1. Introduction
The COVID-19 pandemic has uncovered numerous immediate and long-term health challenges, most commonly being Long COVID, or post-acute sequelae of SARS-CoV-2 infection (PASC) (Sisó-Almirall et al., 2021). This condition, defined by the long-term persistence of symptoms lasting over 12 weeks post-recovery and not explained by an alternative diagnosis, has been officially recognized by the World Health Organization (Gültekin & Özçelik, 2022). Long COVID is elucidated by symptoms persisting or arising for a time period of at least 2 months following the onset of the viral infection and not otherwise explained. Long COVID has been described with a wide variety of symptoms, from severe fatigue and breathing difficulties to important neurological problems, reducing the quality of life, and overloading the burden on persons and healthcare systems (S. T. Liu et al., 2023) (Kamamuta et al., 2022). The proportion of individuals developing Long COVID after mild-to-moderate COVID-19 infection is approximately between 10% and 30%, with the majority experiencing prolonged health complications and suffering from high resource use in healthcare (O’ Mahony et al., 2022).
Amid these challenges, the largest gene family encoded in the genome, G-protein-coupled receptors (GPCRs) offer new hope in finding working drugs. GPCRs transduce extracellular signals to mediate cellular actions and are important mediators of a host of physiological processes: immune responses, neurotransmission, and cellular metabolism. The human genome encodes over 800 GPCRs, mediating nearly every form of human physiology, including diseases (Shen et al., 2023). Over one-third of currently FDA-approved drugs target such receptors, and this is not unfounded, given the tremendous potential in therapeutics (D. Yang et al., 2021).
Given the complex symptomatology of Long COVID, cutting across multiple organ systems and biological functions, GPCRs are uniquely placed for exploitation toward therapeutic benefit. It is in this regard that the possibility of repurposing existing drugs targeting GPCRs finds special mention, as it offers a means of rapid translation from bench to bedside using known pharmacodynamics and pharmacokinetics toward rapid availability to the Long COVID patient (X. Li et al., 2021) (Rodrigues et al., 2022). It also gives a pragmatic approach to the management of these complex and diverse presentations in Long COVID, saving time and investment in bringing new therapies to the market. On the other hand, focusing on GPCRs allows the creation of a multitarget strategy to decrease such general effects of Long COVID that would result in relief and improvement in the quality of life of millions of sufferers. This becomes imperative since the newly found interdisciplinary collaborative innovations need to tackle these widespread and long-lasting impacts of the COVID-19 pandemic.
In their computational analysis, Das and Kumar have identified key GPCR genes linked to the diverse symptoms of long COVID. The researchers found that hub genes such as GNGT1, GNG12, GNB3, GNB4, GNG13, GNG8, GNG3, GNG7, GNG10, and GNAI1, all associated with GPCRs, were strongly connected to the 255 different long COVID symptoms examined. Their study suggests that persistent SARS-CoV-2 infection affects various organ systems and promotes chronic inflammation by dysregulating GPCR signaling pathways. Specific GPCR-related genes like CTLA4, PTPN22, KIT, KRAS, NF1, RET, and CTNNB1 were identified as common regulators of T-cell immunity, contributing to the diverse long COVID symptoms, including autoimmune, cardiovascular, dermatological, gastrointestinal, and other effects.
Additionally, a review by Cell Press, titled "RNA Therapeutics in Targeting G Protein-Coupled Receptors," highlights that GPCRs are a highly tractable class of drug targets. Modulating GPCR signaling could be a promising approach for treating long COVID. The authors note that GPCR-targeted RNA therapeutics are an active area of research that may lead to new treatments for long-term COVID-19 complications.
Recent research strongly implicates the dysregulation of GPCR signaling pathways as a key mechanism underlying the diverse and persistent symptoms observed in long COVID patients. Targeting these GPCR-related genes and pathways may be a fruitful avenue for developing effective long COVID therapies.
The objective of this study is to investigate computationally the gene expression profiles associated with SARS-CoV-2 infection and long COVID symptoms. This involves identifying differentially expressed genes and examining their functional similarities to uncover key genes that could serve as biomarkers for long COVID. Additionally, the study aims to examine the role of G-protein coupled receptors (GPCRs) in long COVID, with a particular focus on their involvement in T-cell immunity and viral infection responses. Ultimately, the goal is to provide a comprehensive understanding of the molecular mechanisms underlying long COVID, paving the way for the development of effective diagnostic and therapeutic interventions.
3. Results
Long COVID is a multifaceted condition that affects numerous biological systems, underscoring the complexity of its pathology. This post-viral syndrome can manifest in a wide array of symptoms, impacting the respiratory, cardiovascular, neurological, and immune systems, among others. Researchers have been diligently investigating the underlying mechanisms to identify effective treatments and management strategies. A significant focus has been on genes associated with G protein-coupled receptors (GPCRs) and related pathways. GPCRs play a crucial role in various physiological processes, including immune response, inflammation, and cellular signaling. By examining the expression and regulation of GPCR-related genes, scientists have been able to uncover important insights into the pathophysiology of Long COVID.
Notably, a study by Das and Kumar (2023) recently found a complex linkage among 331 Long COVID genes and 255 associated symptoms and conditions. This gene-symptom mapping is crucial for understanding the genetic influences on the risk and severity of Long COVID. The importance of these mappings has been previously emphasized, highlighting their role in providing a deeper understanding of the condition’s genetic underpinnings. Among others, 2949 GPCR genes sourced from the GtoPDB database have been instrumental in revealing the genetic and pharmacological treatment mechanisms of Long COVID and offering insights into symptoms and possible therapeutic targets.
The analysis of functional enrichment for Long COVID with the GPCR genes found a statistically significant relationship for both groups, focusing on signal transduction over four major pathways (
Table 1). The ToppGene Suite analysis underlines the highly significant functional enrichment of “WP G Protein Signaling Pathways” and “Reactome G Protein-Mediated Events,” suggesting a close relationship between Long COVID genes and GPCR signaling components. The genes HRAS, GNA11, GNAQ, KRAS, and PRKACA were over-represented in Long COVID relative to the GPCRs, indicating possible overlapping GPCR-mediated signaling by symptom severity and persistence (
Table 2).
Further scrutiny of 275 initially mapped GPCR genes with 30 Long COVID-associated genes across four pathways showed a refined focus on 111 GPCR genes of higher biological significance after applying a 60% sequence similarity threshold (Table3). This selective approach revealed major reductions in gene counts, pointing to specific pathways where only a few genes had potential disease mechanisms. The high sequence conservation among critical genes such as the RAS and GNA families and the protein kinase family indicated functional interactions crucial in Long COVID (
Table 4).
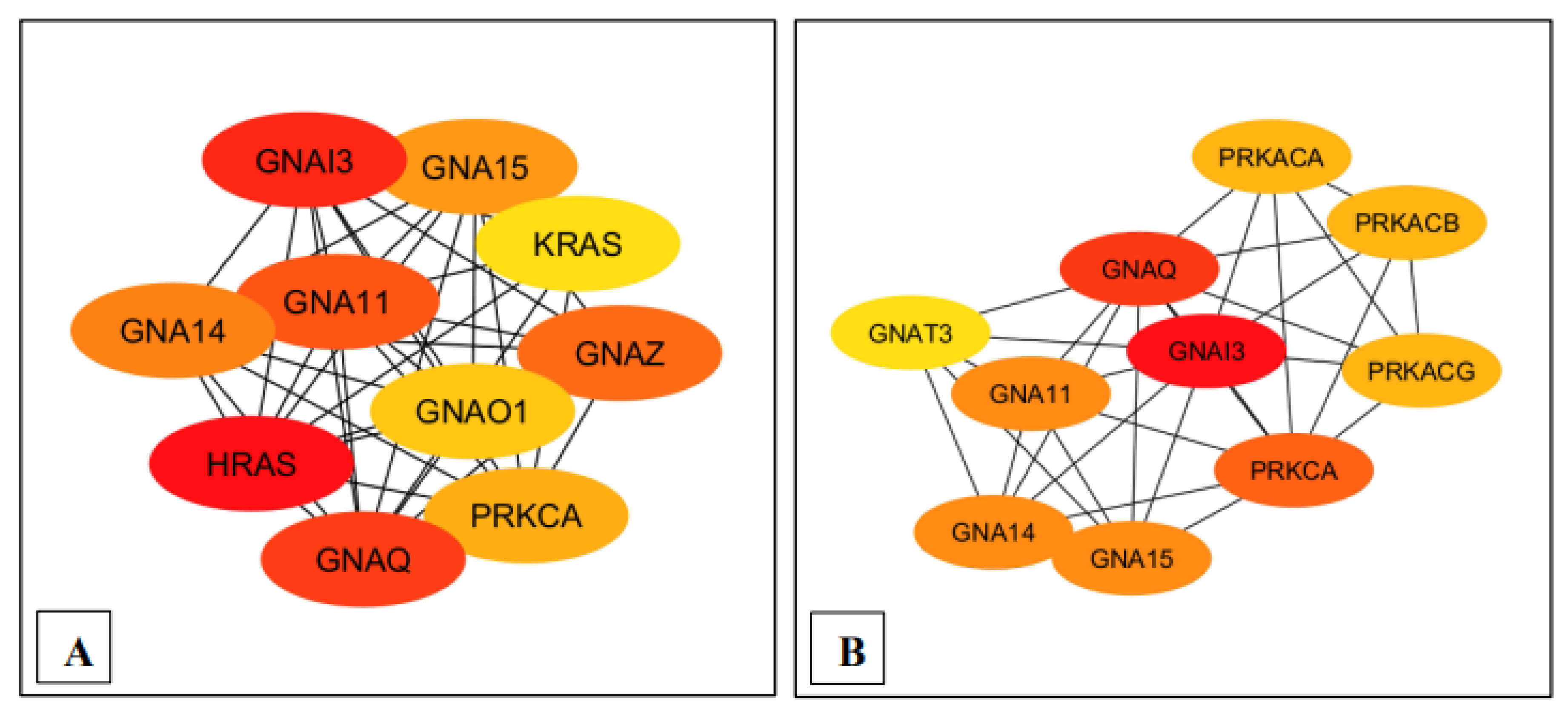
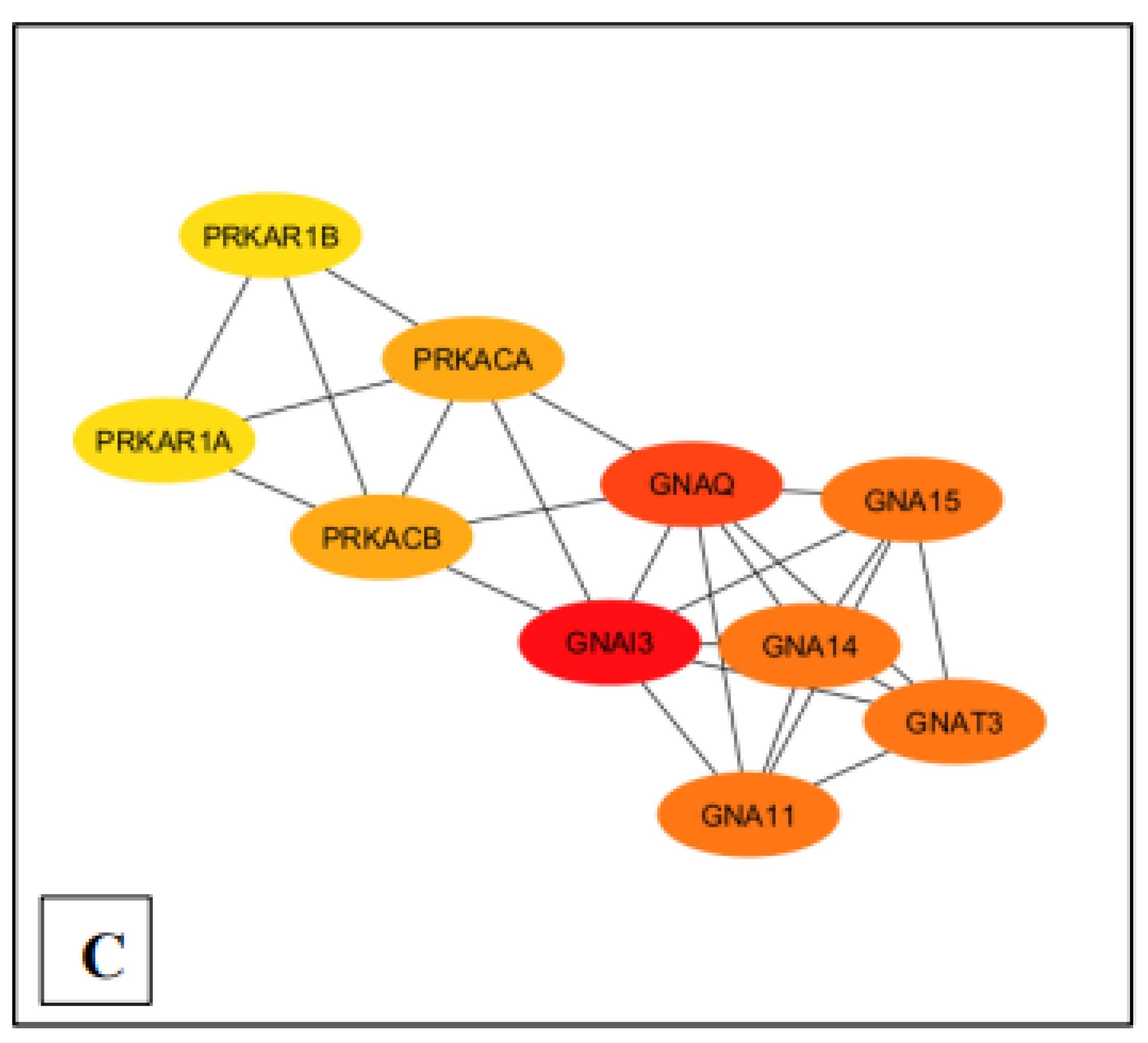
Moreover, Cytoscape software, the MCC module, and the CytoHubba plug-in further assisted in identifying the statistically significant genes GNA11 and GNAQ, both of which played pivotal roles in the GPCR signaling pathway associated with the pathobiology of Long COVID (
Figure 2 and
Figure 3). Additionally, HRAS and KRAS were identified within the WP G protein signaling pathway, attributed to roles in cell growth and cell survival, while PRKAR1A and PRKAR1B, the protein kinase A regulatory subunits, were found to affect numerous pathways modulating cellular functions impacting Long COVID pathology (
Table 5,
Figure 2 and
Figure 3).
