1. Introduction
Artificial intelligence (AI) is versatile and supports people in various areas, such as intelligent medicine, transportation, education, and entertainment. Brain-control interface (BCI) is a research field in AI that learns and extracts information in human brain waves to control assistive devices such as prosthetic hands [
1], intelligent wheelchairs [
2], and exoskeletons [
3]. In 2013, the Centers for Disease Control and Prevention estimated that about 5.4 million people, which accounted for 1.7% of the United States population, had been living with paralysis [
4]. Regarding the methods of collecting brain waves, BCI can be categorised into invasive, partially invasive, and non-invasive. Electrode devices are implanted directly into human skulls for invasive methods. However, the risk and potential complications after surgery restricted patients to invasive methods. Non-invasive methods place electrodes on the scalp’s surface to record the data. There are many non-invasive modalities such as electroencephalography (EEG) [
5], Positron Emission Tomography (PET) [
6], Magnetoencephalography (MEG) [
7], functional Near-Infrared Spectroscopy (fNIRS) [
8], and Magnetic Resonance Imaging (fMRI) [
9].
Convolutional neural network methods have contributed to most of the significantly advanced studies in brain control interfaces by effectively extracting spatial and temporal features from EEG signals. However, the emergence of Transformer methods, which advanced techniques with powerful self-attention mechanisms, promises to enhance the modelling of long-term dependencies and feature relationships. This study explores the integration of CNN and Transformer techniques, aiming to leverage their combined strengths for improved accuracy and efficiency in motor imagery BCI applications. By extensively evaluating the EEG-TCNet and EEG-Conformer models, including their architecture, hyperparameters, and the impact of bandpass filtering, this research seeks to identify the most effective strategies for enhancing overall performance. Additionally, this study introduces EEG-TCNTransformer, a novel BCI-MI model that incorporates TCN and self-attention and achieves an accuracy of 82.97% without a bandpass filter.
2. Background
Traditional machine learning methods require handcrafted feature extractors and a classification network. In terms of extracting the spatial features, common spatial pattern (CSP) [
10] or extended version of CSP, which is filter bank CSP (FBCSP) [
11], are the most common methods. Moreover, short-time Fourier transform (STFT) [
12] and continuous wavelet transform (CWT)[
13] are used to extract the time-frequency features. Additionally, the feature extractors must be combined with classification, such as support vector machine (SVM) [
14], Bayesian classifier [
11], and linear discriminant analysis (LDA) [
15] to classify the target tasks. However, EEG signals are non-stationary and have a low signal-to-noise ratio, so deep learning methods are researched to improve the weaknesses of traditional machine learning methods.
Regarding traditional deep learning methods, convolutional neural network (CNN) methods are the most common methods that achieve various state-of-the-art studies. Lawhern proposed EEGNet [
16], which is applied to multiple BCI paradigms, including movement-related cortical potentials (MRCP), error-related negativity responses (ERN), sensory-motor rhythms (SMR) and P300 visual-evoked potentials. ShallowNet, proposed by Schirrmeister (2017) [
17], measured the performance of different deep levels in convolution networks, from 5 convolution layers to 31 convolution layer residual networks, using ResNet architecture. EEG-TCNet [
18] incorporates convolution layers in EEGNet and temporal convolutional networks (TCN) [
19], which can exponentially increase the receptive field size by using dilated causal convolution. Moreover, recurrent neural networks (RNN), gated recurrent unit (GRU), and long short-term memory (LSTM) are combined with CNN methods to learn long sequence data [
20].
The transformer method is an advanced method that allows learning features in parallel and improves learning long sequence dependency [
21]. Because of the structure of multi-head attention, the transformer method can efficiently capture the relationships in features through attention mechanisms. Liu proposed a Transformer-based one-dimensional convolutional neural network model (CNN-Transformer), which uses 1D-CNN to extract low-level time-frequency features and a transformer module to extract higher-level features [
22]. Hybrid CNN-Transformer [
23] was proposed as a complex architecture with spatial transformer and spectral transformer to encode the raw EEG signals to attention map, then utilise CNN to explore the local features and temporal transformer to perceive global temporal features. EEG-Conformer [
24], proposed by Song, is a lightweight model with a shallow convolution to extract temporal and spatial features and self-attention mechanisms to learn the relationships in the time domain.
This study aims to research the contributions of different hyperparameters and different bandpass filter parameters to CNN method and Transformer method in MI-BCI. The EEG-TCNet and EEG-Conformer were selected for the study. Therefore, there are two sub-projects as follows
First sub-project: EEG-TCNet and EEG-Conformer are trained and validated with different hyperparameters (i.e. filters, kernel sizes).
Second sub-project: Pre-processing EEG signals with a bandpass filter, then input in training and testing EEG-TCNet and EEG-Conformer.
In this research, a new model is introduced, EEG-TCNTransformer, inspired by EEG-TCNet and EEG-Conformer. EEG-TCNTransformer has the advantage of learning the low-level temporal features and spatial features by the novel convolution architecture from EEG-TCNet and extracting the global temporal correlation from the self-attention mechanism. Moreover, the EEG-TCNTransformer uses the raw EEG data, so the bandpass filter is omitted.
3. Methods
3.1. Dataset
The data studied in this project are from the BCI Competition in 2008 [
25].
Data Group 2A is a set of experimental data on motor imagery for electric wheelchair control. It includes four motor tasks: move the left hand, move the right hand, move both feet and move the tongue, which represents turn left, turn right, move forward and move backward, respectively. This dataset includes 22 EEG channels and 3 EOG channels, collected from nine participants over two sessions on two different days. Each session consists of 6 runs with 48 trials each. The data of each session is stored in one MATLAB file.
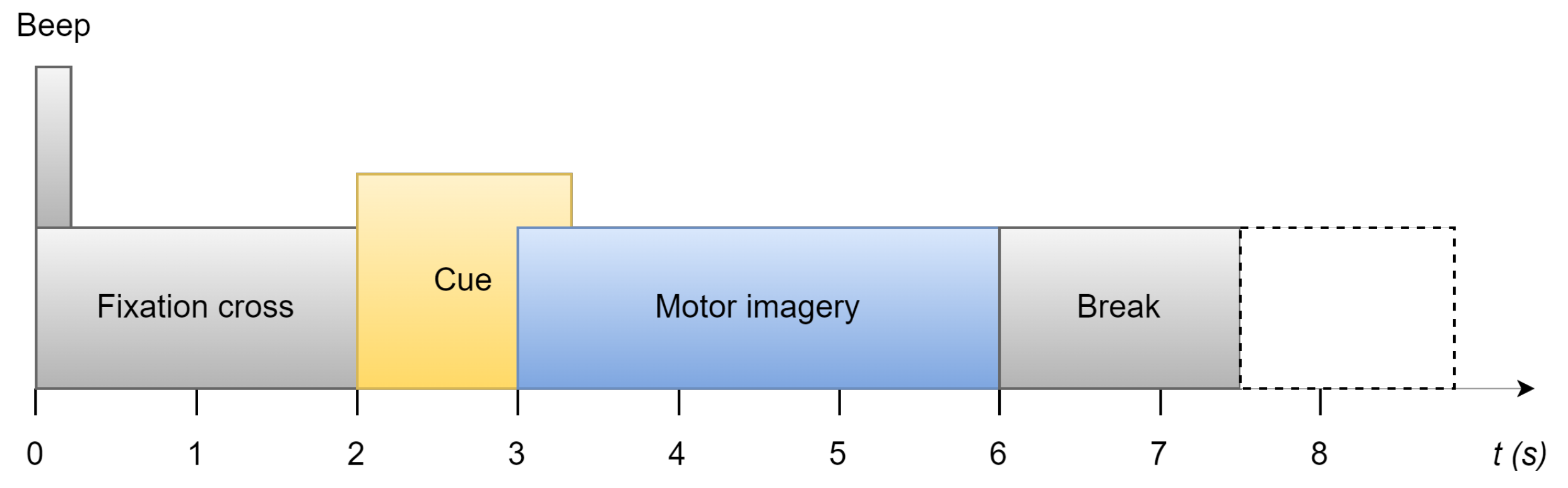
In
Figure 1, each trial is between 7.5 seconds and 8.5 seconds. It starts with a prompt beep and a cross displayed on the screen. Two seconds later, the cue of this trial displays on the screen in the form of an arrow. The four directions it points to represent moving forward, moving backward, turning left, and turning right, respectively. This instruction stays on the screen for 1.25 seconds. The participant is required to perform motor imagery for the next 3 seconds. At the end of the trial, there is a 1.5-second short break before the next trial, which could be increased randomly to 2.5 seconds to avoid adaption.
At the beginning of the data collection, EOG data for each subject was also recorded for the elimination of interference information. These EOG data are removed from this study.
In this study, EEG-LAB is used as the primary tool for data visualisation. EEG-LAB is an add-on for MATLAB that provides various features for analysing electrophysiological data, including processing of EEG, visualisation and integration of algorithms.
