Submitted:
08 August 2024
Posted:
12 August 2024
You are already at the latest version
Abstract
Keywords:
1. Introduction
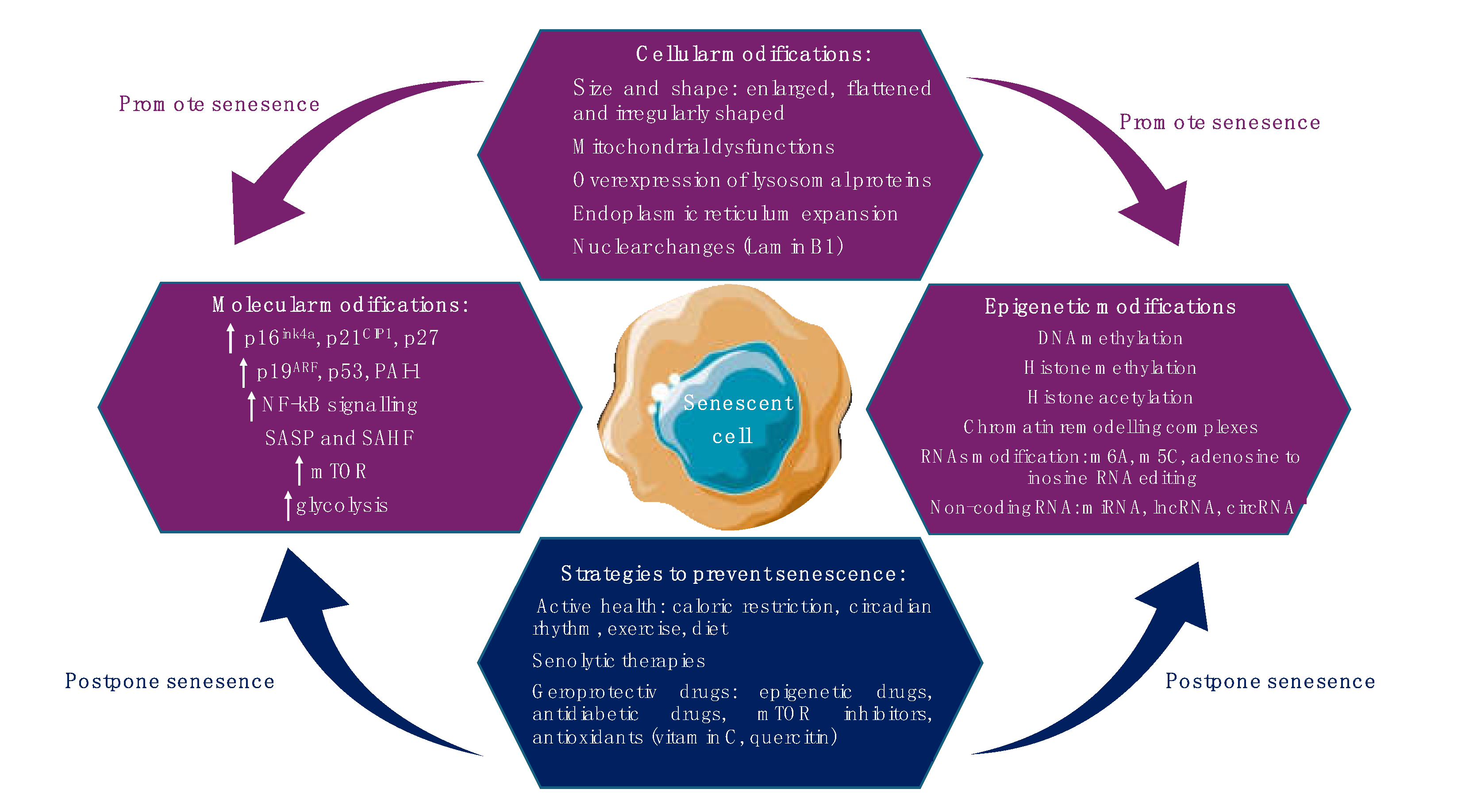
2. Cellular Senescence: Mechanisms and Pathways
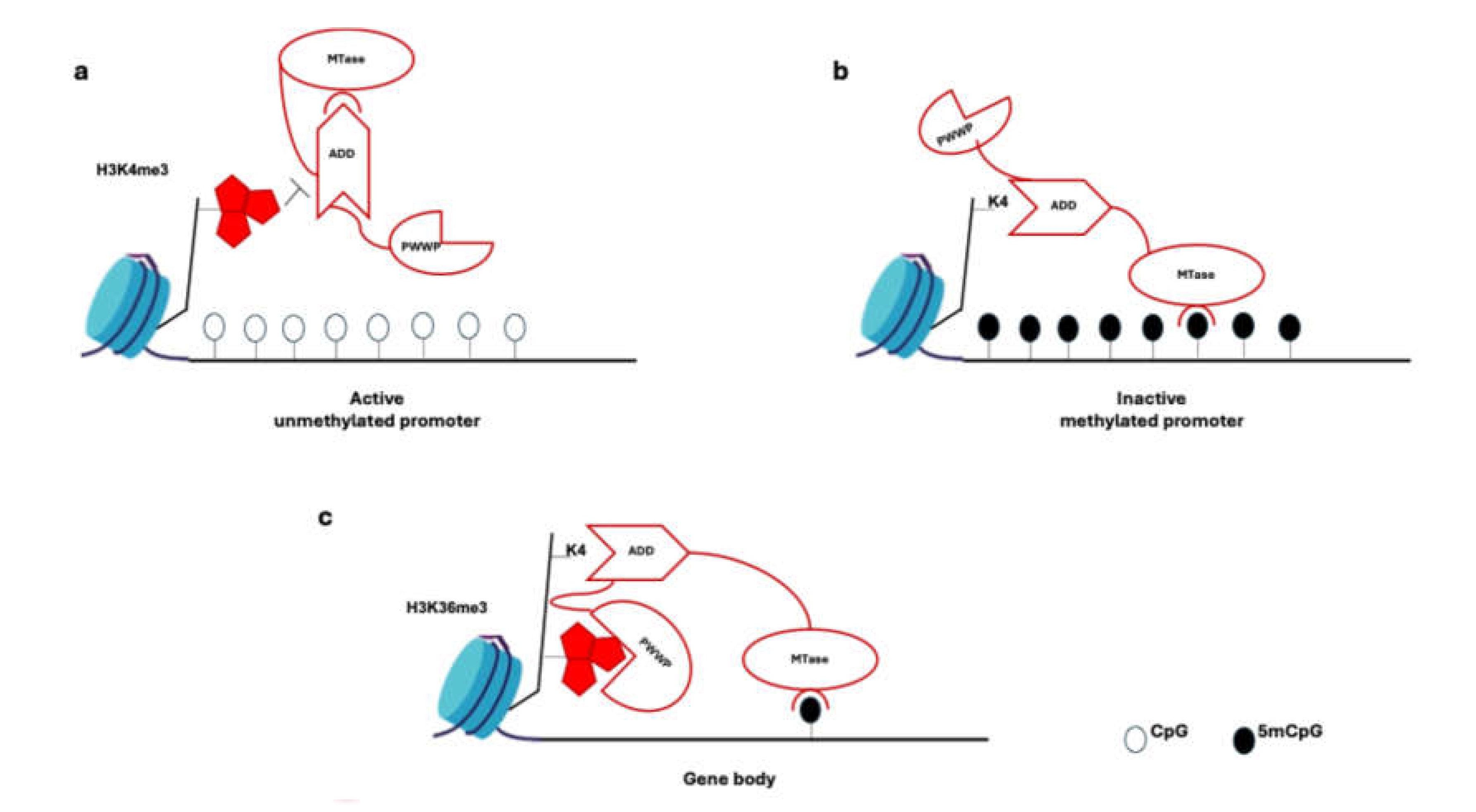
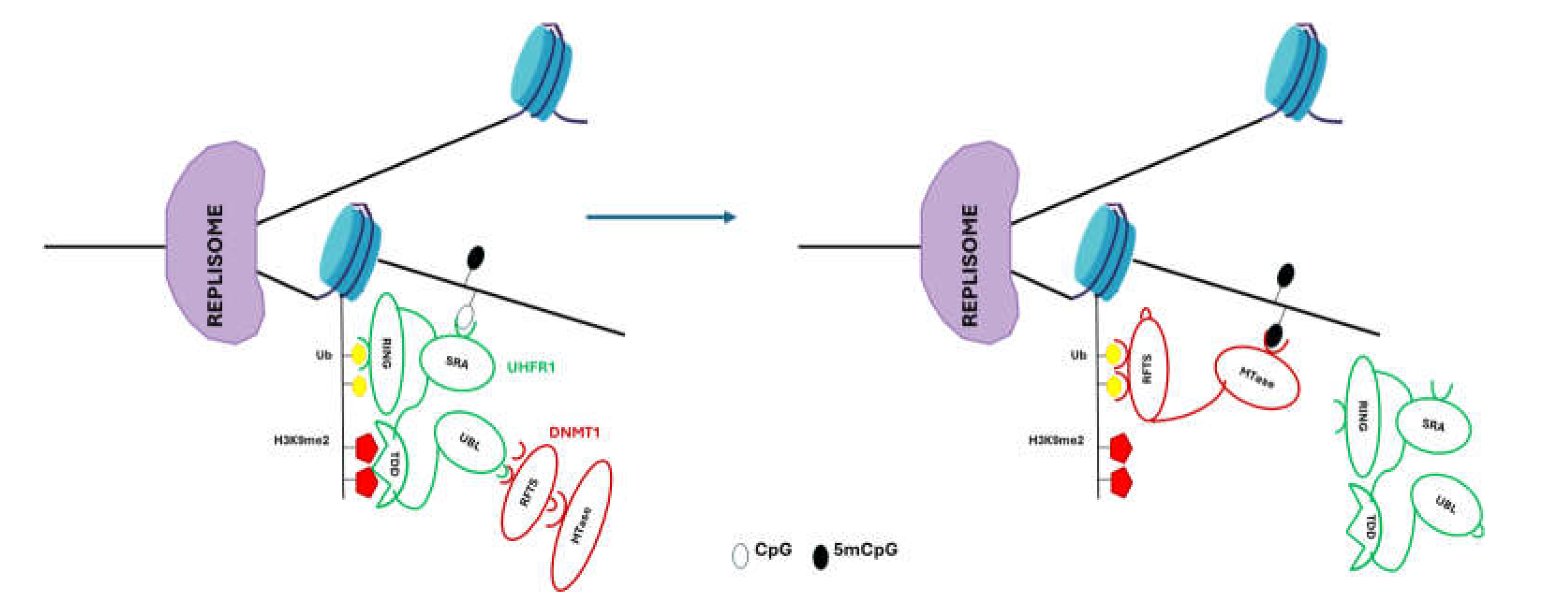
3. DNA Methylation Involved in Stem Cell Senescence
4. Histone Modification and Chromatin Remodeling Complexes Involved in Stem Cell Senescence
4.1. Histone Methylation
4.2. Histone Acetylation/Deacetylation
4.3. Histone Phosphorylation and Ubiquitination
4.4. Chromatin Remodeling Complexes
| Epigenetic modification | Substrate | Enzymes/target | Senescence |
|---|---|---|---|
| DNA methylation DNA demethylation |
CpG sites | DNMT3A, DNMT3B, DMNT1 TET1, TET2, TET3 |
↑ ↓ |
| Histone methylation | H3K4me3 H3K9m2/3 H3K23me3 H3K27me3 H3K27me3 H3K36me2/3 H3R2/17/26me |
KDM5A, ASH1L JMJD-1.2/PHF8, JMJD-3.1/JMJD3 JMJD-1.2/PHF8, JMJD-3.1/JMJD3 JMJD-1.2/PHF8, JMJD-3.1/JMJD3 EZH2 SETD2 CARM1data |
↑ ↓ ↓ ↓ ↑ ↑ ↑ |
| Histone acetylation | H3K9ac H3K56ac K3K27ac H3K14ac H3K9ac H3K9ac |
SIRT6 SIRT6 HDAC4 KAT7 GCN5 (KAT2A) PCAF (KAT2B) |
↓ ↓ ↑ ↑ ↑ ↑ |
| Histone phosphorylation | H3S10p H3T11p H3S28p γH2AX |
Aurora kinase CK2 in Drosophila cells ATM, ATR |
↑ ↑ ↓ ↑ |
| Histone ubiquitination | H2Bub H2Aub |
BRCA1/BARD1 E3 ligase BRCA1/BARD1 E3 ligase |
↑ ↑ |
| Chromatin remodeling complex | SWI/SNF NuRD |