As the final step of this study, analysis on the association between Long COVID and GPCR genes further illustrates the central roles that GPCR genes and their ligands play in the pathogenesis of Long COVID. Some notable characteristics of these genes pertain to their potential roles in boosting SARS-CoV-2 replication or dampening the host’s immune responses (
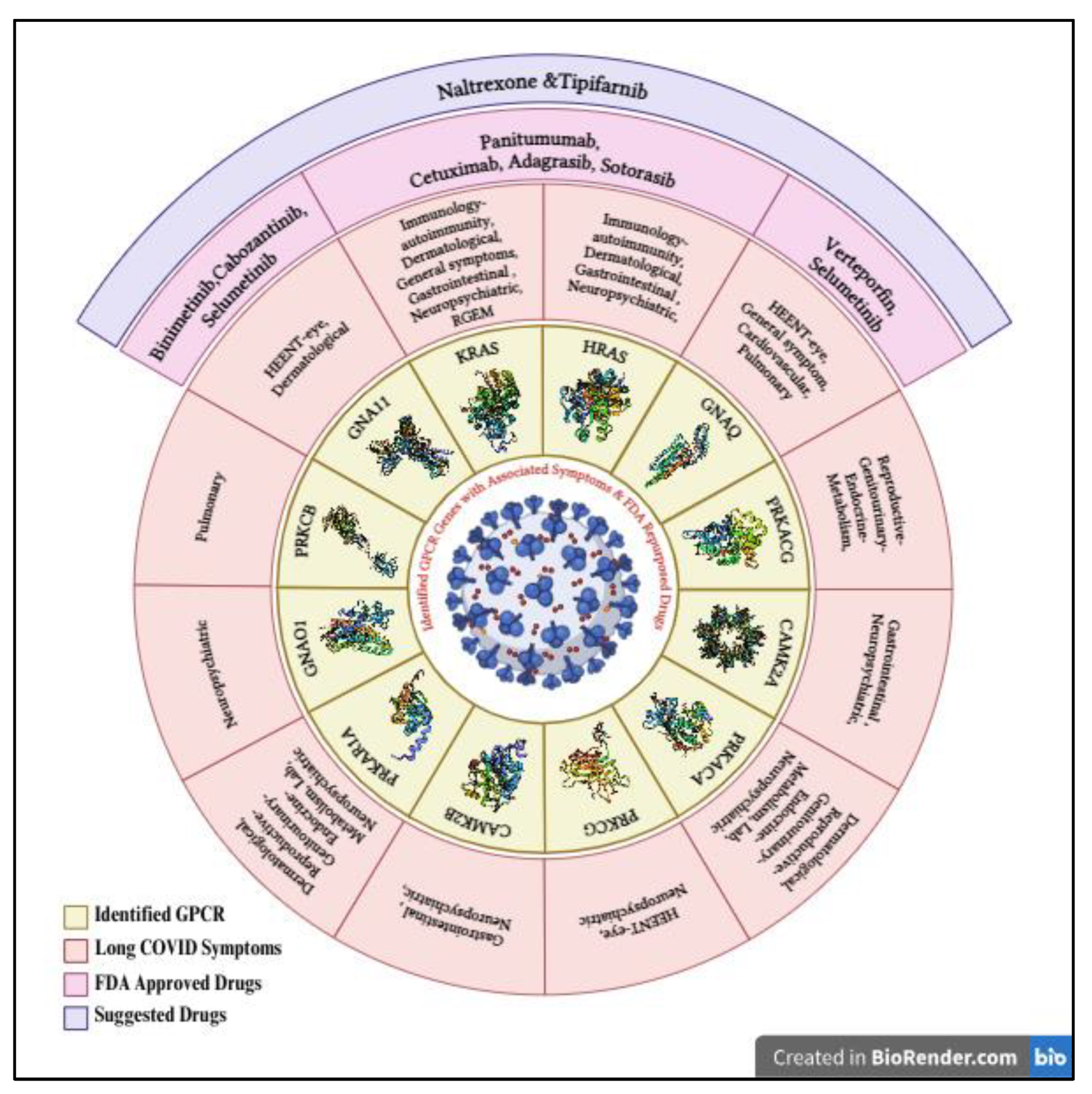
Table 8). The study further associated these genes with clinical symptoms and discussed possible therapeutics, emphasizing the need for continued research and clinical trials to address the long-term effects of Long COVID. Comprehensive research has not only pinpointed critical gene interactions but also suggested repurposing FDA-approved drugs associated with certain genes, such as HRAS, KRAS, GNAQ, and GNA11. Symptoms such as edema, pain, and venous thrombosis could potentially be treated with drugs like Binimetinib, Cabozantinib, Selumetinib, Panitumumab, Cetuximab, Adagrasib, Tipifarnib, and Sotorasib (
Table 6). To further elacudate the association of similarity of genes with GPCR and Long COVID, symptoms of specific GPCR genes and their related signalling pathways were tabulated to explain the effects in the context of long COVID and related questions (
Table 9).
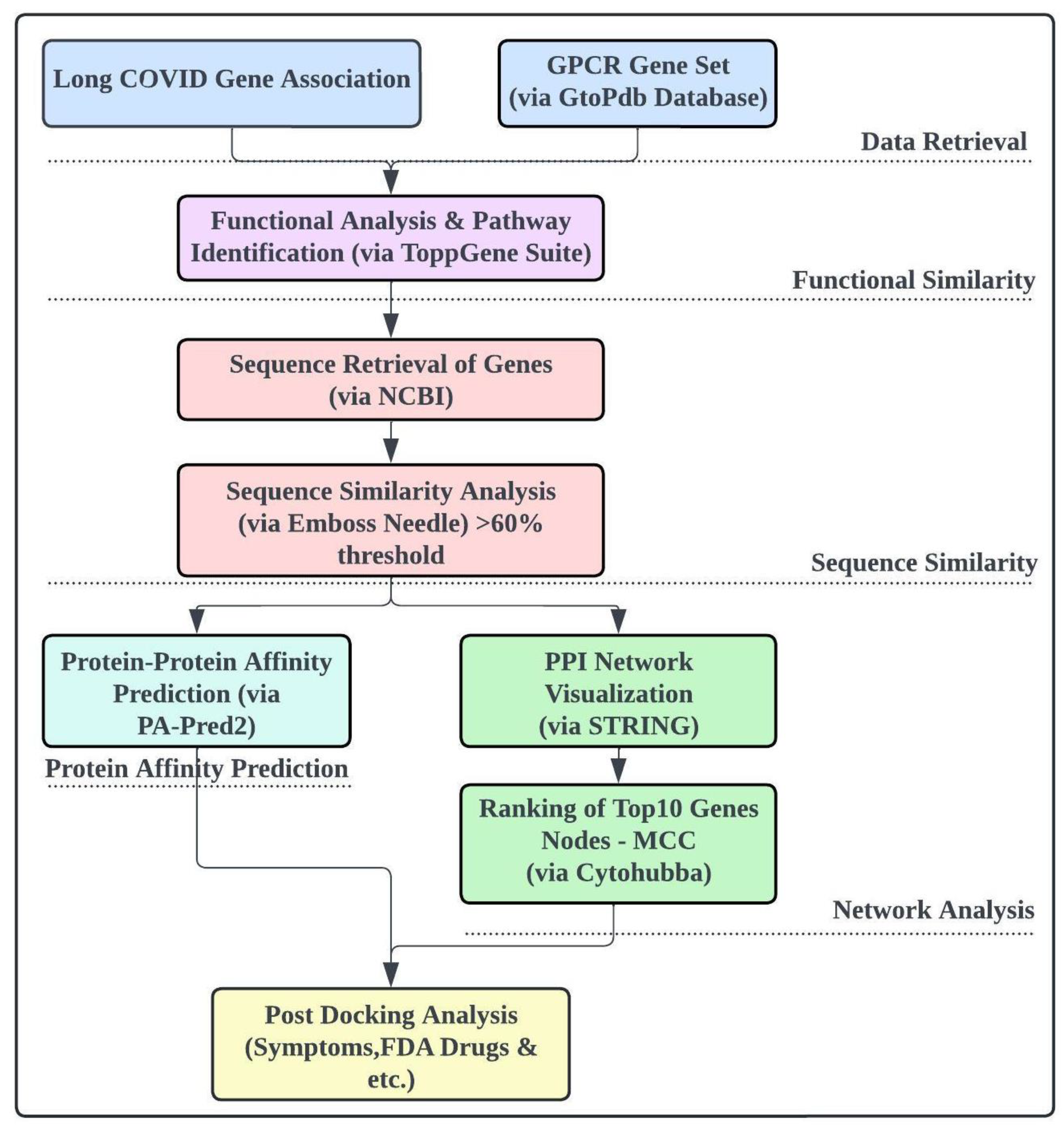
Figure 1.
The overall workflow of the study.
Figure 1.
The overall workflow of the study.
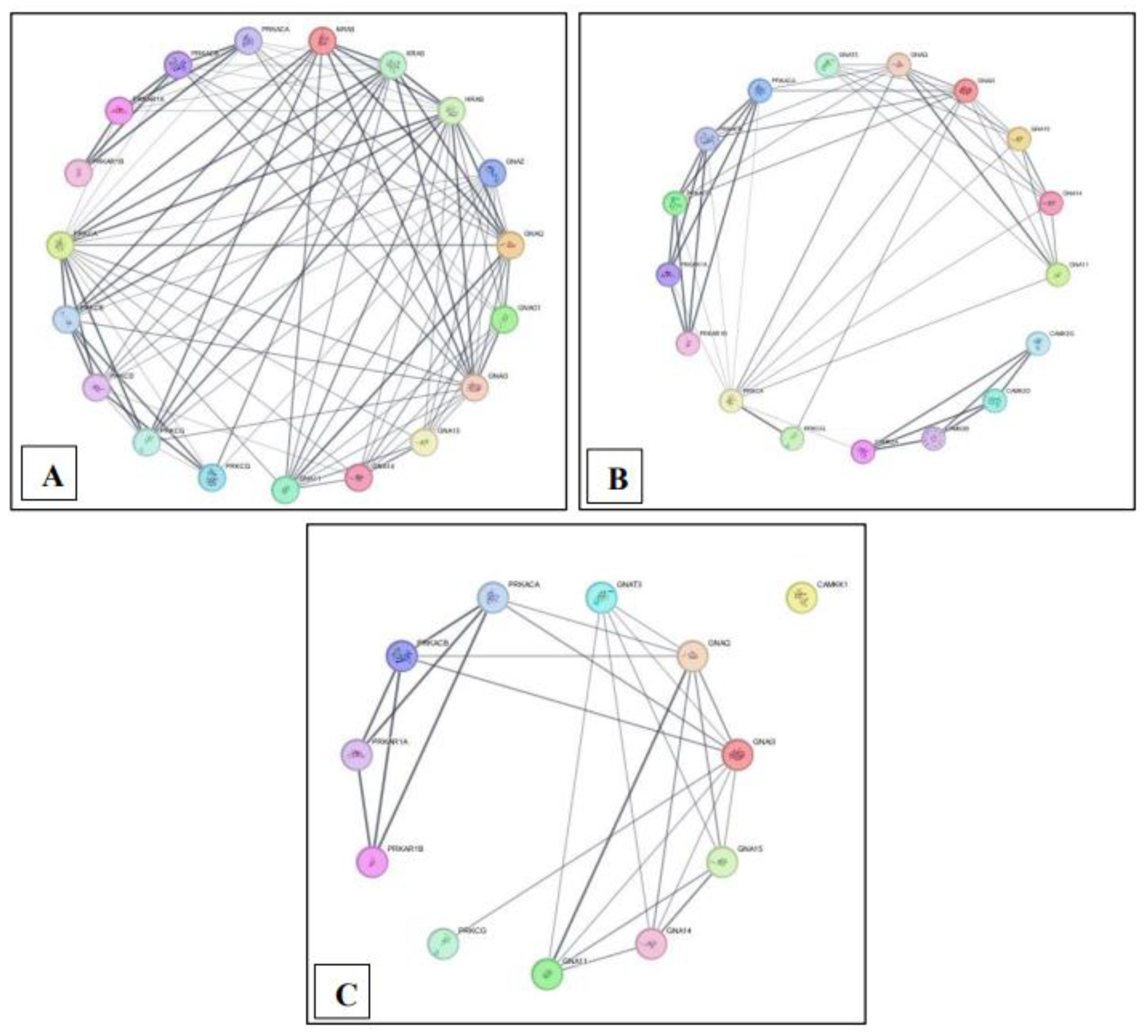
Figure 2.
Protein-protein interaction network overview for each pathway built using STRING in Cytoscape. (A) MM15882 & M39426 (B) M26911 (C) MM14496.
Figure 2.
Protein-protein interaction network overview for each pathway built using STRING in Cytoscape. (A) MM15882 & M39426 (B) M26911 (C) MM14496.
Figure 3.
Ranking of the top 10 nodes for each pathway based on the Maximal Clique Centrality (MCC) with Cytohubba plugin. (A) MM15882 & M39426 (B) M26911 (C) MM14496.
Figure 3.
Ranking of the top 10 nodes for each pathway based on the Maximal Clique Centrality (MCC) with Cytohubba plugin. (A) MM15882 & M39426 (B) M26911 (C) MM14496.
Figure 4.
Identified GPCR genes with associated Long Covid Symptoms & FDA Repurposed Drugs, created with BioRender.com.
Figure 4.
Identified GPCR genes with associated Long Covid Symptoms & FDA Repurposed Drugs, created with BioRender.com.
Table 1.
Summary of Long COVID Genes and Associated GPCR Genes Across Four Identified GPCR Pathways in ToppGene.
Table 1.
Summary of Long COVID Genes and Associated GPCR Genes Across Four Identified GPCR Pathways in ToppGene.
| Pathway ID |
Name |
Gene from Input (Long COVID) |
Gene from Annotations (GPCR) |
| MM15882 |
Wp G Protein Signaling Pathways |
9 |
90 |
| M26911 |
Reactome G Protein Mediated Events |
7 |
54 |
| M39426 |
Wp G Protein Signaling Pathways |
9 |
91 |
| MM14496 |
Reactome G Protein Mediated Events |
5 |
40 |
| |
Total |
30 |
275 |
Table 2.
Long COVID Genes and Associated GPCR Genes Across Four Identified GPCR Pathways.
Table 2.
Long COVID Genes and Associated GPCR Genes Across Four Identified GPCR Pathways.