Figure 2 shows the visualisation of the EGG signals in EEG-LAB.
3.2. Preprocessing
Data preprocessing was conducted to ensure the best and most relevant data for training the models, employing multiple techniques.
The three extra EOG channels measure eye movements and are easily influenced by non-study-induced factors such as attention levels and blinks. Therefore, these channels were excluded from training to avoid introducing noise and confounding variables.
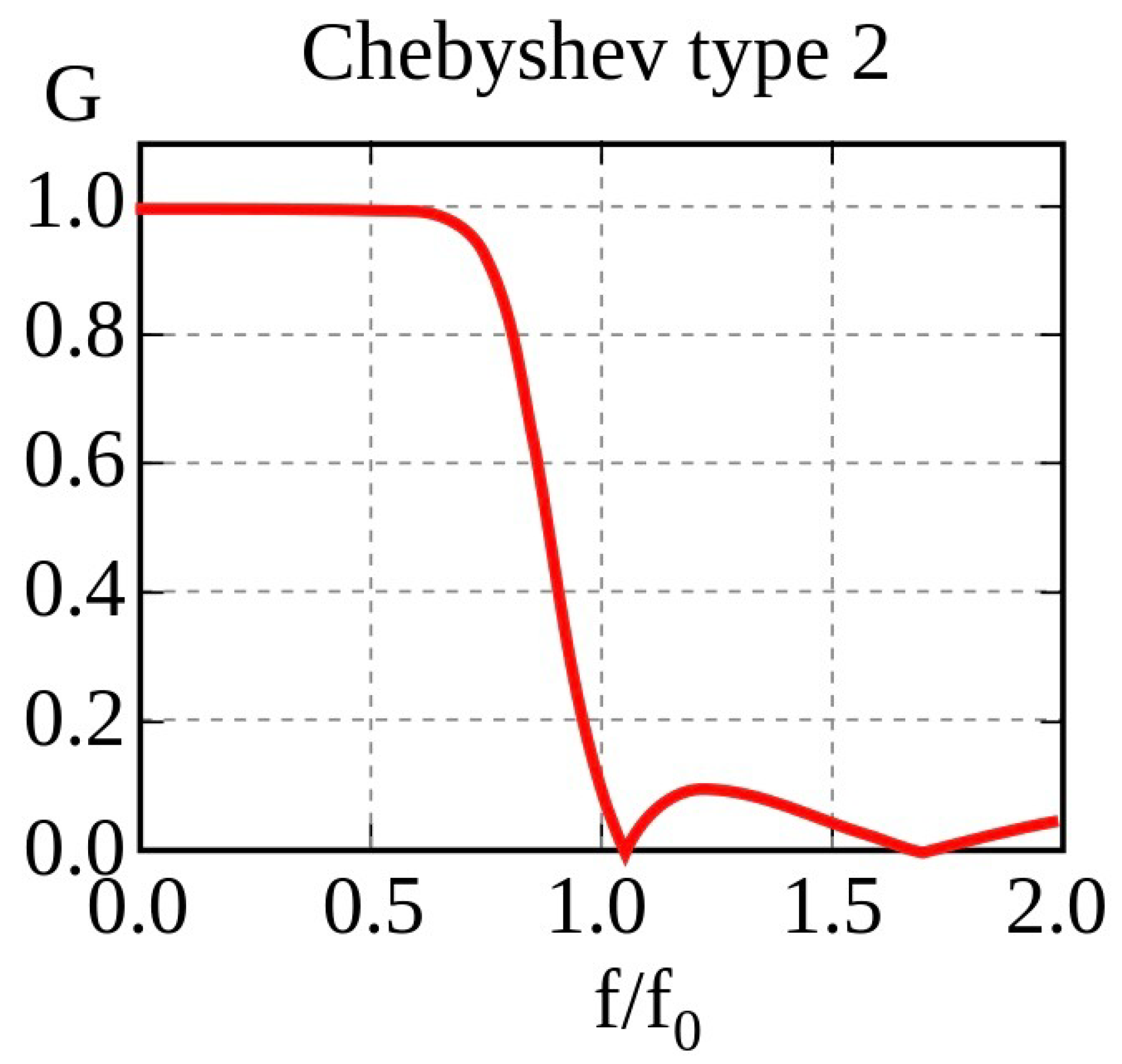
During motor imagery, there is usually a change in neural oscillations indicating Event-Related Desynchronisation (ERD) and Event-Related Synchronisation (ERS), which are mostly observed in the alpha (8–13 Hz) and beta frequency bands (18–24 Hz) [
26]. These provide good features for model training. Signal filtration was employed to increase the signal-to-noise ratio. The sharp transition band between the pass and stop bands, as well as the low ripple in the pass band, made the Chebyshev Type II BandPass Filter (
Figure 3) the filter of choice. The effectiveness of this filter is primarily determined by the order of the filter
N and, to a lesser extent, the stop-band attenuation requirement
The amplitude response is given as
Where
|
A parameter related to the ripples in the stop-band |
|
Is the -order Chebyshev polynomial |
|
Is the stop-band edge frequency |
| f |
Is the frequency variable |
The ripple factor is related to the stop-band attenuation by
These two hyper-parameters, N and were included in the hyper-parameter optimisation process.
In addition, z-score normalisation is applied to the signals to ensure all channels are on the same scale, increasing comparability and handling variability in the dataset. The z-score normalisation is defined as
Where
| z |
standardised value |
| x |
original value |
|
mean of the distribution |
|
standard deviation of the distribution |
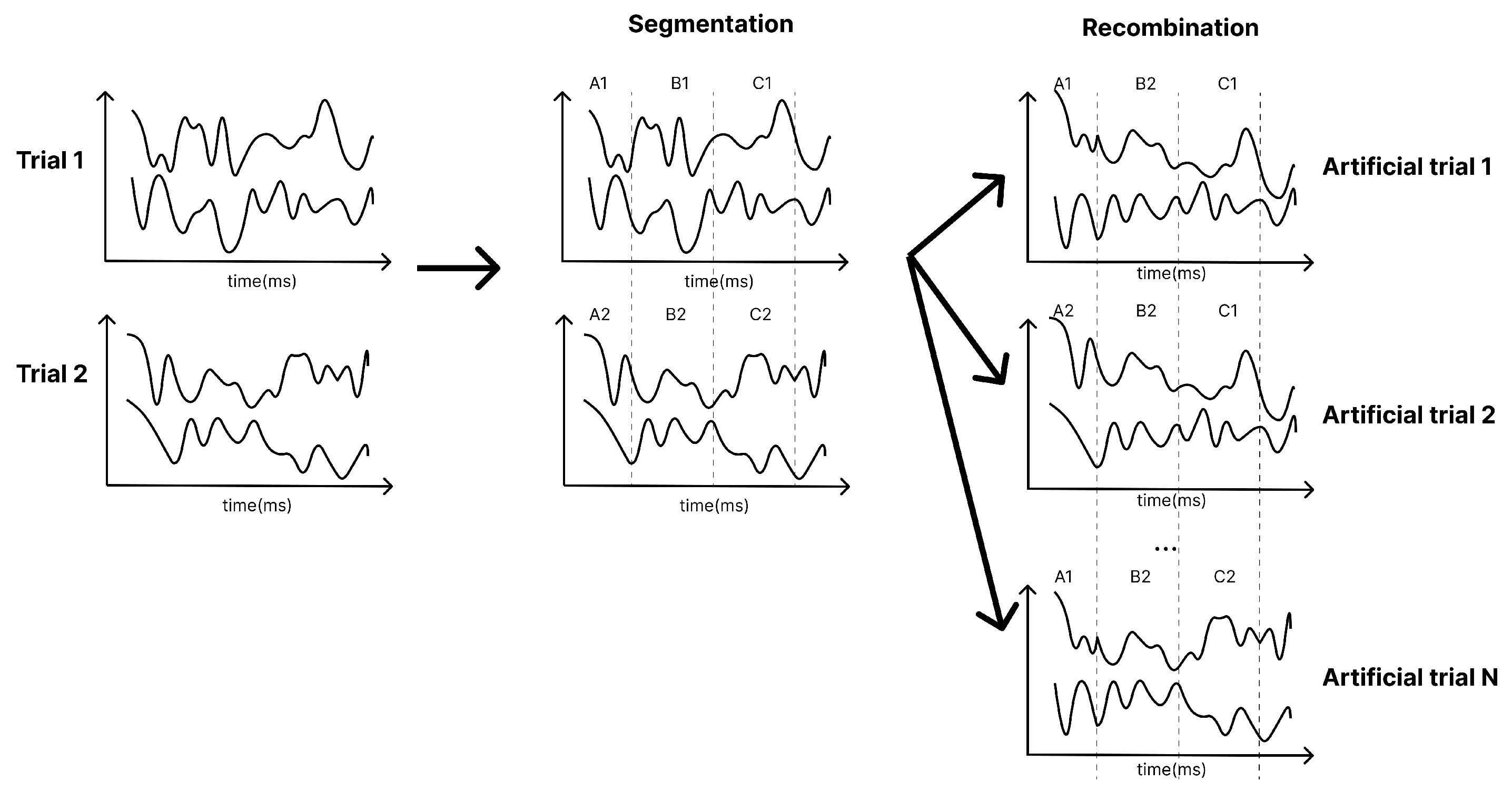
Furthermore, due to the limited size of the training dataset, data augmentation was applied by segmenting and recombining signals in the time domain [
28]. As illustrated in
Figure 4, each training trial is divided into multiple non-overlapping segments. A large amount of new data is generated by randomly concatenating segments from different trials of the same class. This technique was specifically applied to the Conformer model. Further research is needed to analyse the impact of data augmentation across a broader range of models.
