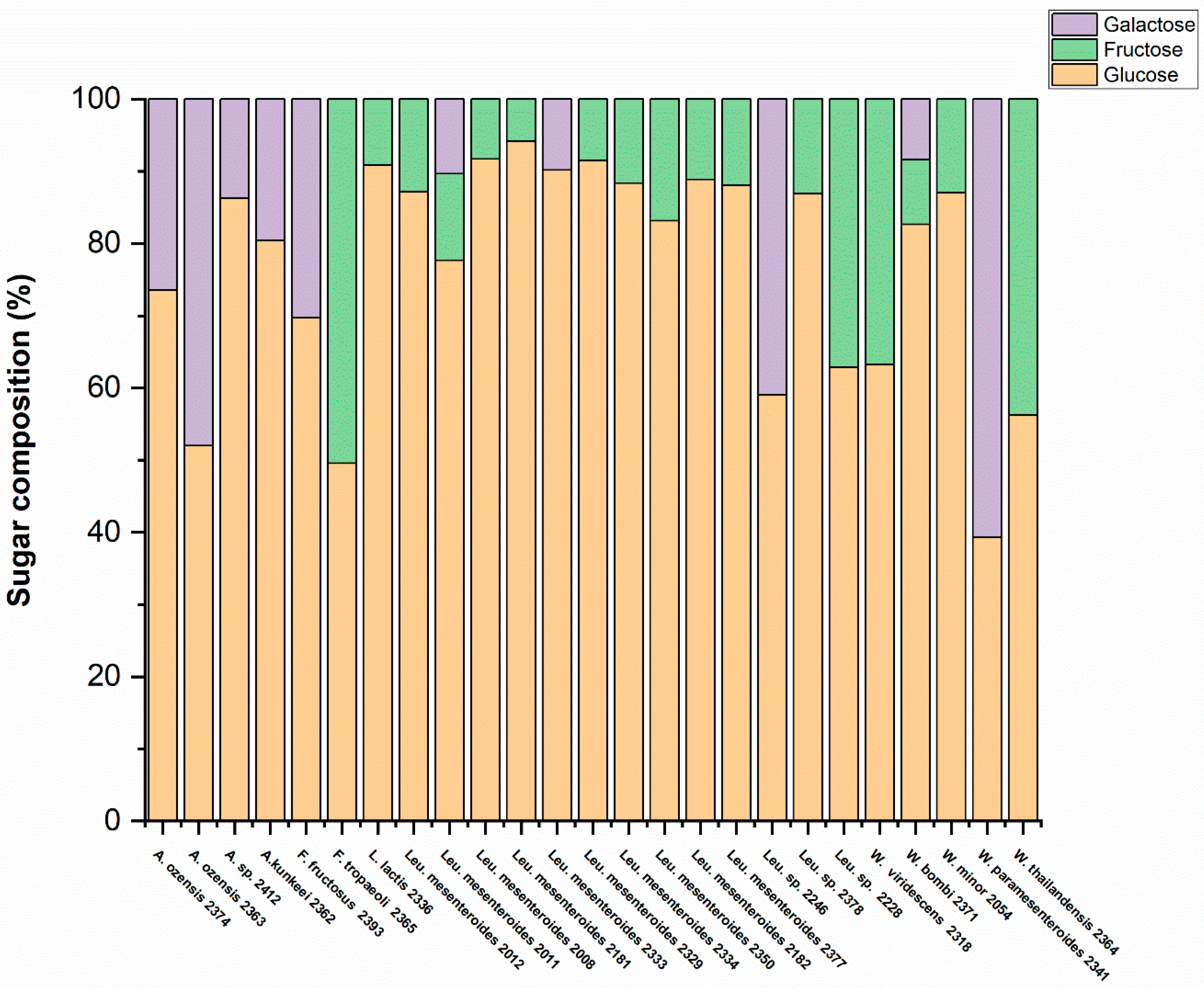
To investigate the monosaccharide composition of exo-cellular polysaccharides, all 40 strains were cultivated in MSL medium supplemented with 2% sucrose. The extraction method used in this study specifically targets EPS, so we focused on analyzing EPS composition. After extraction, 25 strains yielded a satisfactory amount of EPS for compositional analysis. As shown in
Figure 4, the sugar composition of EPS produced by each strain is highly dependent on the species level. Specifically, most
Leuconostoc mesenteroides,
Lactococcus lactis, and
Weissella minor produced EPS dominated by glucose, with a small amount of fructose also detected. In contrast, EPS produced from
Fructoacillus tropaeoli,
Weissella viridescens and
Weissella thailandensis contain both large amounts of glucose and fructose. Furthermore, the EPS produced from
Apilactobacillus kunkeei,
Apilactobacillus ozensis,
Apilactobacillus sp.,
Fructobacillus fructosus, and
Weissella paramesenteroides consisted of a large amount of glucose and galactose. Overall, all strains had glucose monomer in their composition, some of them had fructose or galactose beside the glucose.