| Pathway |
Gene from Input (long covid) |
Gene from Annotations (GPCR) |
| Gene Id |
Symbol |
Gene ID |
Symbol |
| Wp G Protein Signaling Pathways (MM15882 & M39426) |
3265 |
HRAS |
9472 |
AKAP6 |
| 2767 |
GNA11 |
387 |
RHOA |
| 2776 |
GNAQ |
23683 |
PRKD3 |
| 3845 |
KRAS |
3845 |
KRAS |
| 4893 |
NRAS |
5136 |
PDE1A |
| 5566 |
PRKACA |
196883 |
ADCY4 |
| 5573 |
PRKAR1A |
5137 |
PDE1C |
| 5582 |
PRKCG |
6548 |
SLC9A1 |
| 5588 |
PRKCQ |
8852 |
AKAP4 |
| |
|
5141 |
PDE4A |
| |
|
5142 |
PDE4B |
| |
|
5143 |
PDE4C |
| |
|
9495 |
AKAP5 |
| |
|
5144 |
PDE4D |
| |
|
5530 |
PPP3CA |
| |
|
94235 |
GNG8 |
| |
|
4893 |
NRAS |
| |
|
5533 |
PPP3CC |
| |
|
10142 |
AKAP9 |
| |
|
5150 |
PDE7A |
| |
|
10270 |
AKAP8 |
| |
|
9630 |
GNA14 |
| |
|
5151 |
PDE8A |
| |
|
801 |
CALM1 |
| |
|
5153 |
PDE1B |
| |
|
55970 |
GNG12 |
| |
|
805 |
CALM2 |
| |
|
8622 |
PDE8B |
| |
|
10672 |
GNA13 |
| |
|
3760 |
KCNJ3 |
| |
|
9138 |
ARHGEF1 |
| |
|
51764 |
GNG13 |
| |
|
10681 |
GNB5 |
| |
|
5566 |
PRKACA |
| |
|
5567 |
PRKACB |
| |
|
3265 |
HRAS |
| |
|
5573 |
PRKAR1A |
| |
|
10566 |
AKAP3 |
| |
|
5575 |
PRKAR1B |
| |
|
5576 |
PRKAR2A |
| |
|
5577 |
PRKAR2B |
| |
|
5578 |
PRKCA |
| |
|
5579 |
PRKCB |
| |
|
5580 |
PRKCD |
| |
|
5581 |
PRKCE |
| |
|
5582 |
PRKCG |
| |
|
11214 |
AKAP13 |
| |
|
2767 |
GNA11 |
| |
|
11215 |
AKAP11 |
| |
|
5583 |
PRKCH |
| |
|
5584 |
PRKCI |
| |
|
2768 |
GNA12 |
| |
|
11216 |
AKAP10 |
| |
|
2769 |
GNA15 |
| |
|
2770 |
GNAI1 |
| |
|
2771 |
GNAI2 |
| |
|
5331 |
PLCB3 |
| |
|
5587 |
PRKD1 |
| |
|
5588 |
PRKCQ |
| |
|
2773 |
GNAI3 |
| |
|
5590 |
PRKCZ |
| |
|
2774 |
GNAL |
| |
|
2775 |
GNAO1 |
| |
|
2776 |
GNAQ |
| |
|
2778 |
GNAS |
| |
|
2781 |
GNAZ |
| |
|
6237 |
RRAS |
| |
|
2782 |
GNB1 |
| |
|
2784 |
GNB3 |
| |
|
2785 |
GNG3 |
| |
|
2786 |
GNG4 |
| |
|
2787 |
GNG5 |
| |
|
2788 |
GNG7 |
| |
|
8165 |
AKAP1 |
| |
|
2790 |
GNG10 |
| |
|
2791 |
GNG11 |
| |
|
2792 |
GNGT1 |
| |
|
2793 |
GNGT2 |
| |
|
107 |
ADCY1 |
| |
|
27115 |
PDE7B |
| |
|
108 |
ADCY2 |
| |
|
109 |
ADCY3 |
| |
|
111 |
ADCY5 |
| |
|
112 |
ADCY6 |
| |
|
113 |
ADCY7 |
| |
|
114 |
ADCY8 |
| |
|
115 |
ADCY9 |
| |
|
9590 |
AKAP12 |
| |
|
9465 |
AKAP7 |
| |
|
3708 |
ITPR1 |
| Reactome G Protein Mediated Events (M26911) |
2767 |
GNA11 |
5136 |
PDE1A |
| 2776 |
GNAQ |
196883 |
ADCY4 |
| 815 |
CAMK2A |
10768 |
AHCYL1 |
| 816 |
CAMK2B |
5137 |
PDE1C |
| 5566 |
PRKACA |
10645 |
CAMKK2 |
| 5573 |
PRKAR1A |
156 |
GRK2 |
| 5582 |
PRKCG |
9630 |
GNA14 |
| |
|
84254 |
CAMKK1 |
| |
|
5153 |
PDE1B |
| |
|
801 |
CALM1 |
| |
|
814 |
CAMK4 |
| |
|
815 |
CAMK2A |
| |
|
816 |
CAMK2B |
| |
|
817 |
CAMK2D |
| |
|
818 |
CAMK2G |
| |
|
5566 |
PRKACA |
| |
|
5567 |
PRKACB |
| |
|
5568 |
PRKACG |
| |
|
23236 |
PLCB1 |
| |
|
5573 |
PRKAR1A |
| |
|
5575 |
PRKAR1B |
| |
|
346562 |
GNAT3 |
| |
|
5576 |
PRKAR2A |
| |
|
5577 |
PRKAR2B |
| |
|
5321 |
PLA2G4A |
| |
|
5578 |
PRKCA |
| |
|
5580 |
PRKCD |
| |
|
5582 |
PRKCG |
| |
|
2767 |
GNA11 |
| |
|
26960 |
NBEA |
| |
|
2769 |
GNA15 |
| |
|
2770 |
GNAI1 |
| |
|
5330 |
PLCB2 |
| |
|
2771 |
GNAI2 |
| |
|
5331 |
PLCB3 |
| |
|
5332 |
PLCB4 |
| |
|
2773 |
GNAI3 |
| |
|
2774 |
GNAL |
| |
|
2776 |
GNAQ |
| |
|
5594 |
MAPK1 |
| |
|
1385 |
CREB1 |
| |
|
107 |
ADCY1 |
| |
|
108 |
ADCY2 |
| |
|
109 |
ADCY3 |
| |
|
5613 |
PRKX |
| |
|
111 |
ADCY5 |
| |
|
112 |
ADCY6 |
| |
|
113 |
ADCY7 |
| |
|
114 |
ADCY8 |
| |
|
115 |
ADCY9 |
| |
|
3708 |
ITPR1 |
| |
|
3709 |
ITPR2 |
| |
|
3710 |
ITPR3 |
| |
|
3838 |
KPNA2 |
| Reactome G Protein Mediated Events (MM14496) |
2767 |
GNA11 |
23236 |
PLCB1 |
| 2776 |
GNAQ |
5573 |
PRKAR1A |
| 5566 |
PRKACA |
346562 |
GNAT3 |
| 5573 |
PRKAR1A |
5575 |
PRKAR1B |
| 5582 |
PRKCG |
5321 |
PLA2G4A |
| |
|
5577 |
PRKAR2B |
| |
|
5580 |
PRKCD |
| |
|
5582 |
PRKCG |
| |
|
2767 |
GNA11 |
| |
|
5136 |
PDE1A |
| |
|
196883 |
ADCY4 |
| |
|
2769 |
GNA15 |
| |
|
5137 |
PDE1C |
| |
|
5330 |
PLCB2 |
| |
|
2770 |
GNAI1 |
| |
|
2771 |
GNAI2 |
| |
|
5331 |
PLCB3 |
| |
|
5332 |
PLCB4 |
| |
|
2773 |
GNAI3 |
| |
|
10645 |
CAMKK2 |
| |
|
2774 |
GNAL |
| |
|
2776 |
GNAQ |
| |
|
5594 |
MAPK1 |
| |
|
156 |
GRK2 |
| |
|
9630 |
GNA14 |
| |
|
84254 |
CAMKK1 |
| |
|
801 |
CALM1 |
| |
|
5153 |
PDE1B |
| |
|
805 |
CALM2 |
| |
|
808 |
CALM3 |
| |
|
107 |
ADCY1 |
| |
|
108 |
ADCY2 |
| |
|
109 |
ADCY3 |
| |
|
111 |
ADCY5 |
| |
|
112 |
ADCY6 |
| |
|
113 |
ADCY7 |
| |
|
114 |
ADCY8 |
| |
|
115 |
ADCY9 |
| |
|
5566 |
PRKACA |
| |
|
5567 |
PRKACB |
Table 3.
Summary of GPCR and Long COVID Genes after functional similarity analyses.
Table 3.
Summary of GPCR and Long COVID Genes after functional similarity analyses.
| Pathway ID |
Name |
Gene from Input (Long COVID) |
Gene from Annotations (GPCR) |
| MM15882 |
Wp G Protein Signaling Pathways |
9 |
33 |
| M26911 |
Reactome G Protein Mediated Events |
7 |
27 |
| M39426 |
Wp G Protein Signaling Pathways |
9 |
33 |
| MM14496 |
Reactome G Protein Mediated Events |
5 |
18 |
| |
Total |
30 |
111 |
Table 4.
Results of Sequence Similarity Analysis (>60% Threshold) and Protein-Protein Affinity Prediction (3 Lowest Binding Affinities per Gene).
Table 4.
Results of Sequence Similarity Analysis (>60% Threshold) and Protein-Protein Affinity Prediction (3 Lowest Binding Affinities per Gene).
| ToppGene Pathway |
Sequence Similarity |
Protein-protein Affinity |
| Pathway |
Gene from Input (long covid) |
Gene from Annontations (GPCR) |
Similarity Percentage |
Binding Free Energy (kcal/ |
Dissociation Value (M) |
| Gene Id |
Symbol |
Gene ID |
Symbol |
mol) |
| Wp G Protein Signaling Pathways (MM15882 & M39426) |
3265 |
HRAS |
3845 |
KRAS |
94.2 |
-8.55 |
5.33E-07 |
| 4893 |
NRAS |
92.6 |
-8.37 |
7.26E-07 |
| 3265 |
HRAS |
100 |
-8.17 |
1.02E-06 |
| 2767 |
GNA11 |
9630 |
GNA14 |
90.9 |
-6.51 |
1.67E-05 |
| 2767 |
GNA11 |
100 |
-6.39 |
2.05E-05 |
| 2769 |
GNA15 |
71.9 |
-7.25 |
4.81E-06 |
| 2773 |
GNAI3 |
64.5 |
-7.46 |
3.40E-06 |
| 2775 |
GNAO1 |
64.8 |
-7.84 |
1.78E-06 |
| 2776 |
GNAQ |
96.1 |
-6.38 |
2.11E-05 |
| 2781 |
GNAZ |
61.3 |
-7.75 |
2.06E-06 |
| 2776 |
GNAQ |
9630 |
GNA14 |
90 |
-6.49 |
1.73E-05 |
| 2767 |
GNA11 |
96.1 |
-6.38 |
2.11E-05 |
| 2769 |
GNA15 |
70.9 |
-7.21 |
5.12E-06 |
| 2773 |
GNAI3 |
64 |
-7.44 |
3.51E-06 |
| 2775 |
GNAO1 |
64 |
-7.82 |
1.84E-06 |
| 2776 |
GNAQ |
100 |
-6.36 |
2.18E-05 |
| 2781 |
GNAZ |
60.3 |
-7.74 |
2.12E-06 |
| 3845 |
KRAS |
3845 |
KRAS |
100 |
-7.75 |
2.07E-06 |
| 4893 |
NRAS |
94.2 |
-7.22 |
5.05E-06 |
| 3265 |
HRAS |
94.2 |
-8.55 |
5.33E-07 |
| 4893 |
NRAS |
3845 |
KRAS |
94.2 |
-7.57 |
2.82E-06 |
| 4893 |
NRAS |
100 |
-7.37 |
3.96E-06 |
| 3265 |
HRAS |
92.6 |
-8.37 |
7.26E-07 |
| 5566 |
PRKACA |
5566 |
PRKACA |
100 |
-10.75 |
1.32E-08 |
| 5567 |
PRKACB |
77.5 |
-9.89 |
5.61E-08 |
| 2773 |
GNAI3 |
64 |
-9.24 |
1.67E-07 |
| 5573 |
PRKAR1A |
5573 |
PRKAR1A |
100 |
-10.61 |
1.64E-08 |
| 5575 |
PRKAR1B |
90.6 |
-10.23 |
3.14E-08 |
| 5582 |
PRKCG |
5578 |
PRKCA |
80 |
-15.87 |
2.31E-12 |
| 5579 |
PRKCB |
78 |
-16.48 |
8.21E-13 |
| 5582 |
PRKCG |
100 |
-20.77 |
5.84E-16 |
| 5588 |
PRKCQ |
5580 |
PRKCD |
68.5 |
-7.3 |
4.45E-06 |
| 5588 |
PRKCQ |
100 |
-4.54 |
4.68E-04 |
| Reactome G Protein Mediated Events (M26911) |
2767 |
GNA11 |
9630 |
GNA14 |
90.9 |
-6.51 |
1.67E-05 |
| 346562 |
GNAT3 |
63.3 |
-8.08 |
1.19E-06 |
| 2767 |
GNA11 |
100 |
-6.39 |
2.05E-05 |
| 2769 |
GNA15 |
71.9 |
-7.25 |
4.81E-06 |
| 2773 |
GNAI3 |
64.5 |
-7.46 |
4.00E-07 |
| 2776 |
GNAQ |
96.1 |
-6.38 |
2.11E-05 |
| 2776 |
GNAQ |
9630 |
GNA14 |
90.9 |
-6.49 |
1.73E-05 |
| 346562 |
GNAT3 |
63.3 |
-8.06 |
1.23E-06 |
| 2767 |
GNA11 |
100 |
-6.38 |
2.11E-05 |
| 2769 |
GNA15 |
71.9 |
-7.21 |
5.12E-06 |
| 2773 |
GNAI3 |
64.5 |
-7.44 |
3.51E-06 |
| 2776 |
GNAQ |
96.1 |
-6.36 |
2.18E-05 |
| 815 |
CAMK2A |
815 |
CAMK2A |
100 |
-10.21 |
3.27E-08 |
| 816 |
CAMK2B |
67.9 |
-12.35 |
8.72E-10 |
| 817 |
CAMK2D |
88.3 |
-10.06 |
4.19E-08 |
| 818 |
CAMK2G |
82.7 |
-10.9 |
1.02E-08 |
| 816 |
CAMK2B |
815 |
CAMK2A |
67.9 |
-15.45 |
4.68E-12 |
| 816 |
CAMK2B |
100 |
-12.35 |
8.72E-10 |
| 817 |
CAMK2D |
88.3 |
-11.49 |
3.77E-09 |
| 818 |
CAMK2G |
82.7 |
-11.45 |
3.99E-09 |
| 5566 |
PRKACA |
5566 |
PRKACA |
100 |
-10.75 |
1.32E-08 |
| 5567 |
PRKACB |
77.5 |
-9.89 |
5.61E-08 |
| 5568 |
PRKACG |
74.5 |
-9.98 |
4.79E-08 |
| 5573 |
PRKAR1A |
5573 |
PRKAR1A |
100 |
-10.52 |
1.93E-08 |
| 5575 |
PRKAR1B |
90.6 |
-9.99 |
4.68E-08 |
| 5582 |
PRKCG |
5578 |
PRKCA |
80 |
-15.87 |
2.31E-12 |
| 5582 |
PRKCG |
100 |
-20.77 |
5.84E-16 |
| Reactome G Protein Mediated Events (MM14496) |
2767 |
GNA11 |
346562 |
GNAT3 |
63.3 |
-8.08 |
1.19E-06 |
| 2767 |
GNA11 |
100 |
-6.39 |
2.05E-05 |
| 2769 |
GNA15 |
71.9 |
-7.26 |
4.81E-06 |
| 2773 |
GNAI3 |
64.5 |
-7.46 |
3.40E-06 |
| 2776 |
GNAQ |
96.1 |
-6.38 |
2.11E-05 |
| 9630 |
GNA14 |
90.9 |
-6.51 |
1.67E-05 |
| 2776 |
GNAQ |
346562 |
GNAT3 |
63.3 |
-8.08 |
1.23E-06 |
| 2767 |
GNA11 |
96.1 |
-6.38 |
2.11E-05 |
| 2769 |
GNA15 |
70.9 |
-7.21 |
5.12E-06 |
| 2773 |
GNAI3 |
64 |
-7.44 |
3.51E-06 |
| 2776 |
GNAQ |
100 |
-6.36 |
2.18E-05 |
| 9630 |
GNA14 |
90 |
-6.49 |
1.73E-05 |
| 84254 |
CAMKK1 |
91.7 |
-8.15 |
1.06E-06 |
| 5566 |
PRKACA |
5566 |
PRKACA |
100 |
-10.75 |
1.32E-08 |
| 5567 |
PRKACB |
77.5 |
-10.13 |
3.74E-08 |
| 5573 |
PRKAR1A |
5573 |
PRKAR1A |
100 |
-10.61 |
1.64E-08 |
| 5575 |
PRKAR1B |
90.6 |
-10.23 |
3.14E-08 |
| 5582 |
PRKCG |
5582 |
PRKCG |
100 |
-20.77 |
5.84E-16 |
Table 5.
Top 10 GPCR genes associated with Long COVID genes from each pathway identified using network analysis in Cytoscape and CytoHubba plugin.
Table 5.
Top 10 GPCR genes associated with Long COVID genes from each pathway identified using network analysis in Cytoscape and CytoHubba plugin.
| Pathway |
Receptor (Long COVID) |
Ligand (GPCR) |
| Gene Id |
Symbol |
Gene ID |
Symbol |
| Wp G Protein Signaling Pathways (MM15882 & M39426) |
3265 |
HRAS |
3265 |
HRAS |
| 2767 |
GNA11 |
3845 |
KRAS |
| 2776 |
GNAQ |
9630 |
GNA14 |
| 3845 |
KRAS |
2767 |
GNA11 |
| |
|
2769 |
GNA15 |
| |
|
2773 |
GNAI3 |
| |
|
2775 |
GNAO1 |
| |
|
2776 |
GNAQ |
| |
|
2781 |
GNAZ |
| |
|
5578 |
PRKCA |
| Reactome G Protein Mediated Events (M26911) |
2767 |
GNA11 |
9630 |
GNA14 |
| 2776 |
GNAQ |
346562 |
GNAT3 |
| 5566 |
PRKACA |
2767 |
GNA11 |
| |
|
2769 |
GNA15 |
| |
|
2773 |
GNAI3 |
| |
|
2776 |
GNAQ |
| |
|
5566 |
PRKACA |
| |
|
5567 |
PRKACB |
| |
|
5568 |
PRKACG |
| Reactome G Protein Mediated Events (MM14496) |
2767 |
GNA11 |
346562 |
GNAT3 |
| 2776 |
GNAQ |
2767 |
GNA11 |
| 5566 |
PRKACA |
2769 |
GNA15 |
| 5573 |
PRKAR1A |
2773 |
GNAI3 |
| |
|
2776 |
GNAQ |
| |
|
9630 |
GNA14 |
| |
|
5566 |
PRKACA |
| |
|
5567 |
PRKACB |
| |
|
5573 |
PRKAR1A |
| |
|
5575 |
PRKAR1B |
Table 6.