ARID1B/ENTPD7 HDAC1 |
↑ ↑ |
| RNA modifications | m6A m5C A-to-I RNA editing |
methyltransferases: METTL3/14/16, RBM15/15B, ZC3H3, VIRMA, CBLL1, WTAP, and KIAA1429 demethylases: FTO and ALKBH5 tRNA methyltransferase NSUN2 ADAR1, ADAR2 |
↑ ↑ ↑ ↓ |
| miRNA | mir17 family miR-195 miR-486-5p, miR-204 miR-495 miR-141-3p, miR-543, miR-590-3p miR-543, miR-590-3p miR-141 miR-129 miR-188 miR-21 miR-146-5p miR-34a miR-106b family, miR-130b, miR-302a, miR-302b, miR-302c, miR-302d, miR-512-3p, and miR-515-3p |
p12 SIRT1, TERT BMI1 ZMPSTE24 AIMP3/p18 SVCT2, SDF-1 Frizzled-4 MAP3K3 E2F2 TNFα p53 p21 |
↓ ↓ ↑ ↑ ↑ ↓ ↑ ↑ ↑ ↑ ↑ ↑ ↓ |
| LncRNA | NEAT1 APTR LncHSC-1, LncHSC-2 ANRIL GAS5 GUARDIN H19 HCP5 |
CSF1 Dnmt3a p53 -p21 pathway NAMPT, PI3K/AKT pathway PGC1α and LRP130 p16 -p21 pathway, miR-22 miR-128 |
↑ ↓ ↓ ↓ ↑ ↑ ↓ ↑ |
| circRNA | circRNA-0077930 circ-Foxo3 circLARP4 circPVT1 circCCNB1 circACTA2 |
KRAS, p21, p53, p16, miR-622 ID-1, E2F1, FAK, HIF1α miR-761/RUNX3 axis let-7 CCNE2, miR-449a ILF3, CDK4 |
↑ ↑ ↑ ↓ ↓ ↑ |
| ▬ methyltransferase ▬ demethylase ▬ histone deacetylase ▬ histone acetyltransferase | |||
5. RNAs Modification Involved in Stem Cell Senescence
- “writers” m6A methyltransferases: METTL3/14/16, RBM15/15B, ZC3H3, VIRMA, CBLL1, WTAP, and KIAA1429;
- “erasers” demethylases: FTO and ALKBH5;
- “readers” m6A-binding proteins YTHDF1/2/3, YTHDC1/2 IGF2BP1/2/3 and HNRNPA2B1.