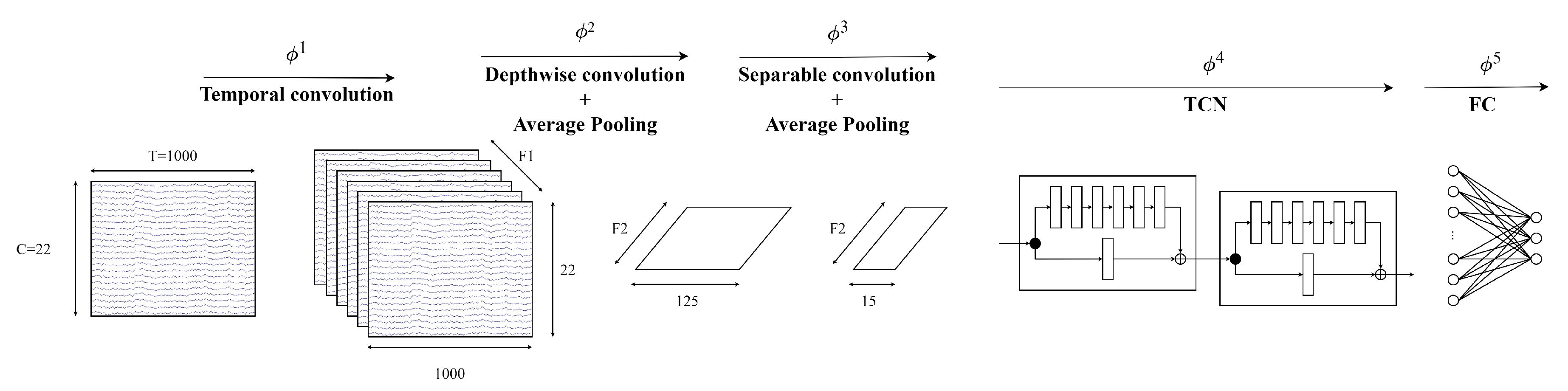
3.3. EEG-TCNet
The first model in this study is EEG-TCNet, which is incorporated from the EEGNet and TCN modules. The EEGNet module in EEG-TCNet is slightly modified, with 2D temporal convolution at the beginning to extract the frequency filters, followed by a depthwise convolution to extract frequency-specific spatial filters. The third part of the EEGNet module is a separable convolution extract temporal summary for each feature map from depthwise convolution and mixes the feature maps before going into the TCN module.
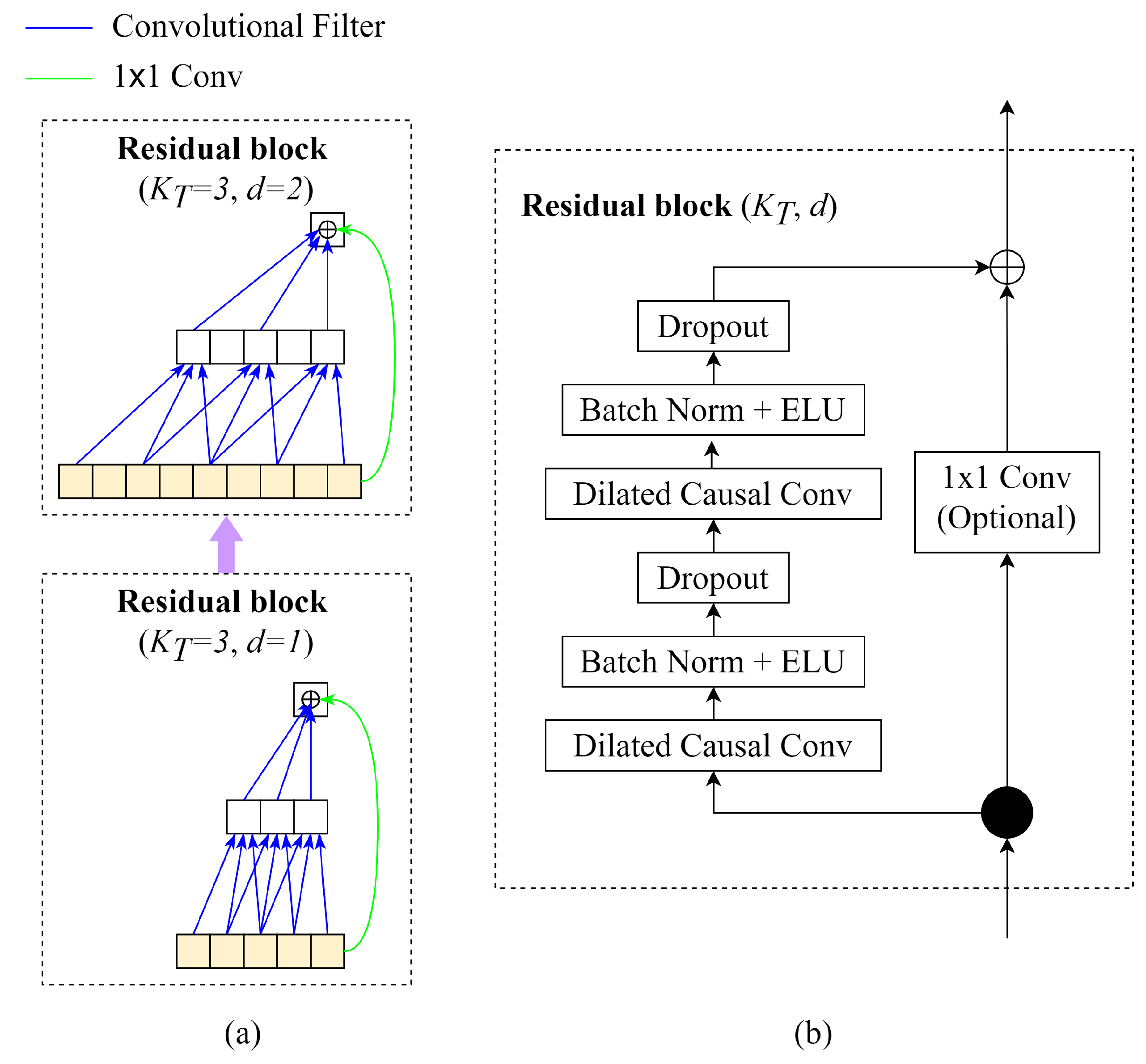
Regarding the TCN module, based on the idea of the temporal convolutional network (TCN), the TCN module in EEG-TCNet could be described as follows (
Figure 5)
- (a)
Causal Convolutions: TCN aims to produce the output of the same size as the input, so TCN uses a 1D fully-convolutional network with each hidden layer having the same size as the input layer. To maintain the length of subsequent layers equal to the previous ones, zero padding of length (kernel size-1) is applied. Moreover, to prevent information leakage from feature to the past in training, only the inputs from time t and earlier determine the output at time t.
- (b)
Dilated Convolutions: The major disadvantage of regular causal convolutions is that the network must be extremely deep or have large filters to obtain a large receptive field size. A sequence of dilated convolutions could increase the receptive field exponentially by increasing dilation factors d.
- (c)
Residual Blocks: In EEG-TCNet, the weight normalisation in the original TCN is replaced by batch normalisation. The residual blocks have a stack of two layers of dilated convolution, batch normalisation with ELU, and dropout. The skip connection 1x1 convolution is applied from the input to the output feature map to ensure the same shape of input and output tensors.
Figure 6 visualises the architecture of EEG-TCNet.