21 Identified GPCR genes with associated symptoms and repurposed FDA drugs.
Table 6.
21 Identified GPCR genes with associated symptoms and repurposed FDA drugs.
| Identified Genes |
Symptoms |
HPO ID |
Symptoms Category |
FDA-Approved Repurposed Drugs |
| HRAS |
Anaphylactic shock |
HP:0100845 |
Immunology-autoimmunity |
/ |
| Anti-thyroid peroxidase antibody positivity |
HP:0025379 |
Immunology-autoimmunity |
| Alopecia |
HP:0001596 |
Dermatological |
| Fragile nails |
HP:0001808 |
Dermatological |
| Gastroesophageal reflux |
HP:0002020 |
Gastrointestinal |
| Sleep apnea |
HP:0010535 |
Neuropsychiatric |
| GNAQ |
Ocular pain |
HP:0200026 |
HEENT-Eye |
Verteporfin, Selumetinib |
| Pain |
HP:0012531 |
General symptom |
| Venous thrombosis |
HP:0004936 |
Cardiovascular |
| Pulmonary embolism |
HP:0002204 |
Pulmonary |
| GNA11 |
Alopecia |
HP:0001596 |
Dermatological |
Binimetinib, Cabozantinib, Selumetinib |
| Ocular pain |
HP:0200026 |
HEENT-Eye |
| GNAO1 |
Hyperkinetic movements |
HP:0002487 |
Neuropsychiatric |
/ |
| KRAS |
Anti-thyroid peroxidase antibody positivity |
HP:0025379 |
Immunology-autoimmunity |
Panitumumab, Cetuximab, Adagrasib, Sotorasib |
| Alopecia |
HP:0001596 |
Dermatological |
| Edema |
HP:0000969 |
Reproductive-Genitourinary-Endocrine-Metabolism |
| Back pain |
HP:0003418 |
General symptom |
| Pain |
HP:0012531 |
General symptom |
| Fatigue |
HP:0012378 |
General symptom |
| Abdominal pain |
HP:0002027 |
Gastrointestinal |
| Anorexia |
HP:0002039 |
Gastrointestinal |
| PRKCB |
Pleural thickening |
HP:0031944 |
Pulmonary-imaging |
/ |
| PRKCG |
Gaze-evoked nystagmus |
HP:0000640 |
HEENT-Eye |
/ |
| Memory impairment |
HP:0002354 |
Neuropsychiatric |
| Dysmetria |
HP:0001310 |
Neuropsychiatric |
| PRKACA |
Alopecia |
HP:0001596 |
Dermatological |
/ |
| Irregular menstruation |
HP:0000858 |
Reproductive-Genitourinary-Endocrine-Metabolism |
| Mania |
HP:0100754 |
Neuropsychiatric |
| Memory impairment |
HP:0002354 |
Neuropsychiatric |
| PRKACG |
Menorrhagia |
HP:0000132 |
Reproductive-Genitourinary-Endocrine-Metabolism |
/ |
| PRKAR1A |
Alopecia |
HP:0001596 |
Dermatological |
/ |
| Irregular menstruation |
HP:0000858 |
Reproductive-Genitourinary-Endocrine-Metabolism |
| Elevated circulating thyroid-stimulating hormone concentration |
HP:0002925 |
Lab |
| Hypofibrinogenemia |
HP:0011900 |
Lab |
| Mania |
HP:0100754 |
Neuropsychiatric |
| Memory impairment |
HP:0002354 |
Neuropsychiatric |
| CAMK2A |
Gastroesophageal reflux |
HP:0002020 |
Gastrointestinal |
/ |
| Abnormal emotion/affect behavior |
HP:0100851 |
Neuropsychiatric |
| Sleep disturbance |
HP:0002360 |
Neuropsychiatric |
| Dystonia |
HP:0001332 |
Neuropsychiatric |
| Gait disturbance |
HP:0001288 |
Neuropsychiatric |
| CAMK2B |
Gastroesophageal reflux |
HP:0002020 |
Gastrointestinal |
/ |
| Sleep disturbance |
HP:0002360 |
Neuropsychiatric |
| GNAQ, GNA13, GNA14, GNA15, GNAT3, PRKCA, PRKACB, PRKAR1, CAMKK1 |
/ |
/ |
/ |
/ |
Table 7.
Common genes identified in more than one Long COVID symptoms.
Table 7.
Common genes identified in more than one Long COVID symptoms.
| Identified Genes |
Occurrences |
Symptoms Category |
| HRAS |
6 |
Immunology-autoimmunity, Dermatological, Gastrointestinal, Neuropsychiatric |
| GNAQ |
4 |
HEENT-eye, General symptom, Cardiovascular, Pulmonary |
| GNA11 |
2 |
Dermatological, HEENT-eye |
| GNAO1 |
1 |
Neuropsychiatric |
| KRAS |
7 |
Immunology-autoimmunity, Dermatological, General symptoms, Gastrointestinal , Neuropsychiatric, Reproductive-Genitourinary-Endocrine-Metabolism |
| PRKCB |
1 |
Pulmonary |
| PRKCG |
3 |
HEENT-eye, Neuropsychiatric |
| PRKACA |
4 |
Dermatological, Reproductive-Genitourinary-Endocrine-Metabolism, Neuropsychiatric |
| PRKACG |
1 |
Reproductive-Genitourinary-Endocrine-Metabolism |
| PRKAR1A |
6 |
Dermatological, Reproductive-Genitourinary-Endocrine-Metabolism, Lab, Neuropsychiatric |
| CAMK2A |
5 |
Gastrointestinal, Neuropsychiatric |
| CAMK2B |
2 |
Gastrointestinal, Neuropsychiatric |
Table 8.
Virus-host interaction table identified through Enrichr tool through COVID-19 related gene sets analysis.
Table 8.
Virus-host interaction table identified through Enrichr tool through COVID-19 related gene sets analysis.
| SARS-CoV-2 Gene |
Human Genes |
Adjusted P-Value |
| SARS coronavirus protein E (gene: E) |
HRAS |
0.3499 |
| GNAO1 |
0.4334 |
| GNAZ |
0.4017 |
| SARS coronavirus P2 envelope protein |
HRAS |
0.3499 |
| GNAO1 |
0.4334 |
| GNAZ |
0.4017 |
| SARS coronavirus Tor2 small envelope E protein |
HRAS |
0.3499 |
| GNAO1 |
0.4334 |
| GNAZ |
0.4017 |
| SARS coronavirus formerly known as growth-factor-like protein (gene: orf1ab) |
HRAS |
0.3499 |
| KRAS |
0.1345 |
| SARS coronavirus nsp7-pp1a/pp1ab (gene: orf1ab) |
HRAS |
0.3499 |
| KRAS |
0.1345 |
| PRKCA |
0.3479 |
| SARS coronavirus hypothetical protein sars7a |
HRAS |
0.3499 |
| KRAS |
0.2006 |
| SARS coronavirus P2 hypothetical protein sars7a |
HRAS |
0.3499 |
| KRAS |
0.2006 |
| SARS coronavirus Tor2 Orf8 |
HRAS |
0.3499 |
| KRAS |
0.2006 |
| SARS coronavirus nsp3-pp1a/pp1ab (gene: orf1ab) |
HRAS |
0.5045 |
| KRAS |
0.157 |
| GNAQ |
0.5409 |
| PRKCA |
0.3286 |
| PRKCG |
0.3064 |
| PRKACB |
0.4816 |
| SARS coronavirus excised_polyprotein 1..4369 (gene: orf1ab) |
HRAS |
0.6646 |
| KRAS |
0.09531 |
| GNAQ |
0.4168 |
| PRKCA |
0.04058 |
| PRKCG |
0.6548 |
| PRKACB |
0.4816 |
| SARS coronavirus P2 full_polyprotein 1..4382 |
HRAS |
0.6713 |
| KRAS |
0.09531 |
| GNAQ |
0.4181 |
| PRKCA |
0.04146 |
| PRKCG |
0.6616 |
| PRKACB |
0.4816 |
| SARS coronavirus nsp13-pp1ab (ZD, NTPase/HEL; RNA (gene: orf1ab) |
KRAS |
0.089 |
| SARS coronavirus RNA-dependent RNA polymerase (gene: orf1ab) |
KRAS |
0.1109 |
| SARS coronavirus nsp4-pp1a/pp1ab (gene: orf1ab) |
KRAS |
0.1316 |
| SARS coronavirus hypothetical protein sars9b |
KRAS |
0.1345 |
| SARS coronavirus Tor2 Orf13 |
KRAS |
0.1345 |
| SARS coronavirus 3C-like proteinase (gene: orf1ab) |
KRAS |
0.1406 |
| SARS coronavirus leader protein (gene: orf1ab) |
KRAS |
0.1428 |
| SARS coronavirus nucleocapsid protein (gene: N) |
KRAS |
0.1714 |
| SARS coronavirus P2 nucleocapsid protein |
KRAS |
0.1714 |
| SARS coronavirus Tor2 nucleocapsid protein |
KRAS |
0.1714 |
| SARS coronavirus nsp8-pp1a/pp1ab (gene: orf1ab) |
KRAS |
0.2234 |
| PRKCA |
0.3479 |
| PRKAR1A |
0.2813 |
| SARS coronavirus P2 spike glycoprotein precursor |
KRAS |
0.3121 |
| GNAQ |
0.4168 |
| GNAO1 |
0.4334 |
| PRKCB |
0.5624 |
| PRKAR1A |
0.3559 |
| SARS coronavirus E2 glycoprotein precursor (gene: S) |
KRAS |
0.3152 |
| GNAQ |
0.4168 |
| GNAO1 |
0.4334 |
| PRKCB |
0.5624 |
| PRKAR1A |
0.3575 |
| SARS coronavirus Tor2 spike glycoprotein |
KRAS |
0.3152 |
| GNAQ |
0.4168 |
| GNAO1 |
0.4334 |
| PRKCB |
0.5624 |
| PRKAR1A |
0.3575 |
| SARS coronavirus excised_polyprotein 1..4369 (gene: orf1ab) |
GNAQ |
0.4168 |
| GNAO1 |
0.4334 |
| GNAZ |
0.4017 |
| PRKCB |
0.5624 |
| PRKCG |
0.6548 |
| PRKACB |
0.4816 |
| SARS coronavirus P2 full_polyprotein 1..4382 |
GNAQ |
0.4181 |
| GNAO1 |
0.4334 |
| GNAZ |
0.4017 |
| PRKCB |
0.5624 |
| PRKCG |
0.6616 |
| PRKACB |
0.4816 |
| SARS coronavirus Tor2 replicase 1AB |
GNAQ |
0.5281 |
| GNAZ |
0.4017 |
| GNA11 |
0.2203 |
| GNA13 |
0.4737 |
| PRKCA |
0.3286 |
| PRKAR1A |
0.4509 |
| SARS coronavirus P2 full_polyprotein 1..7073 |
GNAQ |
0.53 |
| GNAZ |
0.4017 |
| GNA11 |
0.2203 |
| PRKCA |
0.3286 |
| PRKAR1A |
0.4529 |
Table 9.
Identification of Similar Genes Between Long COVID and GPCR Genes and Their Associated Symptoms.
Table 9.
Identification of Similar Genes Between Long COVID and GPCR Genes and Their Associated Symptoms.