5.3. Adenosine to Inosine (A-to-I) RNA Editing
5.4. Non-Coding RNAs
5.4.1. miRNAs
5.4.2. LncRNAs
6. Animal Models of Senescence Research
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Calcinotto, A.; Kohli, J.; Zagato, E.; Pellegrini, L.; Demaria, M.; Alimonti, A. Cellular Senescence: Aging, Cancer, and Injury. Physiol Rev. 2019, 99, 1047–1078. [Google Scholar] [CrossRef] [PubMed]
- Courtois-Cox, S.; Jones, S.L.; Cichowski, K. Many roads lead to oncogene-induced senescence. Oncogene 2008, 27, 2801–2809. [Google Scholar] [CrossRef]
- Hayflick, L.; Moorhead, P.S. The serial cultivation of human diploid cell strains. Exp Cell Res 1961, 25, 585–621. [Google Scholar] [CrossRef] [PubMed]
- d’Adda di Fagagna, F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer 2008, 8, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Lin, A.W.; McCurrach, M.E.; Beach, D.; Lowe, S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997, 88, 593–602. [Google Scholar] [CrossRef]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef]
- Muñoz-Espín, D.; Serrano, M. Cellular senescence: from physiology to pathology. Nat. Rev. Mol. Cell Biol. 2014, 15, 482–496. [Google Scholar] [CrossRef]
- Sharpless, N.E.; Sherr, C.J. Forging a signature of in vivo senescence. Nat. Rev. Cancer 2015, 15, 397–408. [Google Scholar] [CrossRef]
- Hernandez-Segura, A.; de Jong, T.V.; Melov, S.; Guryev, V.; Campisi, J.; Demaria, M. Unmasking Transcriptional Heterogeneity in Senescent Cells. Curr Biol. 2017, 27, 2652–2660. [Google Scholar] [CrossRef] [PubMed]
- Petrova, N.V.; Velichko, A.K.; Razin, S.V.; Kantidze, O.L. Small molecule compounds that induce cellular senescence. Aging Cell. 2016, 15, 999–1017. [Google Scholar] [CrossRef]
- Acosta, J.C.; Banito, A.; Wuestefeld, T.; Georgilis, A.; Janich, P.; Morton, J.P.; Athineos, D.; Kang, T.W.; Lasitschka, F.; Andrulis, M.; Pascual, G.; Morris, K.J.; Khan, S.; Jin, H.; Dharmalingam, G.; Snijders, A.P.; Carroll, T.; Capper, D.; Pritchard, C.; Inman, G.J.; Longerich, T.; Sansom, O.J.; Benitah, S.A.; Zender, L.; Gil, J. A complex secretory program orchestrated by the inflammasome controls paracrine senescence. Nat Cell Biol. 2013, 15, 978–990. [Google Scholar] [CrossRef] [PubMed]
- Price, J.S.; Waters, J.G.; Darrah, C.; Pennington, C.; Edwards, D.R.; Donell, S.T.; Clark, I.M. The role of chondrocyte senescence in osteoarthritis. Aging Cell 2002, 1, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Campisi, J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell 2005, 120, 513–522. [Google Scholar] [CrossRef] [PubMed]
- Bhat, R.; Crowe, E.P.; Bitto, A.; Moh, M.; Katsetos, C.D.; Garcia, F.U.; Johnson, F.B.; Trojanowski, J.Q.; Sell, C.; Torres, C. Astrocyte senescence as a component of Alzheimer’s disease. PLoS One 2012, 7, e45069. [Google Scholar] [CrossRef] [PubMed]
- Jeon, O.H.; Kim, C.; Laberge, R.M.; Demaria, M.; Rathod, S.; Vasserot, A.P.; Chung, J.W.; Kim, D.H.; Poon, Y.; David, N.; Baker, D,J. ; van Deursen, J.M.; Campisi, J.; Elisseeff, J.H. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med 2017, 23, 775–781. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.K.; Tchkonia, T.; LeBrasseur, N.K.; Chini, E.N.; Xu, M.; Kirkland, J.L. Cellular Senescence in Type 2 Diabetes: A Therapeutic Opportunity. Diabetes 2015, 64, 2289–2298. [Google Scholar] [CrossRef]
- Lelarge, V.; Capelle, R.; Oger, F.; Mathieu, T.; Le Calvé, B. Senolytics: from pharmacological inhibitors to immunotherapies, a promising future for patients' treatment. NPJ Aging. 2024, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Raffaele, M.; Vinciguerra, M. The costs and benefits of senotherapeutics for human health. Lancet Healthy Longevity. 2022, 3, 67–77. [Google Scholar] [CrossRef]
- Chaib, S.; Tchkonia, T.; Kirkland, J.L. Cellular senescence and senolytics: the path to the clinic. Nat. Med. 2022, 28, 1556–1568. [Google Scholar] [CrossRef]
- Guerrero, A.; Herranz, N.; Sun, B.; Wagner, V.; Gallage, S.; Guiho, R.; Wolter, K.; Pombo, J.; Irvine, E.E.; Innes, A.J.; Birch, J.; Glegola, J.; Manshaei, S.; Heide, D.; Dharmalingam, G.; Harbig, J.; Olona, A.; Behmoaras, J.; Dauch, D.; Uren, A.G.; Zender, L.; Vernia, S.; Martínez-Barbera, J.P.; Heikenwalder, M.; Withers, D.J.; Gil, J. Cardiac glycosides are broad-spectrum senolytics. Nat Metab. 2019, 1, 1074–1088. [Google Scholar] [CrossRef]
- Poblocka, M.; Bassey, A.L.; Smith, V.M.; Falcicchio, M.; Manso, A.S.; Althubiti, M.; Sheng, X.; Kyle, A.; Barber, R.; Frigerio, M.; Macip, S. Targeted clearance of senescent cells using an antibody-drug conjugate against a specific membrane marker. Sci Rep. 2021, 11, 20358. [Google Scholar] [CrossRef] [PubMed]
- Kurioka, A.; Klenerman, P. Aging unconventionally: γδ T cells, iNKT cells, and MAIT cells in aging. Seminars Immunol. 2023, 69, 101816. [Google Scholar] [CrossRef]
- Amor, C.; Feucht, J.; Leibold, J.; Ho, Y.J.; Zhu, C.; Alonso-Curbelo, D.; Mansilla-Soto, J.; Boyer, J.A.; Li, X.; Giavridis, T.; Kulick, A.; Houlihan, S.; Peerschke, E.; Friedman, S.L.; Ponomarev, V.; Piersigilli, A.; Sadelain, M.; Lowe, S.W. Senolytic CAR T cells reverse senescence-associated pathologies. Nature. 2020, 583, 127–132, Erratum in: Nature. 2024 Mar;627(8004):E9. doi: 10.1038/s41586-024-07197-3.. [Google Scholar] [CrossRef] [PubMed]
- Lear, T.B.; Finkel, T. Senolytic vaccination: a new mandate for cardiovascular health? J. Cardiovasc. Aging. 2022, 2, 17. [Google Scholar] [CrossRef]
- Suda, M.; Shimizu, I.; Katsuumi, G.; Yoshida, Y.; Hayashi, Y.; Ikegami, R.; Matsumoto, N.; Yoshida, Y.; Mikawa, R.; Katayama, A.; Wada, J.; Seki, M.; Suzuki, Y.; Iwama, A.; Nakagami, H.; Nagasawa, A.; Morishita, R.; Sugimoto, M.; Okuda, S.; Tsuchida, M.; Ozaki, K.; Nakanishi-Matsui, M.; Minamino, T. Senolytic vaccination improves normal and pathological age-related phenotypes and increases lifespan in progeroid mice. Nat Aging. 2021, 1, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Raffaele, M.; Vinciguerra, M. The costs and benefits of senotherapeutics for human health. Lancet Healthy Longev. 2022, 3, e67–e77. [Google Scholar] [CrossRef]
- Hernandez-Segura, A.; Nehme, J.; Demaria, M. Hallmarks of Cellular Senescence. Trends Cell Biol. 2018, 28, 436–453. [Google Scholar] [CrossRef]
- Roger, L.; Tomas, F.; Gire, V. Mechanisms and Regulation of Cellular Senescence. International Journal of Molecular Sciences. 2021, 22, 13173. [Google Scholar] [CrossRef]