Leuconostoc mesenteroides (NFICC 2011) and
Weissella bombi (NFICC 2371) had glucose, fructose and galactose in their composition.
LAB strains are well-known for EPS production [
8]. Results of HPLC analysis show that all isolates produced EPS containing glucose, fructose or galactose as sugar monomer in the EPS structure. However, it is unclear if they are producing HePS or mixture of HoPS. Glucans are the main backbone structure with different degrees of branching and binding sites that vary from bacterial strain to bacterial strain. Glucans can be classified as either α-glucans (divided into four groups: dextran, mutan, reuteran, and alternan) or β-glucans, and are produced by a variety of LAB species in the genera
Leuconostoc, Lactobacillus, Streptococcus, and
Weissella [
10,
32]. Fructans (divided into 2 groups: levan and inulin) are produced by strains of
Streptococcus salivarius,
Leuconostoc mesenteroides,
Lactobacillus reuteri,
Lactobacillus johnsonii and
Fructilactobacillus sanfranciscensis [
33]. Galactans are less abundant, and are produced by a few LAB strains belonging to
Weissella confusa,
Lactococcus lactis subsp.
lactis and
Lactobacillus delbrueckii subsp.
Bulgaricus [
8,
10,
16,
33,
34,
35]. In general, LAB that produce HoPS release high amounts of polymers into the environment. However, HePS from LAB are produced in considerably smaller amounts [
3,
10]. We focused on HoPS-producing LAB as we monitored the quantity of EPS and visual characteristics, including slimy and ropy phenotypes. When comparing the 9 genera identified in this study in terms of EPS production efficiency, it was found that
Leuconostoc had the highest amount of homo-EPS production, followed by
Apilactobacillus, Fructobacillus, Weissella, Lactococcus, Lactiplantibacillus, and
Pediococcus, respectively. According to some studies, a high amount of sugar may contribute to an increase in the production of HoPS. Possible explanations for the increased HoEPS synthesis under the stress of high sugar concentration in some strains include osmosis, the unlimited supply of sugar building blocks, and high energy availability [
3,
8].