| Pathway |
Similar Genes Available in Long Covid & GPCR |
Symptoms category |
Symptoms |
HPO ID |
| Gene ID |
Name & Symbol |
| WPG PROTEIN SIGNALING PATHWAYS (MM15882 & M39426) |
3265 |
HRAS (HRas proto-oncogene, GTPase) |
Immunology-autoimmunity |
Anaphylactic shock |
HP:0100845 |
| Immunology-autoimmunity |
Anti-thyroid peroxidase antibody positivity |
HP:0025379 |
| Dermatological |
Alopecia |
HP:0001596 |
| Dermatological |
Fragile nails |
HP:0001808 |
| Gastrointestinal |
Gastroesophageal reflux |
HP:0002020 |
| Neuropsychiatric |
Sleep apnea |
HP:0010535 |
| 3845 |
KRAS (KRAS proto-oncogene, GTPase) |
Immunology-autoimmunity |
Anti-thyroid peroxidase antibody positivity |
HP:0025379 |
| Dermatological |
Alopecia |
HP:0001596 |
| Reproductive-Genitourinary-Endocrine-Metabolism |
Edema |
HP:0000969 |
| General symptom |
Back pain |
HP:0003418 |
| General symptom |
Pain |
HP:0012531 |
| General symptom |
Fatigue |
HP:0012378 |
| Gastrointestinal |
Abdominal pain |
HP:0002027 |
| Gastrointestinal |
Anorexia |
HP:0002039 |
| 4893 |
NRAS (NRAS proto-oncogene, GTPase) |
|
|
|
| 2767 |
GNA11 (G protein subunit alpha 11) |
Dermatological |
Alopecia |
HP:0001596 |
| HEENT-Eye |
Ocular pain |
HP:0200026 |
| 2776 |
GNAQ (G protein subunit alpha q) |
HEENT-Eye |
Ocular pain |
HP:0200026 |
| General symptom |
Pain |
HP:0012531 |
| Cardiovascular |
Venous thrombosis |
HP:0004936 |
| Pulmonary |
Pulmonary embolism |
HP:0002204 |
| 5566 |
PRKACA (protein kinase cAMP-activated catalytic subunit alpha) |
Dermatological |
Alopecia |
HP:0001596 |
| Reproductive-Genitourinary-Endocrine-Metabolism |
Irregular menstruation |
HP:0000858 |
| Neuropsychiatric |
Mania |
HP:0100754 |
| Neuropsychiatric |
Memory impairment |
HP:0002354 |
| 5573 |
PRKAR1A (protein kinase cAMP-dependent type I regulatory subunit alpha) |
Dermatological |
Alopecia |
HP:0001596 |
| Reproductive-Genitourinary-Endocrine-Metabolism |
Irregular menstruation |
HP:0000858 |
| Lab |
Elevated circulating thyroid-stimulating hormone concentration |
HP:0002925 |
| Lab |
Hypofibrinogenemia |
HP:0011900 |
| Neuropsychiatric |
Mania |
HP:0100754 |
| Neuropsychiatric |
Memory impairment |
HP:0002354 |
| 5582 |
PRKCG (protein kinase C gamma) |
Reproductive-Genitourinary-Endocrine-Metabolism |
Menorrhagia |
HP:0000132 |
| 5588 |
PRKCQ (protein kinase C theta) |
|
|
|
| REACTOME G PROTEIN MEDIATED EVENTS (M26911) |
2767 |
GNA11 (G protein subunit alpha 11) |
Dermatological |
Alopecia |
HP:0001596 |
| HEENT-Eye |
Ocular pain |
HP:0200026 |
| 2776 |
GNAQ (G protein subunit alpha q) |
HEENT-Eye |
Ocular pain |
HP:0200026 |
| General symptom |
Pain |
HP:0012531 |
| Cardiovascular |
Venous thrombosis |
HP:0004936 |
| Pulmonary |
Pulmonary embolism |
HP:0002204 |
| 815 |
CAMK2A (calcium/calmodulin dependent protein kinase II alpha) |
Gastrointestinal |
Gastroesophageal reflux |
HP:0002020 |
| Neuropsychiatric |
Abnormal emotion/affect behavior |
HP:0100851 |
| Neuropsychiatric |
Sleep disturbance |
HP:0002360 |
| Neuropsychiatric |
Dystonia |
HP:0001332 |
| Neuropsychiatric |
Gait disturbance |
HP:0001288 |
| 816 |
CAMK2B (calcium/calmodulin dependent protein kinase II beta) |
Gastrointestinal |
Gastroesophageal reflux |
HP:0002020 |
| Neuropsychiatric |
Sleep disturbance |
HP:0002360 |
| 5566 |
PRKACA (protein kinase cAMP-activated catalytic subunit alpha) |
Dermatological |
Alopecia |
HP:0001596 |
| Reproductive-Genitourinary-Endocrine-Metabolism |
Irregular menstruation |
HP:0000858 |
| Neuropsychiatric |
Mania |
HP:0100754 |
| Neuropsychiatric |
Memory impairment |
HP:0002354 |
| 5573 |
PRKAR1A (protein kinase cAMP-dependent type I regulatory subunit alpha) |
Dermatological |
Alopecia |
HP:0001596 |
| Reproductive-Genitourinary-Endocrine-Metabolism |
Irregular menstruation |
HP:0000858 |
| Lab |
Elevated circulating thyroid-stimulating hormone concentration |
HP:0002925 |
| Lab |
Hypofibrinogenemia |
HP:0011900 |
| Neuropsychiatric |
Mania |
HP:0100754 |
| Neuropsychiatric |
Memory impairment |
HP:0002354 |
| 5582 |
PRKCG (protein kinase C gamma) |
Reproductive-Genitourinary-Endocrine-Metabolism |
Menorrhagia |
HP:0000132 |
| REACTOME G PROTEIN MEDIATED EVENTS (MM14496) |
2767 |
GNA11 (G protein subunit alpha 11) |
Dermatological |
Alopecia |
HP:0001596 |
| HEENT-Eye |
Ocular pain |
HP:0200026 |
| 2776 |
GNAQ (G protein subunit alpha q) |
HEENT-Eye |
Ocular pain |
HP:0200026 |
| General symptom |
Pain |
HP:0012531 |
| Cardiovascular |
Venous thrombosis |
HP:0004936 |
| Pulmonary |
Pulmonary embolism |
HP:0002204 |
| 5566 |
PRKACA (protein kinase cAMP-activated catalytic subunit alpha) |
Dermatological |
Alopecia |
HP:0001596 |
| Reproductive-Genitourinary-Endocrine-Metabolism |
Irregular menstruation |
HP:0000858 |
| Neuropsychiatric |
Mania |
HP:0100754 |
| Neuropsychiatric |
Memory impairment |
HP:0002354 |
| 5573 |
PRKAR1A (protein kinase cAMP-dependent type I regulatory subunit alpha) |
Dermatological |
Alopecia |
HP:0001596 |
| Reproductive-Genitourinary-Endocrine-Metabolism |
Irregular menstruation |
HP:0000858 |
| Lab |
Elevated circulating thyroid-stimulating hormone concentration |
HP:0002925 |
| Lab |
Hypofibrinogenemia |
HP:0011900 |
| Neuropsychiatric |
Mania |
HP:0100754 |
| Neuropsychiatric |
Memory impairment |
HP:0002354 |
| 5582 |
PRKCG (protein kinase C gamma) |
Reproductive-Genitourinary-Endocrine-Metabolism |
Menorrhagia |
HP:0000132 |
4. Discussion
Initially, the results indicated 21 significant genes connected with the G protein-coupled receptors, indicative of their potential therapeutic targets for Long Covid. The said genes include members of the RAS family (HRAS, KRAS), the G protein family (GNAQ, GNAZ, GNA11, GNA13, GNA14, GNA15, GNAO1, GNAT3), a series of PRK—protein kinases (PRKCA, PRKCB, PRKCG, PRKACA, PRKACG, PRKACB, PRKAR1A, PRKAR1B) and calcium/calmodulin-dependent protein kinases (CAMK2A, CAMK2B, CAMKK1).
These gene groups are highly involved in general cellular and physiological processes. The Ras family of small GTPases involves molecular switches controlling cell growth, differentiation, and survival activity (Killoran & Smith, 2019). The G proteins bridge signal transduction from receptors to a large variety of effectors to influence processes ranging from sensory perception to hormonal responses (Hepler et al., 1993). Protein kinases are enzymes that modify proteins by phosphorylation, thus keying on and off important cellular functions, such as gene expression, secretion, growth, and apoptosis (Taylor et al., 2021). Lastly, CAMKs play a role in regulating several steps within the cell cycle, memory forming, and muscle contraction—processes very important to the functioning of living organisms—in addition to the many other functions they perform. These gene families dispose of the highly evolved cellular signalling networks of utmost importance in healthy and disease conditions, thus representing highly valuable therapeutic targets (Yasuda et al., 2022).
However, only 12 among these genes have been identified as pertinent to Long COVID symptoms since they participate in the receptor G protein-coupled cascade. These include HRAS, KRAS, GNAQ, GNA11, GNAO1, PRKCB, PRKCG, PRKACA, PRKACG, PRKAR1A, CAMK2A, and CAMK2B. As a whole, these genes account for the varied and long-lasting COVID symptoms in several systems. For example, an HRAS mutation can lead to anaphylactic shock and alopecia, while KRAS disruption leads to chronic pain and the presence of autoimmune problems. GNAQ and GNA11, on the other hand, lead to ocular pain and other vascular issues while GNAO1 can lead to neurotransmission problems, hence neuropsychiatric complications. On the other hand, PRKCB and PRKCG play important roles in inflammation and synaptic signalling, which can account for pleural thickening and neurologic symptoms, respectively. On the other hand, PRKACA and PRKACG also play an important role in the process of cellular signalling and control of reproductive functions, while PRKAR1A is implicated in metabolic and cognitive dysfunctions, and CAMK2A and CAMK2B are involved in neuropsychiatric and gastroesophageal disorders. It is through these interrelations that the potential to target these genes for Long COVID interventions is realized.
Consequently, the symptoms of Long COVID persist, because of immune dysregulation, in which multiple proteins and tissue regions are targeted by autoantibodies, leading to gross inflammation of multiple tissues and, ultimately, organ failure. With multisystem activity, these antibodies can cause anaphylactic shock or gut-related symptoms by targeting G-protein-coupled receptors, neuronal proteins such as CAMK2A and CAMK2B, which ultimately leads to neuropsychiatric disorders, or hormonal regulators such as PRKACA and PRKAR1A and disrupt endocrine function and cause metabolic dysregulation. Such symptom persistence and severity are features of chronic fatigue syndrome and rheumatoid arthritis, amongst autoimmune illnesses, that clarify the imperative of precise therapy targeting immune system modulation to enhance outcomes in the spectrum of long COVID and other post viral conditions.
Furthermore, these identified GPCRs genes are highly involved in immune modulation, inflammation, and respiratory functions, which are very key in the pathology and recovery of Long COVID. To give examples, some of these identified genes are associated with the RAS/MAPK signaling pathways, such as HRAS and KRAS, central in immune cell proliferation and survival against viral infections, including the causative agent of COVID-19. Meanwhile, all three members of the Protein Kinase C family belong to the immune and inflammatory response mediators: PRKCA, PRKCB, and PRKCG. Members of the GNAQ family—GNA11 and GNA13—modify the movement of immune cells and the production of cytokines. GPCR pathways also regulate some very important functions in the respiratory system, including the level of airway muscle contraction and mucosal secretion. These are central to the management of Long COVID with respiratory symptoms. CAMK2A and CAMK2B regulate calcium signaling in inflammation processes, which is central in the characteristic chronic inflammation of Long COVID. Although these identified functions regarding these genes are important for the development of targeted treatments to enhance the management of Long COVID by improving symptoms such as cognitive impairments and fatigue, some gaps need to be shown on the specific impacts of some genes.
Based on this, the Drug repurposing for the Long COVID symptoms is targeting certain genes with the FDA-approved drugs for the management of the symptoms (
Figure 4). Precisely, the medications Verteporfin and Selumetinib are found to target the gene GNAQ, influencing symptoms of ocular and cellular proliferation, whereas Binimetinib, Cabozantinib, and Selumetinib target GNA11 for dermatologic and ocular issues. For the gene KRAS, there is Panitumumab, Cetuximab, Adagrasib, and Sotorasib. All these medications are effective for autoimmunity and cellular growth. Another set of rich promise came through the targets Tipifarnib for immune modulation and tissue repair, whereas G-protein coupled receptor signaling is important for immune and inflammatory responses (Odeniyide et al., 2022). Moreover, the anti-inflammatory and immune-regulatory effects of Naltrexone, which was originally used to treat addiction, are currently a new therapeutic avenue in easing symptoms associated with long COVID by modifying opioid receptors and important signaling pathways during the therapy (Choubey et al., 2022). It must be noted that not all genes have drugs of high value/interest because of the specificity of the gene action concerning disease mechanisms. High clinical testing is needed the guarantee safety and efficacy since, as discussed, Long COVID is a complex problem, and personalized medicine approaches are needed.
An observation that should be made from these results is noting that identification of the genes such as HRAS, KRAS, GNAQ, and GNA11 has been consistent across the studies, testifying to the credibility of methodologies and suggesting a central role in the underlying pathology and therapy of Long COVID. These four genes also represent the only genes compared among the rest which are linked to FDA-approved drugs, underlining the enormous potential these genes have in clinical applications.
4.1. Overview of Long COVID Symptoms
The Long COVID symptoms is a wide spectrum of complications that include respiratory, dermatologic, gastrointestinal, neurologic, and other systemic persistent symptoms following a SARS-CoV-2 infection.
One of the pivotal hallmarks of Long COVID include substantial respiratory symptoms such as dyspnea, fatigue, and sarcopenia, and those resulting in increased vulnerability to mental problems like anxiety or depression, and other cardiovascular problems, which increase the probability of hospital readmissions (Das et al., 2022). Besides these usual symptoms, the long COVID cases, which had no prior respiratory diseases, may present with symptomatic airway diseases such as asthma. Radiological imaging might allow a clearer depiction of possible post-viral damage or remodelling by detecting structural abnormalities within the airways (Garg et al., 2021) (Taquet, Geddes, et al., 2021).
The range of cardiovascular symptoms and manifestations that persist appears due to incorrect remodelling of the structure of the heart, fluctuation of blood pressure rates, increased marks of troponin, inflammation of the myocardium, and chest pains (angina). Some patients develop postural orthostatic tachycardia syndrome (POTS) suggesting autonomic dysfunction (Crook et al., 2021). Studies have shown that remaining viral elements or immune imbalances resulting from acute infection could exacerbate the already hyperactive immune responses, leading to other cytokine storms that can initiate an autoimmune course which ultimately will manifest into severe cardio dysfunctions. Some genetic mutations such as SCN5A, MYBPC3 & MYH6 are associated with Long COVID cardiac health problems such as Brugada Syndrome, Atrial Fibrillation, Sick Sinus Syndrome, Dilated Cardiomyopathy among others (Das et al., 2022).
Dermatological effects of Long COVID may range from those that are easily observed, like brittle nails and inflammation of the skin, to severe or pronounced conditions, like alopecia and petechiae. It has, however, been shown that common post-COVID-19 symptoms during the first 1–2 months after the onset of acute illness include more hair loss than usual, skin rashes, with an increased severity at the outset of the infection. The severity of these side effects may mirror the severity of the acute illness the patient had been suffering. On the other side, other research works have pointed that gender is among the main factors that influence the susceptibility of a person to dermatological manifestations of which females are at higher risk (Thuangtong et al., 2021). Autoimmune disorders also present the possibility of causing inflammation in skin cells towards similar outcomes in existing patients.
Furthermore, gastrointestinal symptoms pose a possible manifestation of Long COVID, based on the fact that their occurrence has been reported in infected patients at varying rates from 11.4% to 61.1%. These include anorexia, diarrhoea, nausea, and vomiting, along with abdominal discomforts that can lead to gastric ulcers and malnutrition, as well as liver or spleen enlargement. On the other part, the onset time for these gastrointestinal manifestations seems variable; it may occur early on in some instances while in others, later in the course of the disease (Kariyawasam et al., 2021).
Additionally, the impact of COVID-19 could lead to a series of symptoms and complications associated with the Head, Ears, Nose, Throat, and Eyes (HEENT). However, they have not been as well documented as respiratory symptoms. The main sites of infection and diagnosis are usually within the tissues of the nose, nasopharynx, or oropharynx. Symptoms in these categories will present with ear problems, ENT issues, as well as eye-related conditions, which may manifest with conjunctivitis or vision that blurred (El-Anwar et al., 2021). Moreover, the brainstem inflammation brought about by the SARS-CoV2 virus has an association with many abnormalities that relate to the auditory system, leading toward sensory impairments such as hearing loss, hyperacusis, tinnitus, vertigo, ocular involvement, and motor deficits (Jafari et al., 2022).
Meanwhile, clinical and laboratory abnormalities, include cytokine storm and multi-organ dysfunction, raised levels of inflammatory markers like D-dimer, and activation of immune cells. Unfavourable outcomes in COVID-19, such as cytokine storm syndrome, happen simultaneously with an increase in systemic cytokines and immune cell activation. Studies prove that SLC2A3, TTC26, and CASR genes are found to be important in case of prolonged symptoms of COVID-infected patients, which contribute to Fanconi-Bickel Syndrome Type 2 Diabetes Mellitus hydrocephalus or hypocalcemia Autosomal Dominant as reported by Das et al. (2022). Some of the findings in patients with the post-COVID syndrome include unusual concentrations of hormones, signifying lymphocytopenia, the disproportionate raising of other laboratory findings such as the abnormal platelet count lower levels, creatinine, and kinase, lactate dehydrogenase (LDH), C-reactive protein (CRP), apart from the proportionate rise of transaminases (ALT and AST); as found among long-haulers, signifying anterior responses that could result in problems of blood clotting (a major contributor to multiple organ failure), while other abnormalities include high blood concentrations of circulating D-dimers. Abnormal liver functions are highlighted by raised ALT, and AST (Low R, Low R & Akrami, 2023).
Severe COVID-19 is characterized by the pathology of notable immunological-autoimmunity manifestations that may be contributory to multi-system organ failure observed in long COVID patients. Histologically, in pulmonary microangiopathy, fibrin thrombi, active platelets, and neutrophil extracellular traps are demonstrated largely suggesting autoimmunity affecting areas other than the lung. Furthermore, cases of evolving autoimmune diseases, like arthritis and type 1 diabetes, occurring in the wake of acute SARS-CoV-2 infection, point to a possible continuum between de novo autoimmunity and further sequelae of this disease (Knight et al., 2021).
On the other hand, patients affected by COVID-19 often present numerous neuropsychiatric symptoms, such as changes in speech and language capabilities, cognitive defects, and frequent headaches, among many others. Patients usually present with disorders in olfactory and gustatory sensitivity, emotional lability, and memory and sleep impairments, along with changes in behaviour patterns. These are typical signs that are experienced by patients who develop long COVID (Taquet, Geddes, et al., 2021). According to Das et al. (2022)., SARS-CoV-2 is effectively infecting the brain through different routes. This can lead to mild abnormalities such as changes in smell or taste, but it can also lead to a more serious condition like encephalopathy or seizures. Patients themselves have reported other neurological diseases such as epilepsy, intellectual disability, autism spectrum disorder, developmental delays, schizophrenia, etc. The cognitive issues among the infected are now prevalent, the hypoxemia, and inflammation furthering their negative impact on the motor function, attention, social cognition, language, and memory.