- Wiley, C.D.; Campisi, J. From Ancient Pathways to Aging Cells-Connecting Metabolism and Cellular Senescence. Cell Metab. 2016, 23, 1013–1021. [Google Scholar] [CrossRef]
- Faraonio, R. Oxidative Stress and Cell Senescence Process. Antioxidants (Basel). 2022, 11, 1718. [Google Scholar] [CrossRef]
- Kuilman, T.; Peeper, D.S. Senescence-messaging secretome: SMS-ing cellular stress. Nat. Rev. Cancer. 2009, 9, 81–94. [Google Scholar] [CrossRef] [PubMed]
- Coppé, J.P.; Desprez, P.Y.; Krtolica, A.; Campisi, J. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu Rev Pathol. 2010, 5, 99–118. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Correia-Melo, C.; Marques, F.D.M.; Anderson, R.; Hewitt, G.; Hewitt, R.; Cole, J.; Carroll, B.M.; Miwa, S.; Birch, J.; Merz, A.; et al. Mitochondria are required for pro-ageing features of the senescent phenotype. EMBO J. 2016, 35, 724–742. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhu, H.; Yan, X.; Gu, H.; Gu, Z.; Liu, F. Endoplasmic reticulum stress participates in the progress of senescence and apoptosis of osteoarthritis chondrocytes. Biochem Biophys Res Commun. 2017, 491, 368–373. [Google Scholar] [CrossRef] [PubMed]
- Ohno-Iwashita, Y.; Shimada, Y.; Hayashi, M.; Inomata, M. Plasma membrane microdomains in aging and disease. Geriatr Gerontol Int. 2010, 10, S41–52. [Google Scholar] [CrossRef] [PubMed]
- Burkewitz, K.; Zhang, Y.; Mair, W.B. AMPK at the nexus of energetics and aging. Cell Metab. 2014, 20, 10–25. [Google Scholar] [CrossRef] [PubMed]
- van Vliet, T.; Varela-Eirin, M.; Wang, B.; Borghesan, M.; Brandenburg, S.M.; Franzin, R.; Evangelou, K.; Seelen, M.; Gorgoulis, V.; Demaria, M. Physiological hypoxia restrains the senescence-associated secretory phenotype via AMPK-mediated mTOR suppression. Mol Cell. 2021, 81, 2041–2052. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, N.; Zhang, X.; Horng, T. Mitochondrial metabolism regulates macrophage biology. J Biol Chem. 2021, 297, 100904. [Google Scholar] [CrossRef] [PubMed]
- Wenz, T. Mitochondria and PGC-1α in Aging and Age-Associated Diseases. J Aging Res. 2011, 810619. [Google Scholar] [CrossRef]
- Korolchuk, V.I.; Miwa, S.; Carroll, B.; von Zglinicki, T. Mitochondria in Cell Senescence: Is Mitophagy the Weakest Link? EBioMedicine. 2017, 21, 7–13. [Google Scholar] [CrossRef]
- Evangelou, K.; Lougiakis, N.; Rizou, S.V.; Kotsinas, A.; Kletsas, D.; Muñoz-Espín, D.; Kastrinakis, N.G.; Pouli, N.; Marakos, P.; Townsend, P.; Serrano, M.; Bartek, J.; Gorgoulis, V.G. Robust, universal biomarker assay to detect senescent cells in biological specimens. Aging Cell. 2017, 16, 192–197. [Google Scholar] [CrossRef] [PubMed]
- Knaś, M.; Zalewska, A.; Krętowski, R. ; Niczyporuk, M.; Waszkiewicz, N.; Cechowska-Pasko, M.; Waszkiel, D.; Zwierz, K. The profile of lysosomal exoglycosidases in replicative and stress-induced senescence in early passage human fibroblasts. Folia Histochem Cytobiol. 2012, 50, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Matos, L.; Gouveia, A.M.; Almeida, H. ER Stress Response in Human Cellular Models of Senescence. J Gerontol A Biol Sci Med Sci. 2015, 70, 924–935. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Ruiz, P.D.; McKimpson, W.M.; Novikov, L.; Kitsis, R.N.; Gamble, M.J. MacroH2A1 and ATM Play Opposing Roles in Paracrine Senescence and the Senescence-Associated Secretory Phenotype. Mol Cell. 2015, 59, 719–731. [Google Scholar] [CrossRef] [PubMed]
- Hoskins, V.E.; Smith, K.; Reddy, K.L. The shifting shape of genomes: dynamics of heterochromatin interactions at the nuclear lamina. Curr Opin Genet Dev. 2021, 67, 163–173. [Google Scholar] [CrossRef]
- Aird, K.M.; Zhang, R. Detection of senescence-associated heterochromatin foci (SAHF). Methods Mol Biol. 2013, 965, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Kumari, R.; Jat, P. Mechanisms of Cellular Senescence: Cell Cycle Arrest and Senescence Associated Secretory Phenotype. Front Cell Dev Biol. 2021, 9, 645593–doi10. [Google Scholar] [CrossRef]
- Buj, R.; Leon, K.E.; Anguelov, M.A.; Aird, K.M. Suppression of p16 alleviates the senescence-associated secretory phenotype. Aging (Albany NY). 2021, 13, 3290–3312. [Google Scholar] [CrossRef]
- Liu, X.; Wan, M. A tale of the good and bad: Cell senescence in bone homeostasis and disease. Int Rev Cell Mol Biol. 2019, 346, 97–128. [Google Scholar] [CrossRef]
- Zhu, L.; Lu, Z.; Zhao, H. Antitumor mechanisms when pRb and p53 are genetically inactivated. Oncogene. 2015, 34, 4547–4557. [Google Scholar] [CrossRef]
- Narasimha, A.M.; Kaulich, M.; Shapiro, G.S.; Choi, Y.J.; Sicinski, P.; Dowdy, S.F. Cyclin D activates the Rb tumor suppressor by mono-phosphorylation. Elife, e: 3, 0287. [Google Scholar] [CrossRef]
- Inoue, K.; Fry, E.A. Aberrant expression of p16INK4a in human cancers - a new biomarker? Cancer Rep Rev. 2018, 2, 10–15761. [Google Scholar] [CrossRef] [PubMed]
- Safwan-Zaiter, H.; Wagner, N.; Wagner, K.D. P16INK4A-More Than a Senescence Marker. Life (Basel). 2022, 12, 1332. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, A.M.; Attardi, L.D. Deconstructing networks of p53-mediated tumor suppression in vivo. Cell Death Differ. 2018, 25, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Kastenhuber, E.R.; Lowe, S.W. Putting p53 in Context. Cell. 2017, 170, 1062–1078. [Google Scholar] [CrossRef]
- Al Bitar, S.; Gali-Muhtasib, H. The Role of the Cyclin Dependent Kinase Inhibitor p21cip1/waf1 in Targeting Cancer: Molecular Mechanisms and Novel Therapeutics. Cancers. 2019, 11, 1475. [Google Scholar] [CrossRef] [PubMed]
- Georgakilas, A.G.; Martin, O.A.; Bonner, W.M. p21: A Two-Faced Genome Guardian. Trends Mol Med. 2017, 23, 310–319. [Google Scholar] [CrossRef]
- Campisi, J.; d’Adda di, F.F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007, 8, 729–740. [Google Scholar] [CrossRef]
- Fernandez-Rebollo E, Franzen J, Goetzke R, Hollmann J, Ostrowska A, Oliverio M, Sieben T, Rath B, Kornfeld JW, Wagner W. Senescence-Associated Metabolomic Phenotype in Primary and iPSC-Derived Mesenchymal Stromal Cells. Stem Cell Reports. 2020 Feb 11;14(2):201-209. [CrossRef]
- Franzen, J.; Georgomanolis, T.; Selich, A.; Kuo, C.C.; Stöger, R.; Brant, L.; Mulabdić, M.S.; Fernandez-Rebollo, E.; Grezella, C.; Ostrowska, A.; Begemann, M.; Nikolić, M.; Rath, B.; Ho, A.D.; Rothe, M.; Schambach, A.; Papantonis, A.; Wagner, W. DNA methylation changes during long-term in vitro cell culture are caused by epigenetic drift. Commun Biol. 2021, 4, 598. [Google Scholar] [CrossRef]