3.4. High-Throughput Screening for Texturing Strains
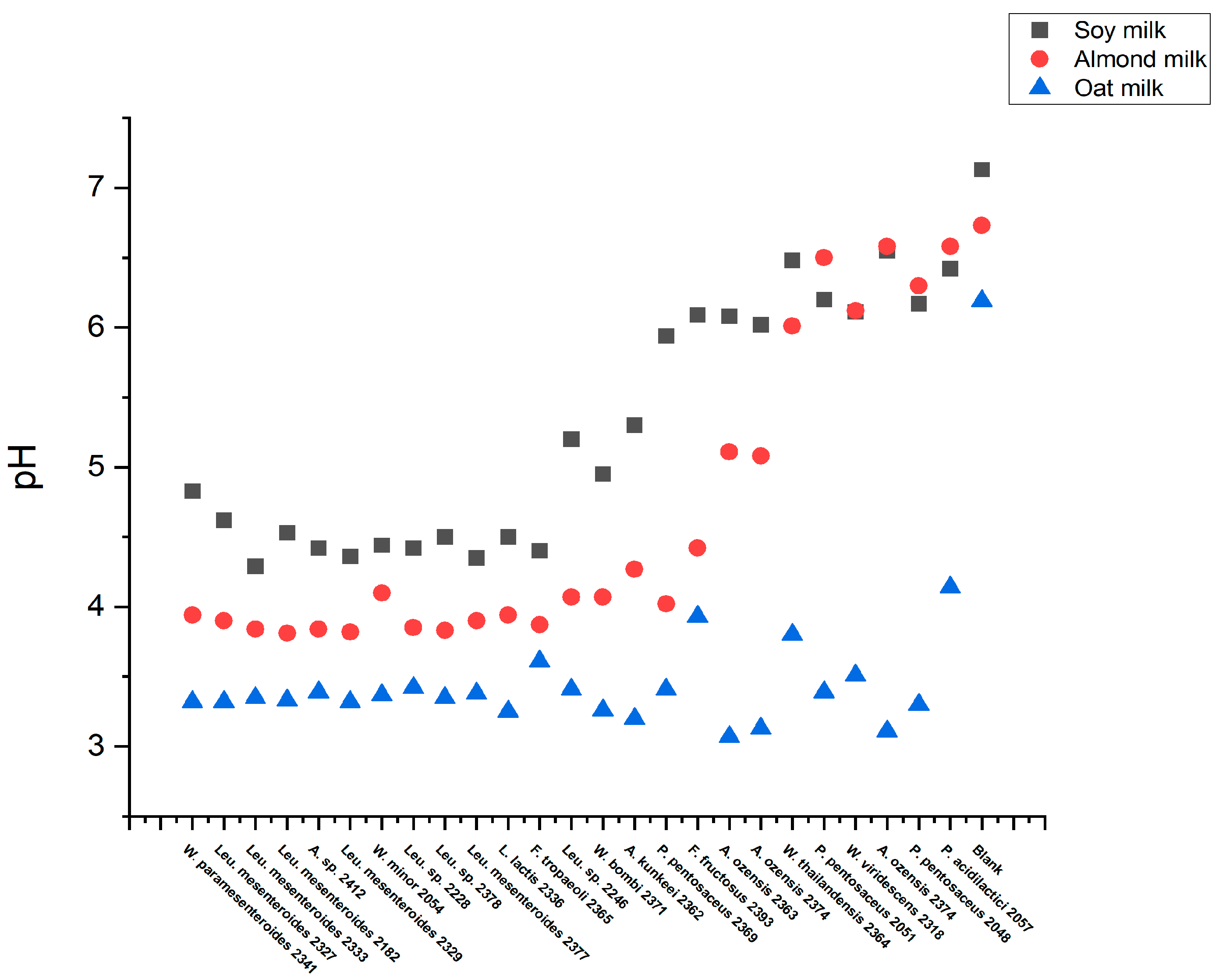
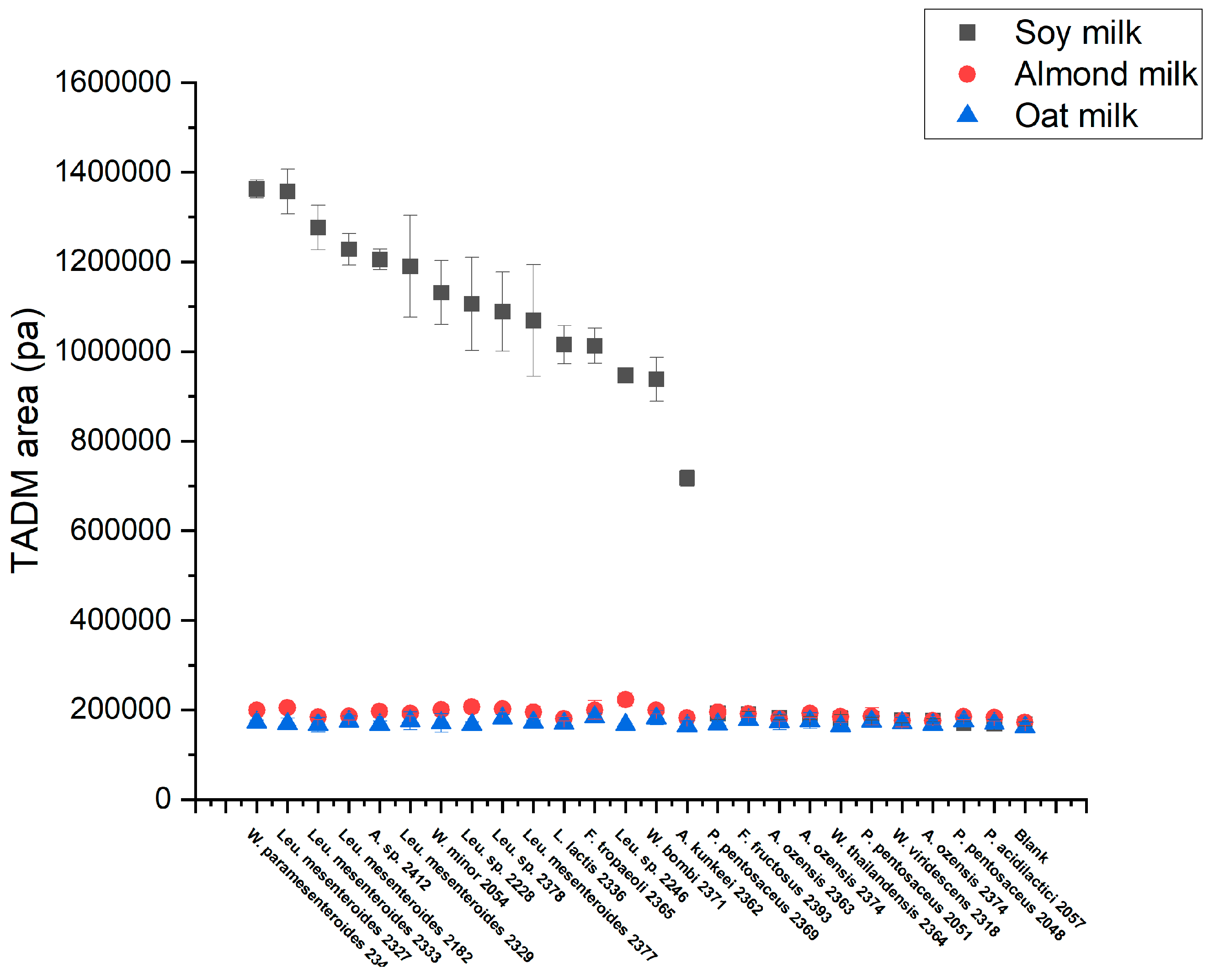
To characterize whether the strains could texturize plant-based drink, oat, almond, and soy drink were used without additional sugars (
Table 1). Here, 25 strains were selected based on their exo-cellular polysaccharide production on the MLS plate. The fermentation was carried out using 96 deep-well plates at 30℃ for 1 day. The starting pH was about 7. At the end of fermentation, the endpoint pH and texture in each sample were investigated. The endpoint pH less than 5.5 is regarded as acidification and TADM areas of above 800,000 Pa × ms is regarded as texturing. The results showed that all strains could acidify oat, but none achieved texturing. In contrast, when using soy and almond drink, several strains exhibited a similar acidification tendency in both substrates depending on the strain used (
Figure 5). Interestingly, only soy drink observed texturing. This is likely because of the relatively high protein content of soy, opposite to oat and almond. Specifically, 15 strains species texturing, including 7
Leuconostoc mesenteroides, 1
Leuconostoc sp., 1
Weissella cibaria, 1
Weissella minor, 1
Weissella paramesenteroides, 1
Apilactobacillus kunkeei, 1
Apilactobacillus sp., 1
Fructobacillus tropaeoli, and 1
Lactococcus lactis. In addition, texturing was found to be highly dependent on acidification. In acidified samples, TADM areas varied from 800,000 to 1,400,000 Pa × ms, indicating significant differences in texturing ability among strains (
Figure 6).