Patients with long COVID, therefore, may have clinical manifestations or symptoms related to Reproductive, Genitourinary, Endocrine, and Metabolism (RGEM) that represent quite a wide and possible spectrum. For example, menstrual irregularities may include the duration of the cycle intensity of flow, and total absence of menstruation. The long-term effect may present in the victim facing challenges in conceiving and holding a pregnancy from any fertility-related complications. The effect is also endocrine based; the dysregulation derives from the malfunctions of several glands, such as thyroid dysfunction (hypothyroidism/hyperthyroidism), insufficiency of the adrenal gland, or diabetes mellitus (C. Liu et al., 2021). According to Das et al. (2022), onset can result in great fatigue, complete exhaustion, and mood swings, along with secondary changes in weight likely to influence the appetite/thirst regulating mechanisms. Furthermore, alterations in the genitourinary system can lead to potential damage to the testes in males, while inflammation affecting the ovaries in females also leads to similar outcomes in their efforts for procreation (fertility rate). Additionally, in the presence of comorbidities, the control of glucose exhibits unusual defects, which largely characterize cardiac health adversity influencing the metabolic streamlining thereof. This includes defects such as abnormalities in lipid levels contributing significantly toward the metabolism alteration, which causes phenomena for cardiovascular risks.
Some common symptoms of long COVID linked with are weight loss, joint inflammation, dry mouth (xerostomia), chest, and muscle pain. High-grade fever can stay for a while, but normally it subsides after about 60 days. Due to its general nature, this characteristic of pervasive fatigue has been subjected to a lot of attention in most clinical research studies as its manifestation tends to negatively affect levels of motivation and faculties for concentration. Research such as Crook et al. (2021) and Das et al. (2022) has shown that patients suffering from this kind of infection, admitted and non-admitted alike, within seventy-nine days from the onset of the acute infection, have shown prolonged episodes of fatigue, which is an indicator for diseases related to neuropsychiatric illnesses that resemble myalgia encephalomyelitis/chronic fatigue syndrome (ME/CFS). Overlapping syndromes include pain treatment and movement skills difficulties, as well as management techniques.
In short, the wide variety of the Long COVID symptoms, which are way beyond the initial virus infection, affect whole-body systems and hence significant health challenges, physiologically and physically.
4.2. Hypothesis on Long COVID Cause
There have been multiple hypotheses put forward to account for the factors that cause Long COVID, as below:
4.2.1. Viral Presistence
This hypothesis is that some remains of the virus, like viral RNA, stay in the body for quite some time after the acute infection is over. An example of it is that the viral material keeps on stimulating the immune system, thus causing continuous triggering of inflammation and persistent damage to tissues. These receptors, which are responsible for sensing immune responses, upon continuous triggering of the viral particles, might mediate chronic immune activation throughout the tissues. The ongoing presence of viral RNA and the immune response generated give ample reason for the prolongation of symptoms and damage to the tissues.
4.2.2. Immune Dysregulation
This hypothesis suggests that the continual activation of immunological cells impairs responses, and eventually, immunity is weakened because the normal operation of the immune system is a necessity for it to function properly. T-cells, which are white blood cells responsible for immune functions, might become exhausted with constant triggering and become inefficient in their responses.
4.2.3. Latent Virus Reactivation
According to this hypothesis, Long COVID could trigger the reactivation of dormant viruses - infections that have remained inactive in the body. This is because stress on our immune system can cause viruses like Epstein-Barr virus (EBV) and herpes simplex virus (HSV) to become active again. When this happens, it can result in more symptoms and complications which intensify the impact of Long COVID.
4.2.4. Autoimmunity
In autoimmunity, the immune system uses autoantibodies to target the body's cells, mistakenly recognizing them as invaders, leading to inflammation and a tissue dilemma. This autoimmune response can manifest as persistent symptoms of joint pain, fatigue, and organ dysfunction contributing to the long-term effects of long COVID.
4.2.5. Microclots
This is the theory behind the formation of microthrombi in the vasculature that can block small blood vessels, leading to less oxygenation and delivery of nutrients to tissues. Incongruous blood delivery can result in the presentation of symptoms like fatigue, shortness of breath, and pain, thus some of the ongoing features associated with Long-COVID, and maybe will have more important sequelae associated with it.
4.2.6. Dysfunctional Neurological Signaling
Neurological dysfunction results from an injury, whether intentional or unintentional, in the nervous system. Communication of the signaling of nerves can be tampered with by the destruction of myelin, the protective cover on the fibers. Such dysfunction may happen and present with cognitive problems such as brain fog, memory problems, and a lack of concentration—the most common significant symptoms of Long COVID.
4.2.3. Disruption of the Microbiome
The final hypothesis is that the disruption of the microbiome leads to an inflammatory response and weakens mucosal barriers predisposing patients to infections and adaptation of inflammation. An imbalanced microbiome impairs the state of health and can be contribute of the various symptoms and conditions seen with Long COVID.
4.3. Key Genes & Pathways of Long COVID
4.3.1. Pathways in Long COVID
In our study, we identified several genes and symptoms associated with long COVID that follow the WPG (Gq/11) Protein Signaling Pathways and G-protein mediated events pathway. These pathways involve signaling cascades mediated by the Gq/11 protein family, which activate phospholipase C (PLC) upon stimulation by G-protein coupled receptors (GPCRs), leading to the production of second messengers such as IP3 (inositol trisphosphate) and DAG (diacylglycerol), initiating downstream signaling events.
4.3.1.1. WPG (Gq/11) Protein Signaling Pathways
The WPG (Gq/11) protein signaling pathways encompass a complex network of interactions involving G proteins, second messengers, and various downstream effectors, playing a pivotal role in regulating vital cellular processes. These pathways primarily involve the activation of specific G proteins (GNA11 and GNAQ) through G Protein-Coupled Receptors (GPCRs). Upon ligand binding, these Gα subunits exchange GDP for GTP, dissociating from the Gβγ dimer to activate downstream effectors. Key components include the activation of Phospholipase C-β (PLC-β), where GNA11 and GNAQ hydrolyze PIP2 into IP3 and DAG. IP3 triggers Ca²⁺ release from the endoplasmic reticulum, while DAG activates Protein Kinase C (PKC). This calcium signaling cascade leads to various cellular responses, such as muscle contraction and secretion. Additionally, PKC activation by DAG and Ca²⁺ results in the phosphorylation of target proteins, influencing gene expression, cell proliferation, and differentiation.
The MAPK/ERK pathway, involving small GTPases like HRAS, KRAS, and NRAS, can also be activated by PKC, promoting cell proliferation and differentiation. Moreover, the cAMP/PKA pathway, including PRKACA and PRKAR1A, intersects with Gq signaling, highlighting the intricate cross-talk between these pathways. Specific proteins such as HRAS, KRAS, NRAS in the MAPK/ERK pathway, GNA11, GNAQ as Gq alpha subunits, PRKACA, and PRKAR1A in cAMP-dependent PKA signaling, and PRKCG and PRKCQ as PKC isoforms, are crucial in regulating various physiological processes.
The detailed examination of long COVID symptoms reveals their potential connections to the WPG (Gq/11) Protein Signaling Pathways, as outlined in table 9. Symptoms such as anaphylactic shock, anti-thyroid peroxidase antibody positivity, alopecia, fragile nails, gastroesophageal reflux, sleep apnea, edema, back pain, fatigue, abdominal pain, anorexia, ocular pain, venous thrombosis, pulmonary embolism, irregular menstruation, mania, memory impairment, elevated TSH concentration, hypofibrinogenemia, and menorrhagia, along with conditions like hypertension, cardiac arrhythmias, diabetes mellitus, osteoporosis, dermatitis, asthma, chronic obstructive pulmonary disease (COPD), psoriasis, infertility, and depression, highlight the broad spectrum of health issues potentially linked to GPCR signaling, particularly those involving Gq/11 proteins.
Key genes and proteins implicated in these pathways, including GNA11, GNAQ, HRAS, KRAS, NRAS, PRKCG, PRKCQ, and PRKACA, play roles in immune responses, inflammation, hormone regulation, neurotransmitter signaling, vascular function, smooth muscle contraction, glucose metabolism, bone metabolism, skin inflammation, bronchoconstriction, and mood regulation. For instance, GPCR signaling through Gq/11 proteins regulates vascular smooth muscle tone and heart function in hypertension and cardiac arrhythmias. In diabetes mellitus, G protein signaling affects insulin secretion and action, while Gq/11 signaling influences bone resorption and formation in osteoporosis. Inflammatory skin conditions involve GPCR signaling through PKC isoforms and RAS genes, and Gq/11 signaling in airway smooth muscle cells regulates bronchoconstriction and inflammatory responses in asthma and COPD. Additionally, hormonal regulation of reproductive processes through GPCR signaling pathways can impact infertility, and GPCR pathways play crucial roles in neurotransmitter signaling related to mood regulation in depression.
The involvement of WPG (Gq/11) protein signaling pathways in such a diverse array of diseases underscores their critical role in maintaining physiological homeostasis and their potential as targets for therapeutic interventions. Understanding these connections provides valuable insights into diagnosing and targeting treatments for conditions involving WPG protein signaling pathways and the associated genes. Future research should aim to elucidate further the molecular mechanisms underpinning these pathways and their role in disease progression, particularly in the context of long COVID. This understanding could pave the way for developing novel therapeutic strategies aimed at modulating these signaling pathways to treat or mitigate the symptoms of long COVID and other related conditions.
4.3.1.2. G-Protein Mediated Events Pathway
The G-protein mediated events pathway involves key proteins GNA11 and GNAQ, which are activated by GPCRs and subsequently activate phospholipase C-beta (PLCβ). PLCβ hydrolyzes PIP2 into IP3 and DAG, triggering calcium release and activating protein kinase C (PKC). The released calcium ions, through calmodulin, activate calcium/calmodulin-dependent protein kinase II (CaMKII), encoded by CAMK2A and CAMK2B. These kinases phosphorylate various substrates involved in cellular responses. Protein kinase A (PKA), composed of catalytic (PRKACA) and regulatory (PRKAR1A) subunits, is activated by cAMP, which can be influenced by GPCR signaling. PKA phosphorylates target proteins, affecting metabolic pathways and gene expression. Protein kinase C gamma (PKCγ), encoded by PRKCG, is activated by DAG and calcium, playing crucial roles in neuronal signaling and cell growth. This complex pathway begins with GPCR activation of GNA11/GNAQ, leading to a cascade of events including calcium release, activation of various kinases (CaMKII, PKA, PKC), and ultimately resulting in diverse cellular responses through substrate phosphorylation. This intricate network of interactions underscores the complexity of G-protein mediated cellular signaling and its response to external stimuli. The genes involved in this pathway (GNA11, GNAQ, CAMK2A, CAMK2B, PRKACA, PRKAR1A, and PRKCG) are linked to various symptoms through specific physiological and pathological mechanisms. For instance, these genes influence hair follicle development (alopecia), pain perception (ocular pain and general pain), vascular function (venous thrombosis and pulmonary embolism), gastrointestinal motility (gastroesophageal reflux), neurotransmitter signaling (abnormal emotion/affect behavior, sleep disturbance, mania, and memory impairment), muscle control (dystonia and gait disturbance), hormonal regulation (irregular menstruation, elevated TSH, and menorrhagia), and blood clotting (hypofibrinogenemia). Understanding these connections helps elucidate the complex mechanisms underlying diverse symptoms, ranging from sensory disturbances to hormonal and cognitive dysfunctions, and highlights the far-reaching impact of G-protein-mediated signaling on human physiology and pathology.
4.4. Pathways & Their Identified Key Genes in Long COVID
4.4.1. G-Protein Signaling
G protein signaling pathways embody the cellular response to a plethora of extracellular signals, and some of them include GNAQ, GNA11, GNAO1, and GNAZ, coding for different alpha subunits of the G protein. They all contribute to the signaling relayed by the G protein-coupled receptors. Stimulation of G protein-coupled receptors mediates a variety of physiological processes that include immune response and inflammation. Aberrant G protein activity within this signaling pathway is known to cause abnormalities and be accountable for sustained inflammation and immune dysregulation, typical of Long COVID. Altered GPCR operation could impair the body's capability to regulate inflammation properly and could allow for extended and worsened symptoms particular to long COVID, such as chronic fatigue and joint pain.
4.4.2. RAS-MAPK
The downstream pathway from the RAS-MAPK pathway involves major players in the uptake of cellular responses to growth signals and inflammatory signals. The proteins of the HRAS and KRAS gene products respectively, through their activation of the MAPK pathway, of course play their role in mediating an inflammatory response. Aberrant activation of the MAPK pathway, particularly through mutation in HRAS or KRAS, is accompanied by a persistent inflammatory response in Long COVID. This aberrant activation may sustain a chronic inflammatory state, furthering the long-term symptoms of Long COVID, such as muscle pain and chronic fatigue. The inflammation that the continuously dysregulated signaling would drive would thus put an additional load on the body over time and could possibly also complicate the recovery of the overall disease burden.
4.4.3. Protein Kinase
Protein kinases are a group of enzymes that add phosphate groups to proteins, a process central to the control of many cellular processes. PRKCB, PRKCG, PRKACA, and PRKACG encode different protein kinases, each involved in various regulatory functions. Gene mutations may lead to abnormal protein kinase activity and hence disrupt normal cellular processes. This will then manifest as various symptoms seen during long COVID, such as incapacitating chronic fatigue, muscle pain, or cognitive impairments. Altered kinase activity acts at the level of cellular signaling and homeostasis to possibly modulate the persistence of Long COVID symptoms and their severity.
4.4.3. Calcium/Calmodulin-Dependent Protein Kinase (CaMK) Pathway
Specifically, the CaMK pathway, more precisely the CaMKII isoforms encoded by CAMK2A and CAMK2B genes, play critical roles in neuronal functions and synaptic plasticity. Although the direct link between CaMKII activity and Long COVID is less well established, abnormalities in CaMKII function could underlie neurological symptoms commonly reported by Long COVID patients, such as brain fog and cognitive impairments. Disruptions in CaMKII activity could thus misregulate neuronal signaling to impact cognitive function in a manner that underlies some of the neurological manifestations seen in Long COVID. Knowing the role of CaMKII in such a context could help in the elucidation of mechanisms underlying these persistent neurological symptoms.
4.5. Association of the Key Genes with Long Covid
4.5.1. HRAS
HRAS is a gene that encodes a member of the Ras family of GTPases, which plays a critical role in the transmission of signals that promote cell growth, differentiation, and survival. Despite the attention of a plethora of scientific articles having been focused on HRAS in relation to cancer, recent works have implicated this gene's significant contribution to COVID-19 and its long consequence, known as long COVID.
In the context of COVID-19, HRAS appears to perform a crucial role in both viral entry and viral replication steps. One would thus ensure the modulation of cellular signaling pathways interacting with the ACE2 receptors, one of the prime entry routes for SARS-CoV-2 into the human cell system. Such an alteration in the HRAS-regulated pathway would not only affect signaling but would also subsequently modulate the level of viral entry and replication. In addition, HRAS is a critical factor involved in immune response regulation, and its dysfunction could be one of the regulators of immune system failure. This dysregulation is often seen in severe COVID-19 cases characterized by hyperinflammation and cytokine storms, leading to severe complications such as ARDS and multiple organ failure.
In the context of long COVID, HRAS mediates its action through chronic inflammation and tissue remodeling. The sustained activation of HRAS would continue to drive chronic inflammation, with continuous tissue injury that might eventually manifest throughout the body to give rise to the symptomatic smorgasbord of long COVID. Furthermore, fibrosis and tissue-remodeling pathways of HRAS signal aberrations, which may lead to problematic fibrosis, particularly in the lungs, with an attendant perpetuity of respiratory issues related to reduced organ functionality. Last but not the least, considering the involvement of HRAS protein in the functioning as well as the survival of neural cells, it is going to affect the outcome of neurological symptoms, which have been recorded quite frequently in the cases of long COVID, therefore including brain fogging and memory problems associated with disrupted cellular activity in the neural tissues.