3.4. EEG-Conformer
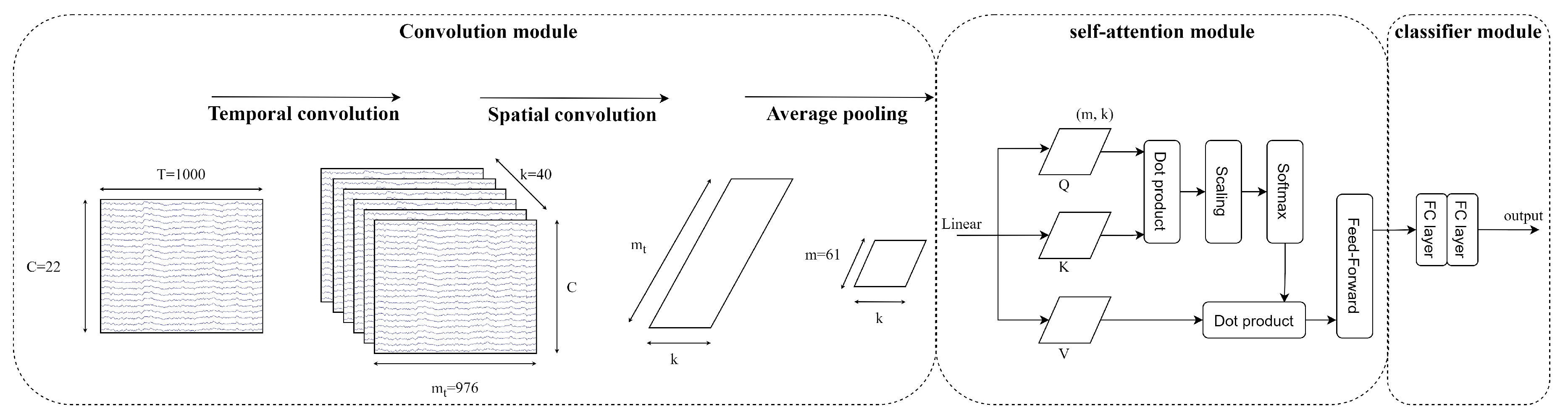
The second model in this study is EEG-Conformer, proposed by Song [
24]. Based on the concept of using CNN to learn the low-level features of temporal and spatial in EEG signals combined with self-attention to learn the global temporal dependencies in EEG feature maps. The EEG-Conformer could be described in detail as follows
- (a)
Convolution module: The EEG signals after pre-processing and augmenting are learned through two 1D temporal and spatial convolution layers. The first temporal convolution layer learns the time dimension of input, and then the spatial convolution layer learns the spatial features. The spatial features are the information between the electrode channels. Then, the feature maps go through batch normalisation to avoid overfitting and the nonlinearity activation function ELU. The average pooling layer smooths the feature maps and rearranges to put all feature channels of each temporal point as a token into the self-attention module.
- (b)
-
Self-Attention module: The self-attention module has advantages in learning global features and long sequence data, which improve the CNN weaknesses. The self-attention module calculates as below
Where Q is a matrix of stacked temporal queries, K and V are temporal matrices that denote key and value, and k is the length of tokens.
Multi-head attention (MHA) is applied to strengthen representation performance. The tokens are separated into the same shape
h segments, described as follows
Where MHA is the multi-head attention indicate the query, key and value acquired by a linear transformation of segment token in the head, respectively.
- (c)
-
Classifier module: The features from the self-attention module go through two fully connected layers. The Softmax function is utilised to predict M labels. Cross-entropy loss is utilised as a cost function
Where is the batch size, M is the number of target labels (M=4), y is the ground truth and is the prediction.
Figure 7 visualises the architecture of EEG-Conformer architecture, including the convolution module, self-attention module and classifier module.
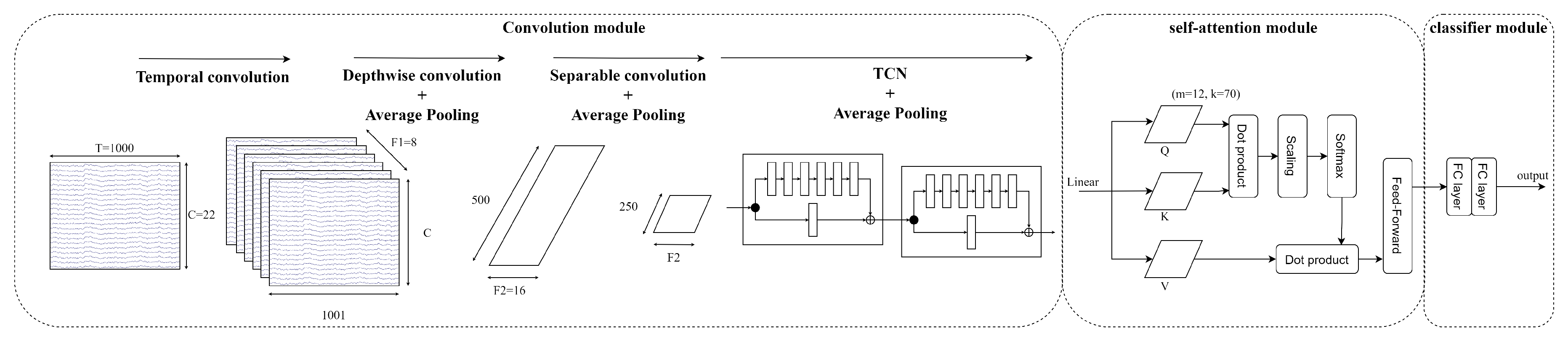
3.5. EEG-TCNTransformer
The concept of EEG-TCNTransformer is inspired by the EEG-TCNet and EEG-Conformer. EEG-TCNet performs well in learning the local features in EEG signals because of the EEGNet and TCN modules. Besides that, EEG-Conformer solved the problem of global feature dependencies, which improved the learning of long sequence inputs. In EEG-TCNTransformer has a convolution module, self-attention module and classifier module
- (a)
Convolution module: The sequence of 2D convolution layer, depthwise convolution layer, separable convolution layer and average pooling layer extract the temporal and spatial information in EEG signals. After that, a sequence of TCN blocks continues to learn large receptive fields efficiently with small parameters because of dilated causal convolution and residual block structure. In this study, the different numbers of filters k in TCN block, and numbers of TCN block T are validated to select the best hyperparameter for EEG-TCNTransformer. Then, the average pooling layer smooths the features and rearranges features to shape of channels of each temporal point.
- (b)
-
Self-attention module: Six self-attention blocks with multi-head attention architecture learn the global temporal dependencies in features from the convolution module. The self-attention module calculates as below
Where Q is a matrix of stacked temporal queries, K and V are temporal matrices that denote key and value, and k is the length of tokens.
Multi-head attention (MHA) is applied to strengthen representation performance. The tokens are separated into the same shape
h segments, described as follows
Where MHA is the multi-head attention indicate the query, key and value acquired by a linear transformation of segment token in the head, respectively.
- (c)
-
Classifier module: Two fully-connected layers transform the features into M labels that calculate probability distribution by the Softmax function. The loss function is cross-entropy loss
Where is the batch size, M is the number of target labels (M=4), y is the ground truth, and is the prediction.
Figure 8 visualises the architecture of EEG-TCNTransformer architecture, including the convolution module, self-attention module and classifier module.
4. Experiments and Results
4.1. Experimental Setup
The training environment was set up on a Linux computer with CPU AMD Ryzen 9 5900X, GPU RTX 3080 and CUDA 12.1. EEG-TCNet was implemented in Python 3.6.8 code using TensorFlow. EEG-Conformer and EEG-TCNTransformer were implemented in Python 3.10.14 code using Pytorch. The Conda environment is used in this experiment to change the Python version.
Regarding EEG-TCNet, each subject is trained 1000 epochs with a learning rate of 0.001, and the activation function is ELU.
In terms of EEG-Conformer, each subject is trained 2000 epochs with a learning rate of 0.0002; the activation function in the convolution module and classification module is ELU, and the activation function in self-attention blocks is GELU.
With respect to EEG-TCNTransformer, each subject is trained 5000 epochs with a learning rate of 0.0002; the activation function in EEGNet module, TCN module and classification module is ELU, and the activation function in self-attention blocks is GELU.
The accuracy metric is utilised to measure the performance.
4.2. EEG-TCNet with Different Hyperparameters
The detailed architecture of EEG-TCNet is described in
Table 1.
C and
T denote the number of EEG channels and the number of time points. In the first sub-project,
and
because the time points used in training are 4 seconds from the cue section to the end of the motor imagery section. The baseline hyperparameters of EEG-TCNet are
as same as the authors. Besides that, the dropout values in the EEGNet module and the TCN module are
, the same as the original paper.
Where
| C |
Number of EEG channels |
| T |
Number of timepoints |
|
Number of temporal filters |
|
Number of spatial filters |
|
Number of filters in TCN module |
|
Kernel size in first convolution |
|
Kernel size in TCN module |
|
Dropout in EEGNet module |
|
Dropout in TCN module |
In the first sub-project, EEG-TCNet is trained with different numbers of filters and kernel sizes in the EEGNet module and TCN module, described in
Table 2.
In
Table 3, the contribution of each
to the performance of EEG-TCNet is examined by keeping the baseline value of three parameters and changing one parameter. The average accuracy of EEG-TCNet increases when
and
increase. Increasing
to 32 and
to 6 also improve the accuracy, but the performance declines when increasing
to 48 and
to 8.