Different types of cow milk have a similar profile of proteins, fats, and sugars. In contrast, plant-based milk differs dramatically in nutrient compositions and physicochemical properties. These differences may significantly impact the microbial fermentation process in different plant-based matrices [
5]. Particularly, protein is critical for creating a yogurt-like texture development in fermented drinks. In a food system, proteins are the most significant functional component because of their structuring, texturizing, emulsifying, foaming, hydration, and nutritive properties [
36]. In this study, texture development was observed only in fermented soy milk. We speculate this is due to the high protein content in soy milk. Nevertheless, in oat and almond milk, elevated TADM aspiration pressures were also detected from all strains after fermentation, compared to the blank samples. This could be a clue for the produced extra-cellular polysaccharides but just far from enough for a yogurt-like texture formation, as the protein content in both samples is around 4 times lower than soymilk.
EPS production in LAB is well studied, however, most of the industrial strains are tailored for making yogurt and cheese. Our study showed that several traditional EPS producers such as
Leuconostoc, Weissella, and Lactococcus enabled texturizing soy milk as well. It is worth noting that
Leuconostoc normally grows poorly in milk alone and is often cultured with other LAB like
Lactococcus lactis in dairy fermentation [
37]. In this study, 8
Leuconostoc strains showed better texture development in soy milk, demonstrating their promising role shift from dairy to the plant-based section. In addition,
Apilactobacillus sp. and
Fructobacillus tropaeoli also showed better texture development in soy milk. These LAB species are rarely studied for EPS production and plant-based fermentation. Our results highlight the potential of broadening the LAB diversity used for plant-based fermentation. Furthermore, the plant-based drinks used in this study are free of pre-added sugars from manufacturers. The ability of diverse strains to acidify and texturize plant-based matrices based on their natural nutritional composition highlights their robustness and flexibility in developing plant-based fermented products. To fully understand and harness the fermentation process, comprehensive studies of protein and sugar composition in each plant-based material and genomic analysis for different LAB strains will be essential.
4. Conclusions
This study demonstrates a wide distribution of LAB in Danish flowers along with an innovative and efficient method for isolating LAB from flower samples. Additionally, we highlight the potential use of flower-derived LAB for plant-based food fermentation. The robust acidification and texture development, particularly in soy milk, indicate their significant role in enhancing the quality and diversity of plant-based fermented products. Further genomic study of the strains and compositional studies for different plant-based materials are essential to optimize their application in plant-based food fermentation.
Author Contributions
HX and CHBB conceptualized the study. HX, AKH, APW, MT, EBH, and CHBB designed the study. HX, AKH, APW, EBH, CHBB and VKP contributed to the acquisition of data. AKH and HX made the first draft of the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by a project from Chr. Hansen to HX and CHBB. Additionally, research was conducted as a part of the Innomission pool 1 project within the roadmap (Climate- and environment-friendly agriculture and food production). The granted project REPLANTED 1152-00001B supported the work of CHBB, and EBH.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Acknowledgments
The authors would like to acknowledge the National Food Institute, Technical University of Denmark, (DTU), Kongens Lyngby, Denmark, for their support and facilities provided during the course of this research. Data were generated through the research infrastructure provided by a genius grant from Food and Health Open Innovation Laboratory, Danish Roadmap for Research Infrastructure (FOODHAY). Specifically, the MALDI-TOF Biotyper is located at the DTU National Food Institute.
Conflicts of Interest
The authors EBH, CHBB, AKH, APW, and HX declare no conflict of interest. Additionally, it can be mentioned that VKP and MT are fulltime employee at Novonesis. The grant providers had no influence on the design and interpretation of the results generated and precented in this study.