4.5.2. KRAS
This gene belongs to the RAS oncogene family; its protein product plays a critical role in cell signaling regulating proliferation, differentiation, and survival. Somewhat like HRAS, mutations in KRAS are also related to different kinds of cancers, underscoring its role in pathologies.
KRAS, in the case of COVID-19, has a bearing on cell entry and the replication of the virus. Genetically, it is not directly linked to the ACE2 receptor, but it can alter the cellular environment through its role in regulating the cell cycle and apoptosis, thus impacting the effectiveness of entry or replication of SARS-CoV-2. It also affects immune signaling through dysregulated pathways of KRAS and enhances inflammatory responses, leading to serious COVID-19 symptoms and systemic problems such as cytokine storms.
Meanwhile in Long COVID, sustained or dysregulated KRAS activity could maintain—or even enhance—post-infection inflammatory responses. Tight linking of continued activation with chronic inflammation, previously designated as one of the hallmarks of long COVID, may fuel the aggravation of long-term health sequelae concerning multiple organ systems. KRAS is also involved in fibrosis and tissue remodeling; participation in the latter process, for example, in tissues like the lungs, might drive excessive fibrosis, preventing recovery and organ function due to abnormal activity. Implicating the influence of KRAS on neural cell survival and proliferation, its role in neurological complications from long COVID is neurologically realized. The effects of such dysregulated KRAS signaling on neural pathways and brain heath could drive the cognitive impairments and other neurological symptoms reported in some patients. This underscores the diffuse role of KRAS in both acute and prolonged COVID-19 outcomes.
4.5.3. GNAQ
GNAQ encodes a protein belonging to the Gq class of G proteins that play a critically essential part in different physiologic functions, such as maintaining blood vessel tone, modulating nerve cell activities, and governing inflammatory responses.
In the context of COVID-19, the activities of GNAQ propel cellular responses to which entry and replication of the virus may be able to respond. Although GNAQ does not directly associate with ACE2, the primary entry receptor for SARS-CoV-2, its general role in GPCR signalling may modulate the cell environment in ways that positively or negatively impact SARS-CoV-2's life processes in the host cell. Furthermore, GNAQ modulation of immune-cell signalling is key to eliciting a bodily response to SARS-CoV-2.
It may be that an immune response is either inadequate or overly aggressive in these GNAQ-mediated pathways, thereby affecting the symptoms and severity of COVID-19. Sustained or aberrant GNAQ activity may well drive the continued inflammation of long COVID, maintaining a chronic state of inflammation that interferes with normal tissue function and propagates prolonged symptomatology. In addition, as GNAQ exerts key effects on vascular signalling pathways, it can further lead to vascular anomalies in the course of long COVID, including micro-clotting. These vascular disturbances may make very serious contributions towards long-term organ health and functionality, thus underlining GNAQ's critical role in acute and prolonged features of COVID-19.
4.5.4. GNA11
GNA11 is involved in the same signaling pathways as GNAQ, those activated by GPCRs, although through somewhat different intracellular mechanisms. These are key pathways required for normal cellular responses against a huge variety of exogenous stimuli, including hormonal signals, light, and environmental cues that influence cell behavior.
In the case of COVID-19, though participation by GNA11 in GPCR-mediated signaling pathways is not directly related to the entry and replication of the virus, it performs some key regulatory functions that might indirectly impact the internal cellular environment, thereby influencing viral processes within a host. In addition, GNA11 is a critical regulator of immune responses. Its dysregulated activity gives rise to an underactive as well as overactive immune response, therefore favoring severe manifestations of COVID-19.
However, in Long COVID, probably long-term misassignment of the GNA11 signal contributes to continued inflammatory reactions typical of this condition, which influences recovery and prolongs symptoms. Besides, GNA11 also has effects on various systems of the body; hence it could be contributing to the multisystemic impact seen in Long COVID, such as effects on cardiovascular, neurological, and immunological functions outlining the wide, systemic role of GNA11 in the late processes of disease after acute COVID-19 infection.
4.6. Mechanisms of Long COVID Symptoms
The emergence of Long COVID symptoms is attributed to several linked biological mechanisms:
4.6.1. Chronic Inflammation
Long COVID is driven by persistent activation of inflammatory pathways. In the acute phase of COVID-19, pro-inflammatory cytokines are released, which the immune response uses to clear the virus; in some individuals, the viral urine is not swept up by this process and continues even after the virus has been cleared. High pro-inflammatory cytokines, predominantly IL-6, IL-1β, and TNF-α, sustain chronic inflammation (Kappelmann et al., 2021). Furthermore, chronic inflammation can cause cellular and tissue damage due to the compounded effects of elevated oxidative stress and diminished antioxidant defenses. (Al-Hakeim et al., 2022).
4.6.2. Immune Modulation
There is strong evidence attesting to a state of pronounced immune dysregulation in long COVID, usually characterized by either autoimmune-like responses or ineffective clearance of the virus. This includes changes in T-cell responses, an upsurge in the pro-inflammatory subtypes of T-cells, and indications of a possibility of exhausted T cells that are associated with ongoing immune activation and dysfunction (Yin et al., 2023). This often leads to the attacking of its host's own tissues and presents symptoms such as joint pains, fatigue, and other signs of autoimmunity.
4.6.3. Neurologic Effects
Common neurological symptoms that are coinstantaneous with Long COVID include brain fog, cognitive dysfunction, and fatigue. Some of these symptoms are ascribed to several mechanisms medically. More specifically, neuronal-based inflammation, especially through immune cells like microglia, is responsible for neuroinflammatory responses that disrupt regular functionalities of the brain (Leng et al., 2023). Besides, SARS-CoV-2 might disrupt the blood-brain barrier, leading to the entry of inflammatory cells and molecules in the brain to bring about destruction (Shabani et al., 2023).
4.6.4. Vascular Dysfunction
Another major manifestation of Long COVID involves vascular issues causing complaints like fatigue, cognitive dysfunction, and cardiovascular complications. The endothelial cells lining the blood vessels may be damaged by the SARS-CoV-2 strain, resulting in impairment of vascular function and increased risks in forming clots (Shabani et al., 2023). Microclot formation may also blockade the capillaries, diminishing blood flow and, by extension, oxygen supply to tissues, therefore causing organ failure and long-lasting symptoms (Leng et al., 2023).
4.6.5. Metabolic Disturbances
Metabolic dysfunction is also common in Long COVID states, including mechanisms for both energy production and use. Dysfunction of the mitochondria is often seen in SARS-CoV-2 infection. Impairments in mitochondrial function lead to dysfunctional energy production, resulting in fatigue. This will, in turn, necessitate a compensatory increase in inefficient glycolysis, supporting fatigue and further metabolic symptoms in a vicious cycle. This might be due to vast quantities of dysfunctional mitochondria.
With that being said, the Predisposition through genetic variants and the expression of a specific subset of genes within specific tissues likely has a large hand in the various presentations of Long COVID symptoms. Host variation at immune response, inflammation, and cellular stress pathway genes likely drive the inter-individual differences in disease susceptibility and response that underlie the variation of Long COVID symptom severity or duration (Proal & VanElzakker, 2021). Meanwhile, viral infection can cause cellular epigenetic modifications, which, in return, will turn on changes in gene expression responsible for low-grade chronic inflammation with immune dysregulation (Low et al., 2020).
4.7. Potential Mechanism of Long COVID
Long COVID epitomizes continuous immune dysregulation and presents a complicated challenge to healthcare. It simply means the persistence of chronic inflammation and immune exhaustion well beyond the acute phase of SARS-CoV-2 infection. Regulatory T cells, important for setting a balance in immune responses, are influenced by transcription factors like FOXP3 and probably FOXP4. The dysregulation of Treg function may give way to chronic inflammation and autoimmune responses underlying pathology in long COVID.
Our in-depth analysis identified critical genes that included GNA11, GNAQ, CAMK2A, CAMK2B, PRKACA, PRKAR1A, and PRKCG. These genes are integral to different G-protein coupled receptor signaling pathways that may modulate the T-cell responses. Abnormal signaling via these pathways can be an important cause of immune dysregulation seen in long COVID. Thereafter, FOXP4 might regulate the expression of elements involved in these GPCR signaling pathways, thus impacting T-cell functionality and their responses to viral infections. One may hypothesize that such dysregulation could also occur in neurological functions, which may perhaps be one of the underlying mechanisms associated with the common long COVID symptoms of cognitive impairment and fatigue.
Newer studies put forward the role of chronic activation of innate immune cells and increased pro-inflammatory cytokine signaling, notably via the IL-6 and JAK-STAT pathways, besides disturbances in complement, coagulation, and metabolic pathways. All these pathways point to a systemic state of chronic inflammation and overactive immune system persisting after active infection. Recent studies have further identified sex differences in immune responses, with males exhibiting increased production of TGF-β signaling and decreased production of the DDX3Y gene, while females had increased expression of the XIST gene and increased IL-1 signaling. These differences underline the variability within the manifestations and severity of long COVID between sexes and therefore show a need for personalized therapeutic approaches (Aid et al., 2024; Yin et al., 2024).
Our study has further identified several key genes and pathways of T-cell regulation that likely play a role in the pathogensis of long COVID. There is the implication that targeted intervention strategies, especially those targeting IL-6 and complement pathways, will be potentially crucial in preventing or alleviating long COVID symptoms. Moreover, there are new opportunities for sex-specific, personalized treatment strategies ensuing from the identification of sex-specific immune responses (Hamlin et al., 2024).
This work will increase knowledge of the sophisticated immunological landscape in long COVID and even more so in enabling further research into the long-term consequences of SARS-CoV-2 infection. From these results, one can underline the priority that should be placed on translational research aimed at developing diagnostic tools and therapeutic approaches against common and sex-specific molecular processes. Taken together, future studies will be needed to further translate the findings at a molecular level into clinical applications with the potential to better support care and management in this entity of disease complexity.
4.8. GPCR Signaling Pathway in Long COVID
Proposed Hypothesis and Involvement of GPCR Signaling Pathways
The WPG (Gq/11) Protein Signaling Pathways and G-protein mediated events pathway play crucial roles in various cellular processes and are intricately linked to several hypotheses proposed for long COVID. These pathways, involving key genes such as GNA11, GNAQ, CAMK2A, CAMK2B, PRKACA, PRKAR1A, and PRKCG, are implicated in multiple mechanisms that may contribute to the diverse symptomatology of long COVID.
In the context of viral persistence, persistent activation of GPCR signaling pathways, particularly through Gq/11 proteins (GNA11 and GNAQ), can sustain cellular responses that promote viral persistence. This prolonged signaling may contribute to the ongoing presence of the virus within host cells. Immune dysregulation, another key hypothesis in long COVID, can be influenced by dysregulated GPCR signaling affecting immune cell activation and cytokine production. The involvement of GNA11, GNAQ, and downstream effectors like PKC and MAPK pathways in modulating immune cell function and cytokine release underscores the potential role of these pathways in prolonged inflammatory responses.
Latent virus reactivation, a proposed mechanism in long COVID, may be triggered by fluctuations in GPCR signaling pathways. These pathways, involving GNA11, GNAQ, and associated downstream effectors, could influence cellular states conducive to viral reactivation. Additionally, dysregulated GPCR signaling pathways may contribute to autoimmunity by leading to immune tolerance breakdown and autoantibody production. Proteins like PKC and PKA, activated downstream of GNA11 and GNAQ, can affect immune cell signaling and potentially contribute to autoimmune responses observed in some long COVID patients.
The formation of microclots, another hypothesis in long COVID pathogenesis, may be influenced by GPCR signaling, including calcium signaling via IP3 and DAG pathways. These signaling cascades, activated by PLCβ and involving DAG-mediated PKC activation, can affect vascular function and potentially contribute to thrombotic events. Neurological symptoms in long COVID may be related to dysfunctional neurological signaling, where GPCR signaling pathways, including those involving PKC and MAPK/ERK, play critical roles in neuronal signaling and synaptic plasticity. Proteins like CAMKII in the G-protein mediated events pathway can modulate neuronal function, potentially contributing to the neurological manifestations of long COVID.
Lastly, the disruption of the microbiome, another proposed factor in long COVID, may be influenced by GPCR signaling pathways involved in regulating gastrointestinal functions and microbiome homeostasis. Genes like GNA11, GNAQ, and downstream effectors such as PKA can influence gut motility, secretion, and immune responses that maintain microbiome balance.
In conclusion, the WPG (Gq/11) Protein Signaling Pathways and G-protein mediated events pathway are central to multiple cellular processes implicated in various hypotheses of long COVID. These pathways regulate immune responses, cellular signaling, vascular function, neurological processes, and microbiome homeostasis, potentially influencing the persistence of viral infections, immune dysregulation, autoimmune responses, microclot formation, neurological symptoms, and microbiome disruption observed in Long COVID patients. Further research into the molecular mechanisms involving these pathways and genes could provide valuable insights for developing targeted therapies and interventions to manage Long COVID more effectively.
4.8.1. GPCR Gene Interactions
Our results identified GPCR pathways, including MM15882, M26911, M39426, and MM14496, which were significantly associated with Long COVID genes. Their expression in immune cells modulates their activity concerning T cell activation, differentiation, and response to infections. This is instanced by genes, for example, GNA11 and GNAQ encoding G-protein subunits necessary for GPCR signaling and strongly influencing T-cell function. Furthermore, CAMK2A and CAMK2B encode Ca²⁺/calmodulin-dependent protein kinases that act as downstream effectors in GPCR signaling pathways, modulating T-cell signaling. Again, genes like PRKACA, PRKAR1A, and PRKCG—encoding subunits for PKA and PKC, respectively—engage in the activation of T-cells through GPCR routes and their regulation.
Cyclic adenosine monophosphate, or cAMP, is one important intracellular second messenger that arises from the binding of a variety of hormones, neurotransmitters, and other ligands to their cell-surface receptors. The molecule activates both the canonical PKA pathway and the non-canonical exchange protein activated by cyclic AMP (EPAC) pathway. High levels of endogenous cAMP in regulatory T cells are produced and stored; afterwards, this small molecule is transferred to neighboring target cells through gap junctions. These GJs are formed by the connexin proteins Cx31.1, Cx32, Cx43, Cx45, and Cx46, allowing for intercellular communication and whose expression is increased after Treg activation. The ecto-5′-nucleotidases CD39 and CD73 expressed on the surface of Tregs convert the extracellular ATP to adenosine. Adenosine binds to GPCRs on responding cells, activates the G protein alpha subunit Gsα, which in turns activates adenylyl cyclases to produce cAMP.
These complex interplays of GPCR signaling, T-cell regulation, and manufacture of cAMP need to be understood to define immune responses and mechanisms of diseases such as Long COVID. Their interaction gives insight into the mechanism of the underlying immune-related diseases and opens up targets for potential therapeutic intervention. Gene interactions can now be understood in the context of guiding targeted treatments and improving the overall understanding of the function and malfunctioning of an immune system.
4.8.2. Targeting GPCR Pathways
GPCRs have been in the frontal line of many physiological processes related to modulating immune responses, inflammation, and cell signaling. This makes them basic for the understanding of human biology but extremely needed drug discovery targets. GPCR signaling pathways present promising potential to be targeted for the development of therapeutics, especially in conditions characterized by dysregulated signaling or persistent viral-induced cellular stress.
GPCRs also had a huge impact in the context of immune modulation, especially in the regulation of immune cells and cytokine production. In particular, GNAQ, GNA11, and GNA13 genes are part of a family of G proteins involved in the control of the activity of immune cells and cytokine production. This pathway targeting is relevant to the maintenance of an effective immune response and management of inflammation, both relevant in Long COVID where there are problems with immune dysregulation. These GPCRs can thus be modulated by therapeutic interventions to rectify imbalances in the immune system and alleviate related symptoms.
Moreover, GPCR pathways have a main role in inflammatory processes. Indeed, numerous inflammatory responses are transduced by many proteins of the Protein Kinase C (PRKC) family, notably PRKCA, PRKCB, and PRKCG. Intervention in these kinases to manipulate their activity does open a basis for the treatment of chronic inflammation ensuing from diseases like Long COVID, ultimately helping to alleviate symptoms attributed to a sustained inflammatory response.