- Zemach, A.; McDaniel, I.E.; Silva, P.; Zilberman, D. Genome- wide evolutionary analysis of eukaryotic DNA methylation. Science 2010, 328, 916–919. [Google Scholar] [CrossRef]
- Holliday, R.; Grigg, G.W. DNA methylation and mutation. Mutat. Res. 1993, 285, 61–67. [Google Scholar] [CrossRef]
- Rošić, S.; Amouroux, R.; Requena, C.E.; Gomes, A.; Emperle, M.; Beltran, T.; Rane, J.K.; Linnett, S.; Selkirk, M.E.; Schiffer, P.H.; Bancroft, A.J.; Grencis, R.K.; Jeltsch, A.; Hajkova, P.; Sarkies, P. Evolutionary analysis indicates that DNA alkylation damage is a byproduct of cytosine DNA methyltransferase activity. Nat Genet. 2018, 50, 452–459. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc'his, D. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, L.; Li, J.; Ding, Z.; Xiao, J.; Yin, X.; He, S.; Shi, P.; Dong, L.; Li, G.; Tian, C.; Wang, J.; Cong, Y.; Xu, Y. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature. 2015, 517, 640–644. [Google Scholar] [CrossRef]
- Ishiyama, S.; Nishiyama, A.; Saeki, Y.; Moritsugu, K.; Morimoto, D.; Yamaguchi, L.; Arai, N.; Matsumura, R.; Kawakami, T.; Mishima, Y.; Hojo, H.; Shimamura, S.; Ishikawa, F.; Tajima, S.; Tanaka, K.; Ariyoshi, M.; Shirakawa, M.; Ikeguchi, M.; Kidera, A.; Suetake, I.; Arita, K.; Nakanishi, M. Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance. Mol Cell. 2017, 68, 350–360. [Google Scholar] [CrossRef] [PubMed]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011, 333, 1300–1303. [Google Scholar] [CrossRef] [PubMed]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; Rao, A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed]
- He, Y.F.; Li, B.Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; Sun, Y.; Li, X.; Dai, Q.; Song, C.X.; Zhang, K.; He, C.; Xu, G.L. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011, 333, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Su, Y.; Song, Q.; Tung, B.; Oyinlade, O.; Liu, S.; Ying, M.; Ming, G.L.; Song, H.; Qian, J.; Zhu, H.; Xia, S. Methylated cis-regulatory elements mediate KLF4-dependent gene transactivation and cell migration. Elife. 2017, 6, e20068. [Google Scholar] [CrossRef]
- Bruno, S.; Schlaeger, T.M.; Del Vecchio, D. Epigenetic OCT4 regulatory network: stochastic analysis of cellular reprogramming. npj Syst Biol Appl 2024, 10, 3. [Google Scholar] [CrossRef]
- Schäfer, A.; Mekker, B.; Mallick, M.; Vastolo, V.; Karaulanov, E.; Sebastian, D.; von der Lippen, C.; Epe, B.; Downes, D.J.; Scholz, C.; Niehrs, C. Impaired DNA demethylation of C/EBP sites causes premature aging. Genes Dev. 2018, 32, 11–12. [Google Scholar] [CrossRef]
- He,Y. ; Liu, X.Y.; Gong, R.; Peng, K.W.; Liu, R.B.; Wang, F. NK homeobox 2.2 functions as tumor suppressor in colorectal cancer due to DNA methylation. J Cancer. 2020, 11, 4791–4800. [Google Scholar] [CrossRef]
- Sakaki, M.; Ebihara, Y.; Okamura, K.; Nakabayashi, K.; Igarashi, A.; Matsumoto, K.; Hata, K.; Kobayashi, Y.; Maehara, K. Potential roles of DNA methylation in the initiation and establishment of replicative senescence revealed by array-based methylome and transcriptome analyses. PLoS ONE 2017, 12, e0171431. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, H.; Xu, G.; Liu, L. Tet1 Deficiency Leads to Premature Ovarian Failure. Front. Cell Dev. Biol. 2021, 9, 620. [Google Scholar]
- Huang, G.; Liu, L.; Wang, H.; Gou, M.; Gong, P.; Tian, C.; Deng, W.; Yang, J.; Zhou, T.-T.; Xu, G.L.; et al. Tet1 Deficiency Leads to Premature Reproductive Aging by Reducing Spermatogonia Stem Cells and Germ Cell Differentiation. IScience 2020, 23, 100908. [Google Scholar] [CrossRef] [PubMed]
- Ferrone, C.K.; Blydt-Hansen, M.; Rauh, M.J. Age-Associated TET2 Mutations: Common Drivers of Myeloid Dysfunction, Cancer and Cardiovascular Disease. Int. J. Mol. Sci. 2020, 21, 626. [Google Scholar] [CrossRef]
- Weidner, C.I.; Lin, Q.; Koch, C.M.; Eisele, L.; Beier, F.; Ziegler, P.; Bauerschlag, D.O.; Jöckel, K.-H.; Erbel, R.; Mühleisen, T.W.; et al. Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol. 2014, 15, R24. [Google Scholar] [CrossRef] [PubMed]
- Meer, M.V.; Podolskiy, D.I.; Tyshkovskiy, A.; Gladyshev, V.N. A whole lifespan mouse multi-tissue DNA methylation clock. Elife 2018, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, T.J.; Quarta, M.; Mukherjee, S.; Colville, A.; Paine, P.; Doan, L.; Tran, C.M.; Chu, C.R.; Horvath, S.; Qi, L.S.; et al. Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nat. Commun. 2020, 11, 1545. [Google Scholar] [CrossRef]
- Ocampo, A.; Reddy, P.; Redondo, P.M.; Luengo, A.P.; Hatanaka, F.; Hishida, T.; Li, M.; Lam, D.; Kurita, M.; Beyret, E.; et al. In Vivo Amelioration of Age-Associated Hallmarks by Partial Reprogramming. Cell 2016, 167, 1719–1733.e12. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Brommer, B.; Tian, X.; Krishnan, A.; Meer, M.; Wang, C.; Vera, D.L.; Zeng, Q.; Yu, D.; Bonkowski, M.S.; et al. Reprogramming to recover youthful epigenetic information and restore vision. Nature 2020, 588, 124–129. [Google Scholar] [CrossRef]
- Leon, K.E.; Buj, R.; Lesko, E.; Dahl, E.S.; Chen, C.-W.; Tangudu, N.K.; Kawasawa, Y.I.; Kossenkov, A.V.; Hobbs, R.P.; Aird, K.M. DOT1L modulates the senescence-associated secretory phenotype through epigenetic regulation of IL1A. J. Cell Biol. 2021, 220, 202008101. [Google Scholar] [CrossRef] [PubMed]
- Cakouros, D.; Gronthos, S. Epigenetic Regulation of Bone Marrow Stem Cell Aging: Revealing Epigenetic Signatures associated with Hematopoietic and Mesenchymal Stem Cell Aging. Aging Dis. 2019, 10, 174–189. [Google Scholar] [CrossRef]
- Senthil V Bhoopalan, Ruopeng Feng, Thiyagaraj Mayuranathan, Kalin Mayberry, Yoonjeong Jang, Jonathan S Yen, Yu Yao, Lance Palmer, Yong Cheng, Marcin W Wlodarski, Mitchell J. Weiss. Reduced Polycomb Repressor Complex 2 (PRC2) Activity and Increased TP53 Activity Mediate Hematopoietic Stem Cell Dysfunction in RPS19-Mutated Diamond-Blackfan Anemia. Blood, 2023, 142, Supplement 1, Page 2732, ISSN 0006-4971. [CrossRef]
- Rothenberg, E.V.; Hosokawa, H.; Ungerbäck, J. Mechanisms of Action of Hematopoietic Transcription Factor PU.1 in Initiation of T-Cell Development. Front Immunol. 2019, 10, 228. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Yoo, J.; Jeon, Y.H.; Cho, H.Y.; Lee, S.W.; Kim, G.W.; Lee, D.H.; Kwon, S.H. Advances in Histone Demethylase KDM3A as a Cancer Therapeutic Target. Cancers 2020, 12, 1098. [Google Scholar] [CrossRef]