After training 81 experiments, a fixed result and a variable result are selected. In terms of the fixed result, the best hyperparameters
for all 9 subjects are selected based on the average accuracy. Regarding the variable result, the best hyperparameters
for each subject are selected.
Table 4 shows the hyperparameters of the variable model.
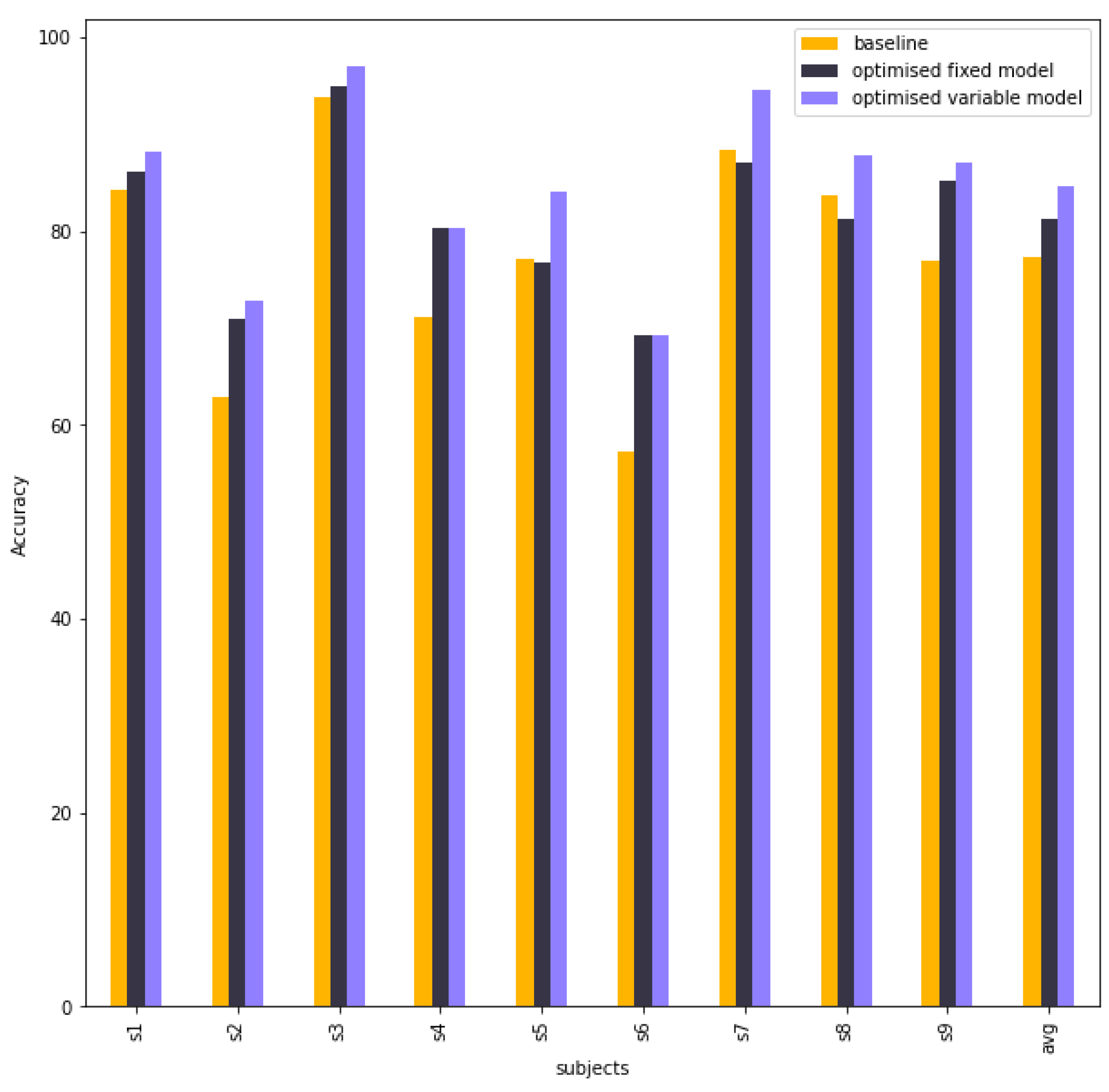
Table 5 compares the accuracy between the baseline model, fixed model and variable model. The fixed model is better than the baseline model in subject 2, subject 4, subject 6, and subject 9, whereas others are nearly equal. When each subject is selected the best result in the variable model, the final performance is much higher than the baseline model, especially subject 2, subject 4, subject 5, subject 6 and subject 9.
Figure 9 visualises the column chart of the accuracy of baseline EEG-TCNet model, fixed EEG-TCNet model and variable EEG-TCNet model.
4.3. EEG-TCNet with Different Bandpass Filter Parameters
In sub-project 2, the EEG-TCNet with baseline hyperparameters is trained with different orders of the filter and the minimum attenuations required in the stop band of Chebyshev type 2, described in
Table 6. The parameters
N and
are defined in
cheby2 of SciPy API for Python.
Where
| N |
The order of the filter |
|
The minimum attenuation required in the stop band |
After training 45 experiments, a fixed result and a variable result are selected. In terms of the fixed result, the best Chebyshev type 2 parameters
for all 9 subjects are selected based on the average accuracy. Regarding the variable result, the best Chebyshev type 2 parameters
N,
for each subject are selected.
Table 7 summarises the variable result.
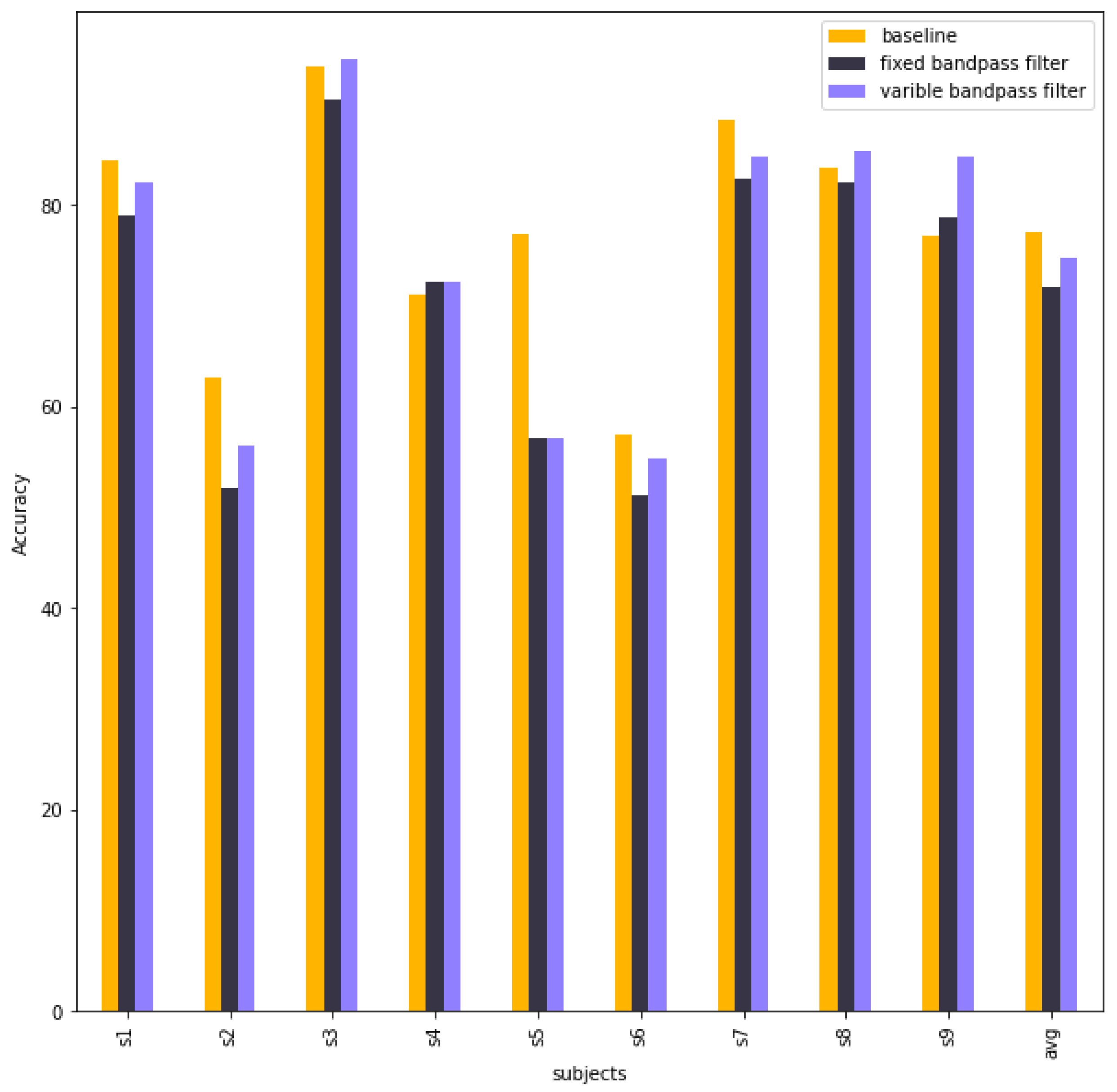
Table 8 compares the accuracy of 9 subjects and the average accuracy between baseline EEG-TCNet, EEG-TCNet with a fixed bandpass filter and EEG-Net with a variable bandpass filter. The performance of EEG-TCNet decreased after using The Chebyshev type 2 in pre-processing in both fixed bandpass filter and variable bandpass filter.