Additionally, GPCR-related pathways, directly implicated with the Ras family—HRAS and KRAS—are essential for cellular growth, differentiation, and survival. Many pathways, like these, might be able to reestablish cellular homeostasis while playing other roles in responding to viral infections like COVID-19. Further dysregulation at this point can result in continuous cellular stress typical of Long COVID. By targeting HRAS and KRAS, possible therapeutics could help attenuate symptoms arising from cellular dysregulation and stress, improving patient outcomes.
In this case, some key genes have so far emerged in Long COVID pathogenesis as critical therapeutic targets for their roles within cellular and physiological processes. For example, targeting GNAQ and GNA11, influencing vascular responses and neuronal function, may alleviate symptoms like ocular pain or neuropsychiatric problems. Moreover, protein kinases such as PRKCB, PRKCG, and PRKACA are involved in the regulation of inflammation and metabolism. Thus, these kinases represent very promising targets for therapies that have already been designed for addressing metabolic dysfunctions and inflammation in Long COVID-19.
This makes targeting specific GPCR pathways and their associated proteins a very promising avenue for the future development of therapeutics that would help manage Long COVID's complex symptoms, among other such diseases, underscoring the critical role GPCRs can play in drug discovery.
4.9. Foxp4 In Long COVID
Role of FoxP4 in Long COVID
FOXP4 belongs to the Forkhead box family of transcription factors. It is a central modulator of development, immune response, and disease pathogenesis. Recent studies have identified a single marker within the FOXP4 locus associated with long COVID, thus designating it as a required genetic factor for the etiology of this disease. Notably, at least the first genome-wide significant association for long COVID resided in the FOXP4 locus. The contribution of FOXP4 does not stop at COVID-19 severity but includes additional lung function and cancer, thus pointing out a general role for this factor in pathophysiological mechanisms representative of long COVID (Ledford, 2023)..
While severe COVID-19 has been established as a causal risk factor for long COVID, the genetic risk associated with the FOXP4 locus cannot be fully explained by its correlation with severe COVID-19 alone. As a transcriptional regulator, FOXP4 may alter the expression of key components in G-protein-coupled receptor signaling cascades. These include critical genes involved in GNA11, GNAQ, and PLCβ signaling, plus all their downstream effectors, like PKC, PKA, and MAPK/ERK pathways. Such manipulation of these pathways by FOXP4 could change cellular responses relevant to long COVID related to immune modulation, vascular functioning, and neurological signaling (Ledford, 2023; Luo et al., 2023).
FOXP4's involvement in immune cell differentiation and function suggests that it might interact with GPCR signaling pathways to influence immune responses in diseases characterized by immune dysregulation. This interaction could affect viral persistence or exacerbate autoimmune responses observed in some long COVID patients. Furthermore, FOXP4's regulatory effects on neuronal function and GPCR signaling pathways could contribute to neurological symptoms associated with long COVID, such as cognitive impairment, fatigue, and mood disorders. Additionally, FOXP4’s role in developmental processes and metabolic regulation may link to developmental abnormalities or metabolic dysregulation seen in long COVID, potentially through modulation of GPCR signaling components. Exploring these interactions could provide valuable insights into the complex pathophysiology of long COVID and lead to novel therapeutic approaches targeting these molecular mechanisms (Luo et al., 2023).
4.9.1. The Interplay between FoxP4, Tregs & GPCR Signalling
One critical point to make in the role of FOXP4 in long COVID is a potential interplay with regulatory T cells and GPCR signaling. FOXP4 is a member of the FOX family of transcription factors, placing it in processes from gene regulation related to immune responses. Although FOXP3 has been rather well known for its role in the development and function of Tregs, less information exists about FOXP4 in Tregs. FOXP4 may transcriptionally regulate other key immune response genes to control the function and differentiation of Tregs. G-protein coupled receptor activities modulate several immune cell functions, including T-cell activation and differentiation wherein some of the encoded key genes—GNA11, GNAQ, CAMK2A, CAMK2B, PRKACA, PRKAR1A, and PRKCG—are for proteins with a central role in T-cell activation and regulation. Although it is known that FOXP4 interacts with these GPCR signaling components, this interaction may have important implications for long COVID immune regulation. While its function, in comparison to FOXP3, in Tregs of crucial significance in the maintenance of immune tolerance and prevention of autoimmunity is not very well understood, FOXP4 could impact Treg function and differentiation through transcriptional regulation of genes involved in immune responses.
4.9.2. Crosstalk Between FoxP4 & Signaling Pathways
The involvement of FOXP4 in long COVID is highlighted by its interaction with G-protein signaling pathways, particularly GNAQ and GNA11. This reveals an additional level of participation, encoding another level related to long COVID. Genes controlling the G-protein signaling pathway, through which immune cells are activated and cytokines produced, include genes encoding ligands that interact with GPCRs, G-proteins themselves, and downstream effectors such as adenylate cyclase or phospholipase C. FOXP4 could be implicated in the regulation of expression of genes in these pathways, influencing downstream effects of HRAS, KRAS, GNAQ, and GNA11 (Lammi et al., 2023).
By contrast, somatic mutations in Ras and G-protein signaling pathways may, conversely, influence FOXP4's transcriptional activity by virtue of post-translational modifications or altered cellular localization. Taking this crosstalk into account, a network acting in a tightly regulated fashion places the FOXP4 transcription factor and these Ras and G-protein-related signaling molecules in the coordinately driven responses to infection and other stressors. This interplay might help explain potential therapeutic targets for diseases like long COVID with dysregulated immune responses. Specifically targeting certain nodes in these pathways could provide new approaches to the management of persistent symptoms of Long COVID. Further research into FOXP4 interactions with the GPCR and Ras signaling pathways will give further insights into how immune regulation can affect disease outcomes (Lammi et al., 2023).
4.10. Viral Protein Interactions
Viral Protein Interaction of COVID-19 in Human Genes
Interactions between several SARS-CoV-2 proteins and human genes are able to have deep implications for viral pathogenesis and clinical manifestation of COVID-19. Each of these viral proteins can interact with specific human proteins, potentially leading to cellular disruptions at a wide range of levels.
The SARS-CoV-2 E protein, the one involved in viral budding and pathogenesis, has been shown to interact with the HRAS gene. This gene is associated with continuing inflammation and abnormal cell growth because of disrupted G-protein signaling. Other G-proteins include GNAO1 and GNAZ. Disrupted links to these might give rise to neurological symptoms in the form of brain fog and changed sensations, reflecting its critical role in multiple cellular pathways.
The Orf1ab proteins of the virus, among others that include non-structural proteins, such as nsp3, nsp4, nsp7, nsp8, and nsp13, and RNA-dependent RNA polymerase, interact with cellular proteins modulating cell growth, KRAS and HRAS, immune function, and inflammatory responses. Those interactions perturb protein kinase C signaling pathways—for instance, PRKCA, PRKCG, PRKACB—thus blazing its role in altering cell division, inflammation, and immune responses.
Binding of the spike glycoprotein of SARS-CoV-2 to ACE2 receptors has been implicated in vascular dysfunction and multiple systemic alterations. Interacts with KRAS, GNAQ, GNAO1, PRKCB, and PRKAR1A; impacts G-protein and protein kinase signaling; leads to immune dysregulation and the prolonged прoвe-inflammatory response that affects a range of organ-based systems.This nucleocapsid protein interaction with KRAS could have implications for cell cycle regulation and apoptosis, as well as host cell gene expression.
Much less is known about functions for SARS-CoV-2 hypothetical proteins and other ORFs, but the interactions suggest modulators of cell-signaling pathways and normal innate immune responses and probably contribute to persistence within the host.
Several non-structural proteins that are of vital significance for the viral replication process result from processing of the excised and full-length polyproteins of the virus. These interactions perturb cellular signaling pathways extensively and, thus, disturb protein synthesis and processing while changing G-protein coupled receptor signaling, interfere with protein kinase C and RAS pathways. This impact can be vast in terms of cell growth, differentiation, and immune response among others, showing the critical involvement of the polyproteins in viral pathogenesis.
The 3C-like proteinase of SARS-CoV-2 plays a crucial role in the processing of its viral polyprotein. Interaction of this proteinase with KRAS can interfere with normal signaling pathways required during cell growth and differentiation. This may result in tissue damage or abnormal repair processes.
Lastly, the leader protein and Tor2 replicase 1AB of SARS-CoV-2 are known to play a role in viral replication, alongside the spike glycoprotein. In addition to potentially affecting host cell transcription, translation, and energy metabolism, this may lead to disruption of G-protein signaling as well as altering interactions with protein kinase C signaling at a molecular level - all impacting immune functions and neuronal signaling across multiple organ systems. The complexity of these effects highlights how COVID-19 influences disease progression while also presenting possible therapeutic approaches that target these mechanisms.
4.10.1. Detailed Analysis of Genes' Connections to COVID-19, Long COVID, and Related Conditions
The function of specific GPCR genes and their related signalling pathways could explain the effects in the context of COVID-19, long COVID, and related questions. The further insight gained from such investigation may provide therapeutic targets or interventions at a deeper level (
Table 9).
The RAS protein family, whose integrants include HRAS, KRAS, and NRAS, is deeply involved in the MAPK/ERK signaling pathway governing cell proliferation and survival. These pathways are presumably modulated during SARS-CoV-2 infection to enhance viral replication and persistence, hence increasing the possibility of survival within host cells. Moreover, this same RAS signaling axis plays a significant role in controlling cytokine production and immune cell function. The pathway dysregulation could be suppressive or over-expressing, which frequently is referred to as cytokine storm in severe cases of COVID-19, leading to wide-ranging tissue damage and inflammation. Stress-induced variation within the RAS signaling can also reactivate latent viruses such as herpesviruses. Furthermore, the possibility that the continuous activation of RAS pathways contributes to COVID-19–induced autoimmunity in relation to long COVID cannot be completely ruled out, besides exerting influence on endothelial function and blood coagulation, thus entertaining the possibility of promoting the formation of microclots and resulting in pronounced neurological symptoms such as brain fog and asthenic syndrome.
Moreover, G-protein subunits Gα, such as GNA11 and GNAQ, which are implicated in GPCR signaling, modulate immune responses and a wide variety of other cellular functions. Dysregulation of these pathways is likely required for viral entry and replication; evidence suggests that SARS-CoV-2 may exploit these mechanisms to help promote persistence in host cells. The dysregulation of GPCR signaling transforms into extreme inflammatory reactions and immune disorders typical of advanced COVID-19 infections. This would then reactivate dormant viruses, such as EBV, and probably trigger autoimmune responses that may also underlie the prolongation of symptoms in long COVID. It will affect vascular functions and neurotransmission, leading to cognitive impairments that will further add on to the formation of microclots.
Besides that, protein kinases A and C are involved in regulating various steps of the viral life cycle, including replication and egress. The alterations mediated in the signaling due to PKA and PKC could result in an exaggerated inflammatory response and might contribute to the cytokine storm observed in severe immune-dysregulated critical COVID-19 cases. In this way, stress- and inflammation-induced activation of PKC could also support the reactivation of latent viruses, thus further complicating clinical manifestations of COVID-19. Furthermore, the autoimmune phenomena developing due to the dysfunctional PKA and PKC signaling can impact platelet function and blood coagulation very strongly, thus predisposing to thrombotic complications.
Finally, CAMK2A and CAMK2B calcium/calmodulin-dependent protein kinase II subunits are involved in controlling calcium signaling that significantly influences various phases of the viral life cycle. Calcium signaling dysregulation would increase the persistence of viruses and their immuno-modulatory effects as a result of the formation of improper inflammatory responses, which, in turn, could lead to latent viruses reactivation of viruses associated with long COVID symptoms. Disruption of CAMK2 signaling may further contribute to autoimmune disorders and neurological symptoms characterized by severe cognitive deficiencies and brain fog.
In summary, the highlighted genes and their pathways are centrally involved in the immune response, viral persistence, autoimmunity, microclot formation, neurological function, and the composition of the microbiome. The dysregulation of these pathways underlies both the acute and long-term effects of COVID-19, emphasizing the need for these pathways as potential therapeutic targets. Such relationships can provide meaningful insight into the development of effective treatments against COVID-19 and management during long COVID, thus underpinning the complexity of COVID-19 as a multisystemic disease requiring comprehensive approaches in research and the development of therapeutics.
4.10.2. Potential Therapeutic Targets for Long COVID
Long COVID is a highly challenging situation due to its persistent dysimmunoregulation and chronic inflammation. Therefore, therapeutic strategies hold the key to targeting molecular pathways involved in the condition. Very important here is that 21 putative genes specific for GPCR that are relevant to symptomatology in Long COVID could raise avenues for targeted therapies that might alter cellular processes and alleviate symptoms. For example, Tipifarnib and Sotorasib can be used to target HRAS and KRAS pathways involved in RAS/MAPK signaling, respectively, which will help modulate immune responses and enhance tissue repair mechanisms (Lee, 2022; Odeniyide et al., 2022). Furthermore, genes related to various G protein subunits, such as GNAO1, GNAQ, and GNAZ, could be targeted by specific inhibitors for the correction of disrupted signaling pathways with an improvement in neurological and inflammatory symptoms.
Other therapeutic potentials were found in genes like GNAT3, the modification of which would enhance the quality of life and nutritional input of the affected patients. Small molecule inhibitors like YM-254890 can be utilised to modulate over-activated immune responses through G protein subunit genes like GNA11, GNA13, GNA14, and GNA15, thereby inhibiting chronic inflammation (Ma et al., 2021). This could also include protein kinase C isoforms, PRKCA, PRKCB, and PRKCG; regulators of the cAMP pathway, PRKACA and PRKACB—both putative enhancers of cellular resilience, therefore mitigating symptoms of metabolic dysregulation such as fatigue and muscle weakness. Hormonal balance and cognitive functions are modulated by PRKAR1A and PRKAR1B regulatory subunits.
Overrepresentation of calcium/calmodulin-dependent protein kinases CAMK2A, CAMK2B, CAMK2G, and CAMK2D that mediate memory and synaptic strength could also contribute to neural restitution and cognitive enhancement. Targeting upstream kinase CAMKK1 as part of an energy conservation strategy in neurons could assuredly produce symptomatic relief in most neurodegenerative diseases that manifest themselves as discomfort.
Moreover, Certain drugs previously approved by the FDA have recently shown potential. For instance, verteporfin and selumetinib target GNAQ and are used in a range of medical conditions, thus showing potential to alleviate a number of categories of Long COVID symptoms. On the other hand, GNA11 targets kinase inhibitors like binimetinib and cabozantinib, which are effective against dermatological and ocular symptoms. This makes the EGFR pathway-targeted drugs, panitumumab and cetuximab, of great utility in keeping immune autoimmunity at bay and controlling multisystem diseases symptomatically, by virtue of their high immunomodulatory effects.
Additionally, naltrexone, an extremely broad-spectrum opioid receptor blocker—well known for playing an anti-inflammatory and immune-regulatory role—is also considered for the treatment of Long COVID. Developed originally for opioid and alcohol addiction, LDN had brightly shown promise in the lowering of proinflammatory cytokines and modulation of immune function—the most relevant aspects underlying the management of chronic symptoms during Long COVID (Srivastava & Gold, 2018).
These therapeutic targets and drug candidates collectively hold out hope to address the complex pathophysiology of Long COVID. Each of these genes and pathways provides opportunities for drug, gene therapy, and biologics—such as monoclonal antibody—development against or to modulate activity and alleviate symptoms. However, these are approaches that need thorough clinical testing for safety and should only be advanced in an individualized approach considering a patient's total genetic background and clinical presentation. Hence, the strategy targeting therapies is underlined in the management of Long COVID, attempting to take outcomes in patients to a better level by addressing disruptions at the molecular level.