References
- Lynch, K.M., A. Coffey, and E.K. Arendt, Exopolysaccharide producing lactic acid bacteria: Their techno-functional role and potential application in gluten-free bread products. Food research international 2018, 110, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Özpınar, F.B.; et al. , Physicochemical and structural characterisation of a branched dextran type exopolysaccharide (EPS) from Weissella confusa S6 isolated from fermented sausage (Sucuk). International Journal of Biological Macromolecules 2024, 130507. [Google Scholar] [CrossRef] [PubMed]
- Iosca, G.; et al. , Anti-Spoilage Activity and Exopolysaccharides Production by Selected Lactic Acid Bacteria. Foods 2022, 11, 1914. [Google Scholar] [CrossRef] [PubMed]
- Ruiz Rodríguez, L.G. , Mohamed, F., Bleckwedel, J., Medina, R., De Vuyst, L., Hebert, E.M. and Mozzi, F. Diversity and functional properties of lactic acid bacteria isolated from wild fruits and flowers present in Northern Argentina. Frontiers in microbiology 2019, 10, 1091. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; et al. , Isolation and characterization of plant-based lactic acid bacteria from spontaneously fermented foods using a new modified medium. LWT 2023, 115695. [Google Scholar] [CrossRef]
- Wu, J.; et al. , Exopolysaccharides synthesized by lactic acid bacteria: Biosynthesis pathway, structure-function relationship, structural modification and applicability. Critical Reviews in Food Science and Nutrition 2023, 63, 7043–7064. [Google Scholar] [CrossRef] [PubMed]
- Verón, H.E.; et al. , Assessment of technological and functional features of Lactiplantibacillus and Fructobacillus strains isolated from Opuntia ficus-indica fruits. NFS Journal 2023, 31, 110–122. [Google Scholar] [CrossRef]
- Yalmanci, D., H. İspirli, and E. Dertli, Identification of Lactic Acid Bacteria (LAB) from pre-fermented liquids of selected cereals and legumes and characterization of their exopolysaccharides (EPS). Food Bioscience 2022, 50, 102014. [Google Scholar]
- Daba, G.M., M. O. Elnahas, and W.A. Elkhateeb, Contributions of exopolysaccharides from lactic acid bacteria as biotechnological tools in food, pharmaceutical, and medical applications. International Journal of Biological Macromolecules 2021, 173, 79–89. [Google Scholar] [CrossRef]
- Jurášková, D., S. C. Ribeiro, and C.C. Silva, Exopolysaccharides produced by lactic acid bacteria: From biosynthesis to health-promoting properties. Foods 2022, 11, 156. [Google Scholar] [CrossRef]
- Zeidan, A.A.; et al. , Polysaccharide production by lactic acid bacteria: From genes to industrial applications. FEMS microbiology reviews 2017, 41 (Suppl. 1), S168–S200. [Google Scholar] [CrossRef] [PubMed]
- Schmid, J., V. Sieber, and B. Rehm, Bacterial exopolysaccharides: Biosynthesis pathways and engineering strategies. Frontiers in microbiology 2015, 6, 496. [Google Scholar] [CrossRef] [PubMed]
- Kavitake, D., P. B. Devi, and P.H. Shetty, Overview of exopolysaccharides produced by Weissella genus–A review. International Journal of Biological Macromolecules 2020, 164, 2964–2973. [Google Scholar]
- Huang, W.; et al. , Evaluation of the fermentation potential of lactic acid bacteria isolated from herbs, fruits and vegetables as starter cultures in nut-based milk alternatives. Food Microbiology 2023, 112, 104243. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; et al. , Microbiological, physicochemical and rheological properties of fermented soymilk produced with exopolysaccharide (EPS) producing lactic acid bacteria strains. LWT-Food Science and Technology 2014, 57, 477–485. [Google Scholar] [CrossRef]
- Poulsen, V.K.; et al. , Screening for texturing Leuconostoc and genomics behind polysaccharide production. FEMS Microbiology Letters 2020, 367, fnaa179. [Google Scholar] [CrossRef] [PubMed]
- Molina, G.E.S.; et al. , Development of a novel lactic acid bacteria starter culture approach: From insect microbiome to plant-based fermentations. LWT 2022, 167, 113797. [Google Scholar] [CrossRef]
- Poulsen, V.K., P. Derkx, and G. Oregaard, High-throughput screening for texturing Lactococcus strains. FEMS microbiology letters 2019, 366, fnz001.