Figure 10 illustrates the column chart of the accuracy of baseline EEG-TCNet, EEG-TCNet with a fixed bandpass filter and EEG-Net with a variable bandpass filter.
4.4. EEG-Conformer with Different Hyperparameters
The architecture of the convolution module of EEG-Conformer is described in
Table 9
Where
| k |
Number of temporal filters in the convolution module |
|
Number of channels in EEG data |
In sub-project 1 with EEG-Conformer, the model is trained with different numbers of temporal filters in convolution module
k and different numbers of self-attention blocks
N, described in
Table 10. The baseline model in the original study has
and
.
In
Table 11, the contribution of each
to the performance of EEG-TCNet is examined by keeping the baseline value of one parameter and changing one parameter. The increasing
k improves the average accuracy, but the performance declines when
k rises from 40 to 60. The average accuracy decreases when
N is from 6 to 7 and then increases slightly when
N is from 7 to 10, but the average accuracy is still lower than
N of 6.
After training 25 experiments, the fixed hyperparameters selected are
and
. The variable hyperparameters are described in
Table 12.
Table 13 and
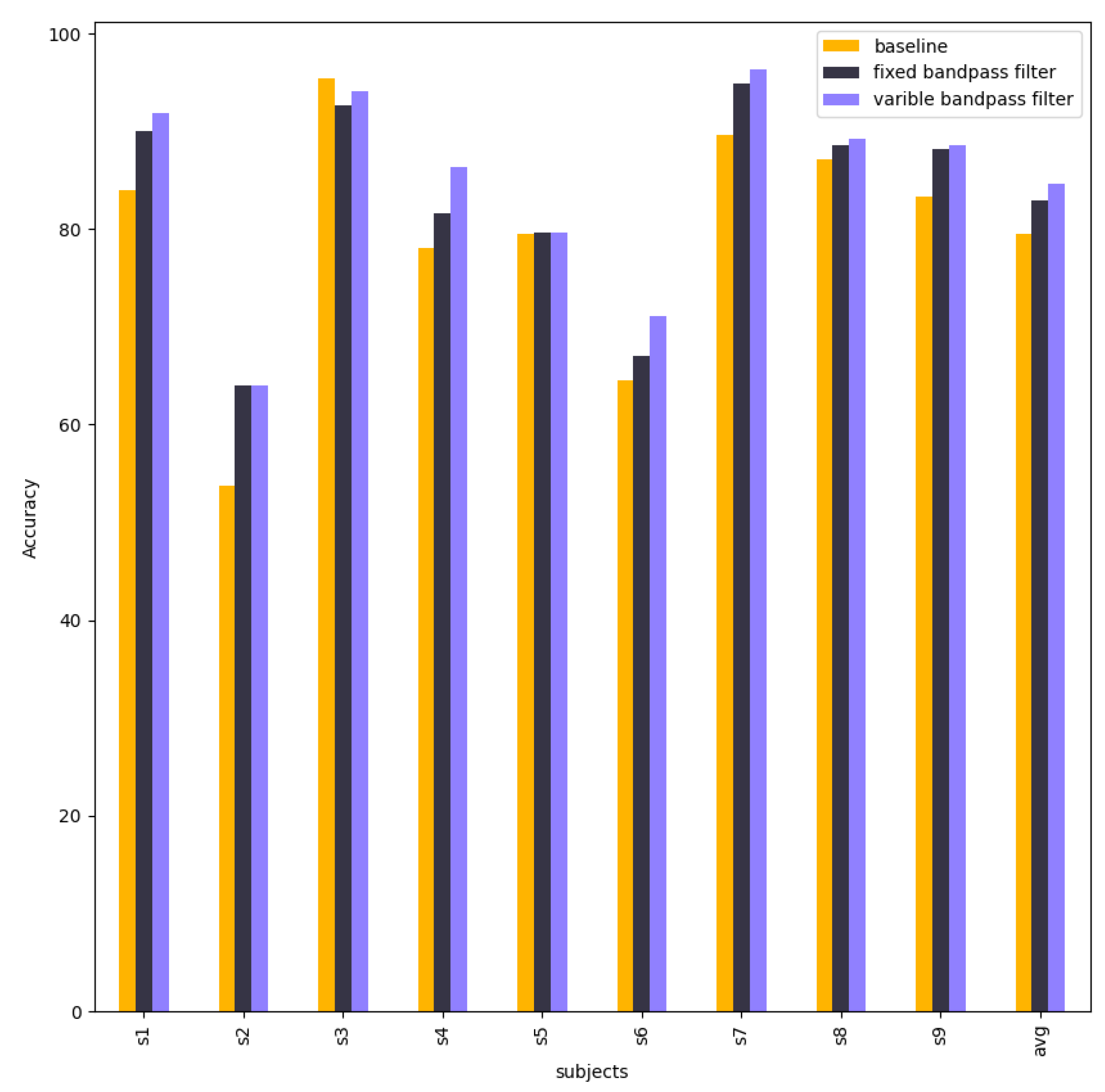
Figure 11 compare and visualise the accuracy of the EEG-Conformer baseline model, fixed model, and variable models. The accuracies of them are nearly equal in each subject. However, subject 2 in the fixed model has higher accuracy than the baseline model.
4.5. EEG-Conformer with Different Bandpass Filter Parameters
In the second sub-project with EEG-Conformer, the baseline model is trained with different orders of the filter and the minimum attenuations required in the stop band of Chebyshev type 2, described in
Table 14.
After training 45 experiments, a fixed result and a variable result are selected. In terms of the fixed result, the best Chebyshev type 2 parameters
for all 9 subjects are selected based on the average accuracy. Regarding the variable result, the best Chebyshev type 2 parameters
for each subject are selected.
Table 15 summarises the variable result.
Table 16 and
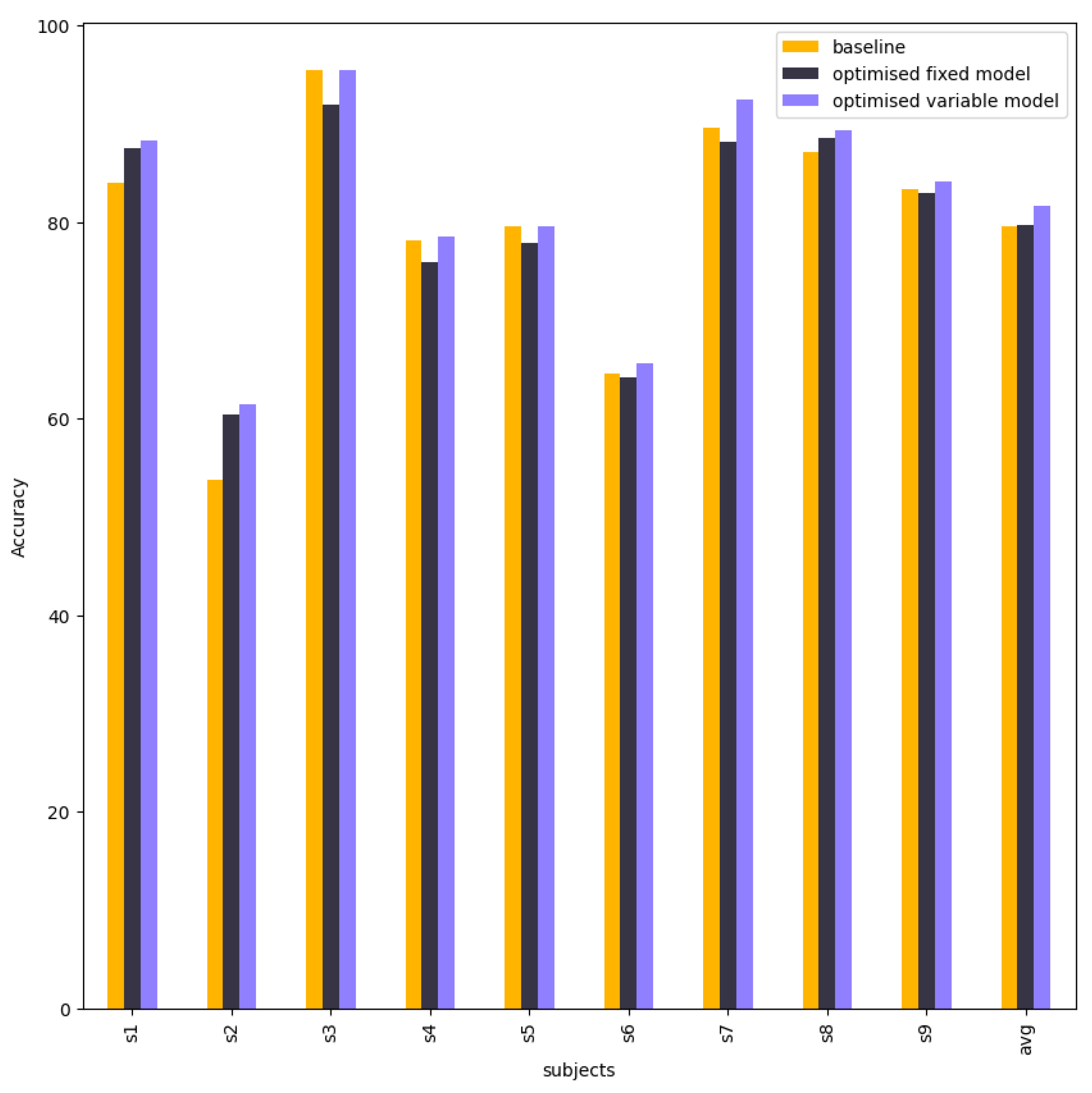
Figure 12 compare and visualise the accuracy of the baseline EEG-Conformer, EEG-Conformer with fixed bandpass filter and EEG-Conformer with variable bandpass filter. The Chebyshev type 2 contributes to improving the accuracy of subject 1, subject 2, subject 4 and subject 7. Other subjects are approximate to the baseline model. The variable result got an average accuracy of 84.61%.