- Zhu, Z.; Wu, X.; Li, Q.; Zhang, J.; Yu, S.; Shen, Q.; Zhou, Z.; Pan, Q.; Yue, W.; Qin, D.; Zhang, Y.; Zhao, W.; Zhang, R.; Peng, S.; Li, N.; Zhang, S.; Lei, A.; Miao, Y.L.; Liu, Z.; Chen, X.; Wang, H.; Liao, M.; Hua, J. Histone demethylase complexes KDM3A and KDM3B cooperate with OCT4/SOX2 to define a pluripotency gene regulatory network. FASEB J. 2021, 35, e21664, Erratum in: FASEB J. 2021 Jul;35(7):e21736. doi: 10.1096/fsb2.21736.. [Google Scholar] [CrossRef]
- Torano, E.G.; Bayon, G.F.; Del Real, A.; Sierra, M.I.; Garcia, M.G.; Carella, A, et al. Age-associated hydroxymethylation in human bone-marrow mesenchymal stem cells. J Transl Med 2016, 14, 207. [Google Scholar] [CrossRef] [PubMed]
- Kouzarides, T. Chromatin modifications and their function. Cell 2007, 128, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Pu, M.; Wang, M.; Wang, W.; Velayudhan, S.S.; Lee, S.S. Unique patterns of trimethylation of histone H3 lysine 4 are prone to changes during aging in Caenorhabditis elegans somatic cells. PLoS Genet. 2018, 14, e1007466. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Wang, W.; Williams, J.B.; Yang, F.; Wang, Z.J.; Yan, Z. Targeting histone K4 trimethylation for treatment of cognitive and synaptic deficits in mouse models of Alzheimer's disease. Sci Adv. 2020, 6, eabc8096. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Luo, M.; Jeong, M.; Rodriguez, B.; Xia, Z.; Hannah, R.; Wang, H.; Le, T.; Faull, K.F.; Chen, R.; Gu, H.; Bock, C.; Meissner, A.; Göttgens, B.; Darlington, G.J.; Li, W.; Goodell, M.A. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014, 14, 673–688. [Google Scholar] [CrossRef] [PubMed]
- Merkwirth, C.; Jovaisaite, V.; Durieux, J.; Matilainen, O.; Jordan, S.D.; Quiros, P.M.; Steffen, K.K.; Williams, E.G.; Mouchiroud, L.; Tronnes, S.U.; Murillo, V.; Wolff, S.C.; Shaw, R.J.; Auwerx, J.; Dillin, A. Two Conserved Histone Demethylases Regulate Mitochondrial Stress-Induced Longevity. Cell. 2016, 165, 1209–1223. [Google Scholar] [CrossRef]
- Jing, H.; Liao, L.; An, Y.; Su, X.; Liu, S.; Shuai, Y.; Zhang, X.; Jin, Y. Suppression of EZH2 Prevents the Shift of Osteoporotic MSC Fate to Adipocyte and Enhances Bone Formation During Osteoporosis. Mol Ther. 2016, 24, 217–229. [Google Scholar] [CrossRef]
- Li, C.; Chai, Y.; Wang, L.; Gao, B.; Chen, H.; Gao, P.; Zhou, F.Q.; Luo, X.; Crane, J.L.; Yu, B.; Cao, X.; Wan, M. Programmed cell senescence in skeleton during late puberty. Nat Commun. 2017, 8, 1312. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Fan, Z.; Yu, B.; Chang, J.; Al Hezaimi, K.; Zhou, X.; Park, N.H.; Wang, C.Y. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell. 2012, 11, 50–61, Erratum in: Cell Stem Cell. 2018 Dec 6;23(6):898-899. doi:10.1016/j.stem.2018.11.002.. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, W.; Chang, L.; Han, Y.; Sun, L.; Gong, X.; Tang, H.; Liu, Z.; Deng, H.; Ye, Y.; Wang, Y.; Li, J.; Qiao, J.; Qu, J.; Zhang, W.; Liu, G.H. Vitamin C alleviates aging defects in a stem cell model for Werner syndrome. Protein Cell. 2016, 7, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Shan, H.; Geng, L.; Jiang, X.; Song, M.; Wang, J.; Liu, Z.; Zhuo, X.; Wu, Z.; Hu, J.; Ji, Z.; Wang, S.; Chan, P.; Qu, J.; Zhang, W.; Liu, G.H. Large-scale chemical screen identifies Gallic acid as a geroprotector for human stem cells. Protein Cell. 2022, 13, 532–539. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zou, Z.; Cai, Y.; Yang, K.; Wang, S.; Liu, Z.; Geng, L.; Chu, Q.; Ji, Z.; Chan, P.; Liu, G.H.; Song, M.; Qu, J.; Zhang, W. Low-dose chloroquine treatment extends the lifespan of aged rats. Protein Cell. 2022, 13, 454–461, Erratum in: Protein Cell. 2024 Apr 1;15(4):313. doi: 10.1093/procel/pwad053. 101. [Google Scholar] [CrossRef]
- Deng, P.; Yuan, Q.; Cheng, Y.; Li, J.; Liu, Z.; Liu, Y.; Li, Y.; Su, T.; Wang, J.; Salvo, M.E.; Wang, W.; Fan, G.; Lyons, K.; Yu, B.; Wang, C.Y. Loss of KDM4B exacerbates bone-fat imbalance and mesenchymal stromal cell exhaustion in skeletal aging. Cell Stem Cell. 2021, 28, 1057–1073.e7. [Google Scholar] [CrossRef]
- Xu, Z.; Wu, W.; Shen, F.; Yu, Y.; Wang, Y.; Xiang, C. Histone Arginine Methylation-Mediated Epigenetic Regulation of Discoidin Domain Receptor 2 Controls the Senescence of Human Bone Marrow Mesenchymal Stem Cells. Stem Cells Int. 2019, 7670316. [Google Scholar] [CrossRef]
- Zhu, Y.; Song, X.; Han, F.; Li, Y.; Wei, J.; Liu, X. Alteration of histone acetylation pattern during long-term serum-free culture conditions of human fetal placental mesenchymal stem cells. PLoS One. 2015, 10, e0117068. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.W.; Lee, S.; Seo, M.S.; Park, S.B.; Kurtz, A.; Kang, S.K.; Kang, K.S. Histone deacetylase controls adult stem cell aging by balancing the expression of polycomb genes and jumonji domain containing 3. Cell Mol Life Sci. 2010, 67, 1165–1176. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, H.; Qiu, T.; Liao, L.; Su, X. Epigenetic regulation of mesenchymal stem cell aging through histone modifications. Genes Dis. 2022, 10, 2443–2456. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Chen, H.; Liu, H.; Zhang, W.; Zhou, J. Emerging roles of SIRT6 in human diseases and its modulators. Med Res Rev. 2021, 41, 1089–1137. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liu, Y.; Jin, C.; Zhang, M.; Tang, F.; Zhou, Y. Histone Acetyltransferase GCN5 Regulates Osteogenic Differentiation of Mesenchymal Stem Cells by Inhibiting NF-κB. J Bone Miner Res. 2016, 31, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Lane, S.I.R.; Jones, K.T. Phosphorylation of histone H3 in 1-and 2-cell embryos. Comment on: Teperek-Tkacz M, et al. Cell Cycle 2010; 9:4674–87. Cell Cycle 2011, 10, 1–15. [Google Scholar] [CrossRef]
- Oh, S.; Suganuma, T.; Gogol, M.M.; Workman, J.L. Histone H3 threonine 11 phosphorylation by Sch9 and CK2 regulates chronological lifespan by controlling the nutritional stress response. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, D.; Avvakumov, N.; Côté, J. Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics 2012, 7, 1098–1108. [Google Scholar] [CrossRef]
- Yi, S.J.; Kim, K. New Insights into the Role of Histone Changes in Aging. International Journal of Molecular Sciences 2020, 21, 8241. [Google Scholar] [CrossRef]