- Ruiz Rodríguez, L.G.; et al. , Diversity and functional properties of lactic acid bacteria isolated from wild fruits and flowers present in Northern Argentina. Frontiers in microbiology 2019, 10, 452267. [Google Scholar] [CrossRef] [PubMed]
- Saleh, G. , Isolation and characterization of unique fructophilic Lactic acid bacteria from different flower sources. Iraqi Journal of Agricultural Sciences 2020, 51, 508–518. [Google Scholar] [CrossRef]
- Anacarso, I.; et al. , Isolation and identification of lactic acid bacteria from plants and other vegetable matrices and microbial recombination with Enterococcus spp. Am. Res. Thoughts 2015, 1, 1503–1515. [Google Scholar]
- Terzić-Vidojević, A.; et al. , Diversity of non-starter lactic acid bacteria in autochthonous dairy products from Western Balkan Countries-technological and probiotic properties. Food Research International 2020, 136, 109494. [Google Scholar] [CrossRef] [PubMed]
- Iorizzo, M.; et al. , Inter-and intra-species diversity of lactic acid bacteria in Apis mellifera ligustica colonies. Microorganisms 2020, 8, 1578. [Google Scholar] [CrossRef] [PubMed]
- Aleklett, K., M. Hart, and A. Shade, The microbial ecology of flowers: An emerging frontier in phyllosphere research. Botany 2014, 92, 253–266. [Google Scholar] [CrossRef]
- Pimentel, T.C.; et al. , Understanding the potential of fruits, flowers, and ethnic beverages as valuable sources of techno-functional and probiotics strains: Current scenario and main challenges. Trends in Food Science & Technology 2021, 114, 25–59. [Google Scholar]
- Ruas-Madiedo, P., N. Salazar, and C.G. de los Reyes-Gavilán, Exopolysaccharides produced by lactic acid bacteria in food and probiotic applications, in Microbial glycobiology. 2010, Elsevier. p. 885-902.
- Fuso, A.; et al. , Feeding lactic acid bacteria with different sugars: Effect on exopolysaccharides (EPS) production and their molecular characteristics. Foods 2023, 12, 215. [Google Scholar] [CrossRef] [PubMed]
- Paulo, E.M.; et al. , Método alternativo de triagem de bactérias láticas produtoras de exopolissacarídeos com confirmação rápida. Food Science and Technology 2012, 32, 710–714. [Google Scholar] [CrossRef]
- Subramanian, S.B.; et al. , Extracellular polymeric substances (EPS) producing bacterial strains of municipal wastewater sludge: Isolation, molecular identification, EPS characterization and performance for sludge settling and dewatering. Water research 2010, 44, 2253–2266. [Google Scholar] [CrossRef] [PubMed]
- van Hijum, S.A.; et al. , Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiology and molecular biology reviews 2006, 70, 157–176. [Google Scholar] [CrossRef] [PubMed]
- Görke, B. and J. Stülke, Carbon catabolite repression in bacteria: Many ways to make the most out of nutrients. Nature Reviews Microbiology 2008, 6, 613–624. [Google Scholar] [CrossRef]
- Gangoiti, J., T. Pijning, and L. Dijkhuizen, Biotechnological potential of novel glycoside hydrolase family 70 enzymes synthesizing α-glucans from starch and sucrose. Biotechnology advances 2018, 36, 196–207. [Google Scholar] [CrossRef]
- Angelin, J. and M. Kavitha, Exopolysaccharides from probiotic bacteria and their health potential. International Journal of Biological Macromolecules 2020, 162, 853–865. [Google Scholar] [PubMed]
- Ayyash, M.; et al. , Physicochemical, bioactive and rheological properties of an exopolysaccharide produced by a probiotic Pediococcus pentosaceus M41. Carbohydrate polymers 2020, 229, 115462. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, M.T.; et al. , Characterisation of dextran AP-27 produced by bee pollen isolate Lactobacillus kunkeei AP-27. Process Biochemistry 2023, 129, 22–29. [Google Scholar] [CrossRef]
- Poulsen, V.K.; et al. , Versatile Lactococcus lactis strains improve texture in both fermented milk and soybean matrices. FEMS Microbiology Letters 2022, 369, fnac117. [Google Scholar] [CrossRef]
- Erkus, O.; et al. , Multifactorial diversity sustains microbial community stability. The ISME journal 2013, 7, 2126–2136. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).