4.6. EEG-TCNTransformer with Different Hyperparameters
The different number of filters
k in TCN block, number of TCN block
T is validated to select the best hyperparameters. In
Table 17, the average of all subjects is summarised with different
k and
T. With each
T, the more filters
k, the higher the average accuracy, but the average accuracy stops increasing and decreases at a level of filters. With
, the average accuracy is highest at k=60. With
, the average accuracy is highest at k=60. With
, the average accuracy is highest at k=70. Overall,
and
produce the best average accuracy of 82.97%.
Table 18 compares the accuracy between EEG-TCNTransformer with
,
, baseline EEG-TCNet, and baseline EEG-Conformer. EEG-TCNTransformer improves the performance of subject 2, which EEG-Conformer is lower than EEG-TCNet. Moreover, EEG-TCNTransformer improves subject 9 by about 5.69% compared to EEG-Conformer. Subjects 1, 4, 5, 6, and 8 also improved from 1.41% up to 3.56%.
4.7. EEG-TCNTransformer with Different Bandpass Filter Parameters
The bandpass filter is also added to the pre-processing to validate the performance of the EEG-TCNTransformer.
Table 19 describes the average accuracy of different orders of filter
N=[1, 2, 3, 4, 5, 6, 7, 8, 9, 10] with the minimum attenuations required in the stop band
=[10, 20, 30]. The highest average accuracy of 9 subjects when training EEG-TCNTransformer with bandpass filter is 80.27% while increasing
reduces the performance. Therefore, the bandpass filter is inefficient for the EEG-TCNTransformer.
5. Discussion
This research aims to study the contribution of changing hyperparameters and changing bandpass filter parameters of Chebyshev type 2 to a CNN method and a Transformer method in Brain-Computer Interface motor imagery. Regarding the CNN method, EEG-TCNet is a novel study for research. The increase in the number of filters and kernel sizes in EEG-TCNet architecture provides better performance. However, the bandpass filter Chebyshev type 2 in data pre-processing is inefficient for training EEG-TCNet. In terms of the Transformer method, the EEG-Conformer is chosen for research. The average accuracy improves when the number of filters is from 20 to 40. The Chebyshev type 2 bandpass filter was found to contribute significantly to the accuracy of each subject, producing an average accuracy of 82.97%. However, EEG-TCNet still has other hyperparameters such as dropout, depth of TCN blocks, depth in EEGNet module or EEG-Conformer still has dropout and head number in multi-head that cannot be examined in this study because of the limit of training resource and time.
In this research, a new model called EEG-TCNTransformer is created, which combines the convolution parts of EEG-TCNet and the sequence of self-attention blocks. The average accuracy of EEG-TCNTransformer improves when the number of TNC blocks increases, which learns better the low-level temporal features in temporal and spatial features. The number of filters also contributes to the performance, but it gets overfitting at specific filters. Moreover, the bandpass filter is inefficient for EEG-TCNTransformer, so the raw EEG data with standardisation is used in training and testing. Therefore, the optimised hyperparameters for EEG-TCNTransformerer generate better accuracy for each subject compared to baseline EEG-TCNet and baseline EEG-Conformer without bandpass filter in pre-processing, and the average accuracy of EEG-TCNTransformer is 82.97%.
6. Conclusion
Brain-computer interface motor imagery is an emerging research topic in Artificial Intelligence. This study explores the contributions of hyperparameters and bandpass filters to the performance of EEG-TCNet and EEG-Conformer. The average accuracy of EEG-TCNet was improved when increasing the number of filters and kernel size, but the bandpass filter is inefficient in training. In contrast, the bandpass filter significantly improves the accuracy of EEG-Conformer, where the performance is unimproved when increasing the number of self-attention blocks. Furthermore, a new model, EEG-TCNTransformer, which incorporates the convolution architecture of EEG-TCNet and a stack of self-attentions with a multi-head structure. This EEG-TCNTransformer outperforms with an average accuracy of 82.97%. In future, various techniques within the transformer architecture will be explored to enhance the global feature correlation for EEG-TCNTransformer.
Author Contributions
Conceptualization, A.H.P.N., O.O.; methodology, A.H.P.N., O.O.; software, A.H.P.N.; validation, A.H.P.N.; formal analysis, A.H.P.N.; investigation, A.H.P.N., O.O.; resources, A.H.P.N.; data curation, A.H.P.N.; writing—original draft preparation, A.H.P.N., O.O.; writing—review and editing, A.H.P.N., O.O., M.A.P, S.H.L; visualization, A.H.P.N.; supervision, M.A.P, S.H.L; project administration, M.A.P, S.H.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding
Institutional Review Board Statement
This study’s EEG dataset is already accessible in the BNCI Horizon 2020 database. For this research, no data were collected from human subjects.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
Conflicts of Interest
The authors declare no conflicts of interest.
Appendix A. TCNet Training Results
Appendix A.1. Experiment EEG-TCNet with Different Hyperparameters
Table A1 shows the accuracy of 9 subjects when changing
in EEG-TCNet architecture.
Table A1.
Accuracy of EEG-TCNet with different
Table A1.