- Rhie, B.H.; Song, Y.H.; Ryu, H.Y.; Ahn, S.H. Cellular aging is associated with increased ubiquitylation of histone H2B in yeast telomeric heterochromatin. Biochem. Biophys. Res. Commun. 2013, 439, 570–575. [Google Scholar] [CrossRef]
- Guo, Y.; Chomiak, A.A.; Hong, Y.; Lowe, C.C.; Kopsidas, C.A.; Chan, W.C.; Andrade, J.; Pan, H.; Zhou, X.; Monuki, E.S.; Feng, Y. Histone H2A ubiquitination resulting from Brap loss of function connects multiple aging hallmarks and accelerates neurodegeneration. iScience 2022, 25, 104519. [Google Scholar] [CrossRef]
- Kim, B.J.; Chan, D.W.; Jung, S.Y.; Chen, Y.; Qin, J.; Wang, Y. The Histone Variant MacroH2A1 Is a BRCA1 Ubiquitin Ligase Substrate. Cell Rep. 2017, 19, 1758–1766. [Google Scholar] [CrossRef]
- Vaicekauskaitė, I.; Sabaliauskaitė, R.; Lazutka, J.R.; Jarmalaitė, S. The Emerging Role of Chromatin Remodeling Complexes in Ovarian Cancer. Int. J. Mol. Sci. 2022, 23, 13670. [Google Scholar] [CrossRef] [PubMed]
- Tordella, L.; Khan, S.; Hohmeyer, A.; Banito, A.; Klotz, S.; Raguz, S.; Martin, N.; Dhamarlingam, G.; Carroll, T.; González Meljem, J.M.; Deswal, S.; Martínez-Barbera, J.P.; García-Escudero, R.; Zuber, J.; Zender, L.; Gil, J. SWI/SNF regulates a transcriptional program that induces senescence to prevent liver cancer. Genes Dev. 2016, 30, 2187–2198. [Google Scholar] [CrossRef] [PubMed]
- Grossi, E. , Raimondi, I., Goñi, E.; Gonzalez, J.; Marchese, F.P.; Chapaparieta, V.; Martin-Subero, J.I.; Guo, S.; Huarte, M. A lncRNA-SWI/SNF complex crosstalk controls transcriptional activation at specific promoter regions. Nat Commun 2020, 11, 936. [Google Scholar] [CrossRef] [PubMed]
- Pegoraro, G.; Kubben, N.; Wickert, U.; Göhler, H.; Hoffmann, K.; Misteli, T. Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol. 2009, 11, 1261–1267. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Shiromoto, Y.; Sakurai, M.; Towers, M.; Zhang, Q.; Wu, S.; Havas, A.; Wang, L.; Berger, S.; Adams, P.D.; Tian, B.; Nishikura, K.; Kossenkov, A.V.; Liu, P.; Zhang, R. ADAR1 downregulation by autophagy drives senescence independently of RNA editing by enhancing p16INK4a levels. Nat Cell Biol. 2022, 24, 1202–1210. [Google Scholar] [CrossRef]
- Sokolova, V.; Fiorino, A.; Zoni, E.; Crippa, E.; Reid, J.F.; Gariboldi, M.; Pierotti, M.A. The Effects of miR-20a on p21: Two Mechanisms Blocking Growth Arrest in TGF-β-Responsive Colon Carcinoma. J Cell Physiol. 2015, 230, 3105–3114. [Google Scholar] [CrossRef]
- Okada, M.; Kim, H.W.; Matsu-Ura, K.; Wang, Y.G.; Xu, M.; Ashraf, M. Abrogation of age-induced microRNA-195 rejuvenates the senescent mesenchymal stem cells by reactivating telomerase. Stem cells 2016, 34, 148–159. [Google Scholar] [CrossRef]
- Kim, Y.J.; Hwang, S.H.; Lee, S.Y.; Shin, K.K.; Cho, H.H.; Bae, Y.C.; Jung, J.S. miR-486-5p induces replicative senescence of human adipose tissue-derived mesenchymal stem cells and its expression is controlled by high glucose. Stem cells and development 2012, 21, 1749–1760. [Google Scholar] [CrossRef]
- Li, X.; Song, Y.; Liu, D.; Zhao, J.; Xu, J.; Ren, J.; Hu, Y.; Wang, Z.; Hou, Y.; Zhao, G. MiR-495 Promotes Senescence of Mesenchymal Stem Cells by Targeting Bmi-1. Cell Physiol Biochem. 2017, 42, 780–796. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Yu, K.R.; Ryu, Y.S.; Oh, Y.S.; Hong, I.S.; Kim, H.S.; Lee, J.Y.; Kim, S.; Seo, K.W.; Kang, K.S. miR-543 and miR-590-3p regulate human mesenchymal stem cell aging via direct targeting of AIMP3/p18. Age (Dordr). 2014, 36, 9724. [Google Scholar] [CrossRef] [PubMed]
- Potter, M.L.; Hill, W.D.; Isales, C.M.; Hamrick, M.W.; Fulzele, S. MicroRNAs are critical regulators of senescence and aging in mesenchymal stem cells. Bone 2021, 142, 115679. [Google Scholar] [CrossRef] [PubMed]
- Borgdorff, V.; Lleonart, M.E.; Bishop, C.L.; Fessart, D.; Bergin, A.H.; Overhoff, M.G.; Beach, D.H. Multiple microRNAs rescue from Ras-induced senescence by inhibiting p21(Waf1/Cip1). Oncogene 2010, 29, 2262–2271. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Baniahmad, A.; Branicki, W.; Taheri, M.; Eghbali, A. Emerging Role of Non-Coding RNAs in Senescence. Front Cell Dev Biol. 2022, 10, 869011. [Google Scholar] [CrossRef]
- Luo, S.; Lu, J.Y.; Liu, L.; Yin, Y.; Chen, C.; Han, X.; Wu, B.; Xu, R.; Liu, W.; Yan, P.; Shao, W.; Lu, Z.; Li, H.; Na, J.; Tang, F.; Wang, J.; Zhang, Y.E.; Shen, X. Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell Stem Cell 2016, 18, 637–652. [Google Scholar] [CrossRef]
- Tan, P.; Guo, Y.H.; Zhan, J.K.; Long, L.M.; Xu, M.L.; Ye, L.; Ma, X.Y.; Cui, X.J.; Wang, H.Q. LncRNA-ANRIL inhibits cell senescence of vascular smooth muscle cells by regulating miR-181a/Sirt1. Biochem Cell Biol. 2019, 97, 571–580. [Google Scholar] [CrossRef]
- Wang, S.; Zhan, J.; Lin, X.; Wang, Y.; Wang, Y.; Liu, Y. CircRNA -0077930 from Hyperglycaemia-stimulated Vascular Endothelial Cell Exosomes Regulates Senescence in Vascular Smooth Muscle Cells. Cell Biochem. Funct. 2020, 38, 1056–1068. [Google Scholar] [CrossRef]
- Du, W.W.; Yang, W.; Chen, Y.; Wu, Z.K.; Foster, F.S.; Yang, Z.; Li, X; Yang, B. B. Foxo3 Circular RNA Promotes Cardiac Senescence by Modulating Multiple Factors Associated with Stress and Senescence Responses. Eur. Heart J. 2017, 38, 1402–1412. [Google Scholar] [CrossRef]
- Du, Y.; Yu, Y.; Hu, Y.; Li, X.W.; Wei, Z.X.; Pan, R.Y.; Li, X.S.; Zheng, G.E.; Qin, X.Y.; Liu, Q.S.; Cheng, Y. Genome-Wide, Integrative Analysis Implicates Exosome-Derived MicroRNA Dysregulation in Schizophrenia. Schizophr. Bull. 2019, 45, 1257–1266. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Houtkooper Riekelt, H. Preface - Animal models for aging. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, 2679; 9. [Google Scholar] [CrossRef]
- Mitchell, S.J.; Scheibye-Knudsen, M.; Longo, D.L.; de Cabo, R. Animal models of aging research: implications for human aging and age-related diseases. Annu Rev Anim Biosci. 2015, 3, 283–303. [Google Scholar] [CrossRef]
- Padmanabhan P, Götz J. Clinical relevance of animal models in aging-related dementia research. Nat Aging. 2023, 3, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Cai, N.; Wu, Y.; Huang, Y. Induction of Accelerated Aging in a Mouse Model. Cells 2022, 11, 1418. [Google Scholar] [CrossRef] [PubMed]
- Conn, P.M. Sourcebook of Models for Biomedical Research. ed 2008, Totowa, NJ.
- Holtze, S.; Gorshkova, E.; Braude, S.; Cellerino, A.; Dammann, P.; Hildebrandt, T.B.; Hoeflich, A.; Hoffmann, S.; Koch, P.; Terzibasi Tozzini, E.; Skulachev, M.; Skulachev, V.P.; Sahm, A. Alternative Animal Models of Aging Research. Front Mol Biosci. 2021, 8, 660959. [Google Scholar] [CrossRef] [PubMed]
- Dammann, P.; Burda, H. Senescence Patterns in African Mole-rats (Bathyergidae, Rodentia). In: Begall, S., Burda, H., Schleich, C.E. Subterranean Rodents. Springer, Berlin, Heidelberg, 2007 (eds). [CrossRef]
- Lee, B.P.; Smith, M.; Buffenstein, R.; Harries, L.W. Negligible senescence in naked mole rats may be a consequence of well-maintained splicing regulation. Geroscience. 2020, 42, 633–651. [Google Scholar] [CrossRef] [PubMed]
- Buffenstein, R. The naked mole-rat: a new long-living model for human aging research. J Gerontol A Biol Sci Med Sci. 2005, 60, 1369–1377. [Google Scholar] [CrossRef] [PubMed]
- Braude, S.; Holtze, S.; Begall, S.; Brenmoehl, J.; Burda, H.; Dammann, P.; Del Marmol, D.; Gorshkova, E.; Henning, Y.; Hoeflich, A.; Höhn, A.; Jung, T.; Hamo, D.; Sahm, A.; Shebzukhov, Y.; Šumbera, R.; Miwa, S.; Vyssokikh, M.Y.; von Zglinicki, T.; Averina, O.; Hildebrandt, T.B. Surprisingly long survival of premature conclusions about naked mole-rat biology. Biol Rev Camb Philos Soc. 2021, 96, 376–393. [Google Scholar] [CrossRef]
- Edrey, Y.H.; Hanes, M.; Pinto, M.; Mele, J.; Buffenstein, R. Successful aging and sustained good health in the naked mole rat: a long-lived mammalian model for biogerontology and biomedical research. ILAR J. 2011, 52, 41–53. [Google Scholar] [CrossRef]
- Bartke, A. Insulin and aging. Cell Cycle 2008, 7, 3338–3343. [Google Scholar] [CrossRef]
- Burdusel, D.; Coman, C.; Ancuta, D.L.; Hermann, D.M.; Doeppner, T.R.; Gresita, A.; Popa-Wagner, A. Translatability of life-extending pharmacological treatments between different species. Aging Cell. 2024, 23, e14208. [Google Scholar] [CrossRef]
- Yousefzadeh, M.J.; Melos, K.I.; Angelini, L.; Burd, C.E.; Robbins, P.D.; Niedernhofer, L.J. Mouse Models of Accelerated Cellular Senescence. Methods Mol Biol. 2019, 1896, 203–230. [Google Scholar] [CrossRef] [PubMed]
- Folch, J.; Busquets, O.; Ettcheto, M.; Sánchez-López, E.; Pallàs, M.; Beas-Zarate, C.; Marin, M.; Casadesus, G.; Olloquequi, J.; Auladell, C.; Camins, A. Experimental Models for Aging and their Potential for Novel Drug Discovery. Curr Neuropharmacol. 2018, 16, 1466–1483. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.M. Aging, Cellular Senescence, and Alzheimer’s Disease. Int. J. Mol. Sci. 2022, 23, 1989. [Google Scholar] [CrossRef] [PubMed]
- Yuan, R.; Tsaih, S.W.; Petkova, S.B.; De Evsikova, C.M.; Xing, S.; et al. Aging in inbred strains of mice: study design and interim report on median lifespans and circulating IGF1 levels. Aging Cell 2009, 8, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.; Horikawa, I.; Harris, C. Cellular Senescence: Mechanisms, Morphology, and Mouse Models. Vet. Pathol. 2020, 57, 747–757. [Google Scholar] [CrossRef] [PubMed]
- Ackert-Bicknell, C.L.; Anderson, L.C.; Sheehan, S.; Hill, W.G.; Chang, B.; Churchill, G.A.; Chesler, E.J.; Korstanje, R.; Peters, L.L. Aging Research Using Mouse Models. Curr Protoc Mouse Biol. 2015, 5, 95–133. [Google Scholar] [CrossRef] [PubMed]
- Murthy, M.; Ram, J.L. Invertebrates as model organisms for research on aging biology. Invertebr Reprod Dev. 2015, 59, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Yue, Y.; Park, Y. A living model for obesity and aging research: Caenorhabditis elegans. Crit Rev Food Sci Nutr. 2018, 58, 741–754. [Google Scholar] [CrossRef]
- Voituron, Y.; de Fraipont, M.; Issartel, J.; Guillaume, O.; Clobert, J. Extreme lifespan of the human fish (Proteus anguinus): a challenge for ageing mechanisms. Biol. Lett. 2011, 7, 105–107. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Wake, D.B. Higher-level salamander relationships and divergence dates inferred from complete mitochondrial genomes. Mol. Phylogenet. Evol. 2009, 53, 492–508. [Google Scholar] [CrossRef]
- Vieira, W.A.; Wells, K.M.; McCusker, C.D. Advancements to the Axolotl Model for Regeneration and Aging. Gerontology. 2020, 66, 212–222. [Google Scholar] [CrossRef]
- Quesada, V.; Freitas-Rodríguez, S.; Miller, J.; Pérez-Silva, J.G.; Jiang, Z.F.; Tapia, W.; et al. Giant tortoise genomes provide insights into longevity and age-related disease. Nat. Ecol. Evol. 2019, 3, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Dodzian, J.; Kean, S.; Seidel, J.; Valenzano, D.R. A Protocol for Laboratory Housing of Turquoise Killifish (Nothobranchius furzeri). J Vis Exp. 2018, 11, 134–57073. [Google Scholar] [CrossRef]
- Cailliet, G.M.; Andrews, A.H.; Burton, E.J.; Watters, D.L.; Kline, D.E.; Ferry-Graham, L.A. Age determination and validation studies of marine fishes: do deep-dwellers live longer? Exp Gerontol. 2001, 36, 4–6. [Google Scholar] [CrossRef] [PubMed]
- Haussmann, M.F.; Winkler, D.W.; Huntington, C.E.; Nisbet, I.C.; Vleck, C.M. Telomerase expression is differentially regulated in birds of differing life span. Ann N Y Acad Sci. 2004, 1019, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Braun, E.J.; Sweazea, K.L. Glucose regulation in birds. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008, 151, 1–9. [Google Scholar] [CrossRef]
- Hickey, A.J.; Jullig, M.; Aitken, J.; Loomes, K.; Hauber, M.E.; Phillips, A.R. Birds and longevity: Does flight driven aerobicity provide an oxidative sink? Ageing Res. Rev. 2012, 11, 242–253. [Google Scholar] [CrossRef]
| Enzyme/factor | Role | Mouse loss-of-function model | Human diseases due to mutations |
|---|---|---|---|
| DNMT3A | De novo DNA methyltransferase |
|
|
| DNMT3B | De novo DNA methyltransferase |
|
|
| DNMT3C | De novo DNA methyltransferase (mice/rats specific isoform) |
|
|
| DNMT3L | DNA methyltransferase cofactor |
|
|
| DNMT1 | Maintenance DNA methyltransferase |
|
|
| UHFR1 | DNMT1 cofactor |
|
|
| TET1 | DNA demethylation via oxidation of methylcytosine |
|
|
| TET2 | DNA demethylation via oxidation of methylcytosine |
|
|
| TET3 | DNA demethylation via oxidation of methylcytosine |
|
|
Histone deacetylase | ||||
|---|---|---|---|---|
| Classification | Localization |
Inhibitors (exemples) |
||
|
Zn+ dependent |
Class I | HDAC 1 HDAC 2 HDAC 3 HDAC 8 |
Mainly nucleus Nucleus Nucleus/cytoplasm Mainly cytoplasm |
Benzamides (MS-275, MCGD0103, CI-994) Cyclic peptide (Depsipeptide, Apicidin) Aliphatic fatty acids (butyrate, valproic acid) Hydroxamate (SAHA, PXD100, LBH589, 4SC-201, Tubacin, ITF2357, PCI2478I) Mercaptoketone (KD5170) |
| Class II | HDAC 4 HDAC 5 HDAC 6 HDAC 7 HDAC 9 HDAC 10 |
Nucleus/cytoplasm Nucleus/cytoplasm Cytoplasm Nucleus/cytoplasm/ mitochondria Nucleus/cytoplasm Nucleus/cytoplasm |
Aliphatic fatty acids (butyrate, valproic acid) Hydroxamate (SAHA, PXD100, LBH589, 4SC-201, Tubacin, ITF2357, PCI2478I) Mercaptoketone (KD5170) |
|
| Class IV | HDAC 11 | Mainly nucleus | Hydroxamate (SAHA, PXD100, IFT2357, 4SC-201) | |
| NAD+ dependent |
Class III | SIRT 1 SIRT 2 SIRT 3 SIRT 4 SIRT 5 SIRT 6 SIRT 7 |
Cytoplasm Cytoplasm/nucleus Mitochondria Mitochondria Mitochondria Nucleus Nucleus |
Hydroxamate (SAHA, PXD100, IFT2357, 4SC-201) Benzamides (MCGD0103 |
|
Histone acetyltransferase | ||||
| Classification | Localization |
Inhibitors (exemples, * non selective) |
||
| Zn+ dependent |
Cytoplasmic | KAT1 (HAT1) HAT4 (NAA60) HAT2 HATB3.1 Rtt109 |
Cytoplasm | *Anacardic acid *Isothiazolones |
| GNAT (bromodomain) | KAT2A (Gcn5) KAT2B (PCAF) ELP3 |
Nucleus | Ischemin Ischemin *Anacardic acid *Isothiazolones |
|
| MYST (acetyl-CoA motif) | KAT5 (TIP60) KAT6A (MOZ, MYST3) KAT6B (MORF,MYST4) KAT7 (HBO1, MYST2) KAT8 (MOF, MYST1) |
Nucleus | TH1834 *Anacardic acid *Isothiazolones |
|
| NAD+ dependent |
P300/CBP | KAT3B (p300) KAT3A (CBP) |
Nucleus | Garcinol, Curcumin, benzylidene barbituric acid, C646, CTPB, TTk21 TTK21, ICG-001, Ischemin |
|
Transcription co-activators |
KAT4 (TAF1, TBP) KAT12 (TIFIIIC90) |
Nucleus | *Anacardic acid *Isothiazolones |
|
|
Steroid receptor co-actovators |
KAT13A (SRC1) KAT13B (SCR3, ACTR) KAT13C (p600) KAT13D (CLOCK) |
Nucleus | *Anacardic acid *Isothiazolones |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