Accuracy of EEG-TCNet with different
|
|
|
|
S1 |
S2 |
S3 |
S4 |
S5 |
S6 |
S7 |
S8 |
S9 |
Average Accuracy (%) |
| 6 |
16 |
10 |
4 |
79.36 |
59.36 |
91.21 |
67.11 |
75.72 |
56.28 |
75.09 |
81.92 |
75.00 |
73.45 |
| 6 |
16 |
10 |
6 |
82.21 |
60.07 |
94.51 |
72.37 |
75.72 |
62.33 |
76.17 |
84.13 |
79.55 |
76.34 |
| 6 |
16 |
10 |
8 |
82.92 |
59.72 |
95.24 |
74.56 |
75.72 |
66.98 |
81.23 |
82.66 |
76.52 |
77.28 |
| 6 |
16 |
12 |
4 |
80.78 |
66.43 |
93.77 |
66.23 |
77.90 |
58.14 |
93.14 |
83.03 |
82.95 |
78.04 |
| 6 |
16 |
12 |
6 |
80.43 |
60.78 |
93.77 |
76.75 |
80.80 |
64.19 |
88.45 |
83.03 |
83.33 |
79.06 |
| 6 |
16 |
12 |
8 |
83.63 |
67.49 |
91.21 |
67.98 |
80.80 |
65.12 |
76.53 |
83.76 |
84.47 |
77.89 |
| 6 |
16 |
14 |
4 |
75.09 |
53.00 |
88.28 |
67.11 |
75.72 |
54.88 |
76.17 |
81.55 |
75.00 |
71.87 |
| 6 |
16 |
14 |
6 |
78.65 |
61.48 |
89.38 |
73.68 |
75.72 |
64.65 |
92.78 |
82.29 |
76.89 |
77.28 |
| 6 |
16 |
14 |
8 |
82.56 |
60.42 |
95.60 |
74.12 |
75.72 |
68.84 |
92.06 |
76.75 |
76.14 |
78.02 |
| 6 |
32 |
10 |
4 |
76.51 |
63.96 |
91.21 |
72.37 |
79.35 |
53.49 |
75.09 |
82.29 |
75.38 |
74.40 |
| 6 |
32 |
10 |
6 |
82.21 |
68.20 |
94.51 |
65.35 |
76.09 |
64.65 |
91.34 |
80.07 |
80.68 |
78.12 |
| 6 |
32 |
10 |
8 |
76.16 |
57.95 |
95.24 |
75.44 |
78.99 |
63.26 |
88.09 |
86.72 |
85.23 |
78.56 |
| 6 |
32 |
12 |
4 |
81.49 |
56.89 |
91.58 |
70.18 |
72.10 |
61.40 |
77.62 |
82.66 |
84.47 |
75.38 |
| 6 |
32 |
12 |
6 |
80.43 |
65.37 |
94.87 |
74.56 |
79.35 |
61.86 |
91.70 |
80.44 |
81.44 |
78.89 |
| 6 |
32 |
12 |
8 |
82.56 |
56.18 |
93.41 |
75.88 |
78.62 |
61.40 |
91.70 |
84.87 |
85.98 |
78.96 |
| 6 |
32 |
14 |
4 |
83.63 |
59.01 |
94.51 |
67.98 |
70.65 |
60.00 |
90.25 |
78.60 |
79.17 |
75.98 |
| 6 |
32 |
14 |
6 |
76.16 |
61.84 |
97.07 |
80.26 |
73.55 |
60.93 |
81.23 |
83.76 |
81.82 |
77.40 |
| 6 |
32 |
14 |
8 |
85.05 |
66.78 |
91.21 |
71.49 |
75.36 |
60.93 |
87.73 |
77.86 |
76.89 |
77.03 |
| 6 |
48 |
10 |
4 |
87.54 |
63.96 |
93.41 |
70.61 |
75.36 |
57.67 |
75.81 |
82.66 |
78.03 |
76.12 |
| 6 |
48 |
10 |
6 |
79.36 |
59.36 |
89.74 |
70.61 |
76.45 |
65.58 |
90.25 |
83.39 |
79.17 |
77.10 |
| 6 |
48 |
10 |
8 |
78.65 |
63.25 |
93.04 |
72.81 |
81.16 |
63.26 |
91.70 |
81.92 |
82.95 |
78.75 |
| 6 |
48 |
12 |
4 |
78.65 |
51.94 |
91.94 |
69.30 |
73.91 |
64.65 |
84.12 |
81.92 |
83.71 |
75.57 |
| 6 |
48 |
12 |
6 |
88.26 |
64.66 |
97.07 |
63.60 |
79.35 |
63.26 |
90.97 |
81.55 |
81.82 |
78.95 |
| 6 |
48 |
12 |
8 |
83.27 |
61.84 |
94.87 |
77.63 |
79.71 |
66.51 |
76.17 |
83.39 |
82.20 |
78.40 |
| 6 |
48 |
14 |
4 |
79.00 |
57.95 |
91.21 |
70.18 |
71.38 |
59.53 |
90.25 |
84.13 |
81.06 |
76.08 |
| 6 |
48 |
14 |
6 |
77.22 |
56.54 |
93.04 |
71.93 |
78.99 |
61.86 |
76.90 |
84.13 |
79.55 |
75.57 |
| 6 |
48 |
14 |
8 |
83.27 |
65.72 |
92.67 |
76.32 |
77.54 |
57.67 |
78.34 |
81.92 |
80.30 |
77.08 |
| 8 |
16 |
10 |
4 |
79.36 |
62.54 |
89.74 |
71.05 |
72.46 |
60.00 |
90.25 |
85.98 |
83.33 |
77.19 |
| 8 |
16 |
10 |
6 |
82.21 |
67.14 |
95.24 |
69.30 |
76.45 |
67.91 |
90.61 |
82.29 |
80.68 |
79.09 |
| 8 |
16 |
10 |
8 |
83.27 |
67.49 |
94.14 |
76.32 |
80.43 |
66.51 |
91.34 |
80.07 |
75.76 |
79.48 |
| 8 |
16 |
12 |
4 |
82.92 |
60.78 |
93.04 |
67.98 |
66.30 |
62.79 |
93.14 |
80.81 |
82.20 |
76.66 |
| 8 |
16 |
12 |
6 |
81.49 |
66.43 |
93.41 |
70.61 |
77.17 |
64.65 |
88.81 |
84.50 |
85.23 |
79.15 |
| 8 |
16 |
12 |
8 |
82.21 |
67.49 |
94.14 |
78.07 |
73.91 |
67.44 |
88.45 |
83.76 |
85.61 |
80.12 |
| 8 |
16 |
14 |
4 |
78.65 |
57.60 |
88.64 |
66.23 |
80.43 |
60.47 |
92.78 |
84.13 |
82.20 |
76.79 |
| 8 |
16 |
14 |
6 |
79.72 |
67.14 |
93.04 |
68.86 |
80.43 |
55.81 |
88.81 |
83.39 |
79.17 |
77.37 |
| 8 |
16 |
14 |
8 |
81.14 |
61.13 |
92.67 |
75.88 |
79.35 |
66.51 |
88.09 |
79.70 |
80.68 |
78.35 |
| 8 |
32 |
10 |
4 |
85.77 |
65.72 |
95.24 |
75.88 |
76.09 |
56.74 |
88.81 |
81.92 |
75.76 |
77.99 |
| 8 |
32 |
10 |
6 |
82.56 |
67.49 |
93.77 |
78.51 |
79.71 |
64.65 |
90.25 |
85.61 |
84.85 |
80.82 |
| 8 |
32 |
10 |
8 |
82.56 |
65.72 |
94.14 |
76.32 |
84.06 |
65.58 |
92.42 |
84.13 |
86.36 |
81.26 |
| 8 |
32 |
12 |
4 |
84.34 |
62.90 |
93.77 |
71.05 |
77.17 |
57.21 |
88.45 |
83.76 |
76.89 |
77.28 |
| 8 |
32 |
12 |
6 |
83.63 |
66.43 |
94.87 |
75.00 |
75.72 |
66.51 |
87.73 |
85.24 |
84.47 |
79.96 |
| 8 |
32 |
12 |
8 |
82.92 |
61.48 |
94.51 |
73.68 |
79.35 |
65.12 |
91.34 |
76.75 |
82.20 |
78.59 |
| 8 |
32 |
14 |
4 |
79.36 |
60.78 |
97.07 |
72.37 |
77.90 |
61.86 |
88.45 |
82.29 |
87.12 |
78.58 |
| 8 |
32 |
14 |
6 |
81.85 |
67.84 |
94.51 |
71.05 |
75.72 |
65.58 |
89.17 |
85.61 |
79.92 |
79.03 |
| 8 |
32 |
14 |
8 |
82.92 |
57.60 |
95.60 |
73.68 |
80.80 |
65.12 |
89.89 |
82.66 |
83.33 |
79.07 |
| 8 |
48 |
10 |
4 |
82.21 |
54.06 |
92.31 |
63.60 |
75.36 |
58.14 |
75.09 |
79.70 |
79.92 |
73.38 |
| 8 |
48 |
10 |
6 |
81.85 |
66.08 |
95.24 |
76.32 |
78.26 |
62.79 |
85.92 |
82.29 |
79.17 |
78.66 |
| 8 |
48 |
10 |
8 |
83.27 |
64.66 |
95.24 |
78.95 |
80.07 |
67.91 |
86.64 |
83.03 |
84.85 |
80.51 |
| 8 |
48 |
12 |
4 |
81.14 |
62.19 |
94.87 |
70.18 |
78.99 |
60.47 |
84.12 |
79.34 |
82.20 |
77.05 |
| 8 |
48 |
12 |
6 |
81.85 |
67.49 |
93.41 |
72.37 |
76.81 |
58.60 |
88.81 |
83.03 |
83.33 |
78.41 |
| 8 |
48 |
12 |
8 |
82.92 |
61.48 |
90.84 |
76.32 |
79.71 |
65.58 |
90.61 |
81.18 |
83.71 |
79.15 |
| 8 |
48 |
14 |
4 |
83.99 |
56.18 |
94.87 |
75.00 |
78.26 |
62.79 |
89.53 |
84.13 |
83.71 |
78.72 |
| 8 |
48 |
14 |
6 |
81.14 |
72.44 |
95.60 |
80.26 |
80.07 |
62.79 |
91.34 |
84.13 |
81.44 |
81.02 |
| 8 |
48 |
14 |
8 |
80.78 |
65.37 |
94.87 |
71.05 |
78.62 |
64.19 |
80.87 |
84.13 |
84.47 |
78.26 |
| 10 |
16 |
10 |
4 |
82.56 |
61.84 |
93.77 |
65.79 |
77.17 |
58.60 |
88.09 |
84.13 |
83.33 |
77.25 |
| 10 |
16 |
10 |
6 |
80.07 |
62.54 |
93.04 |
71.93 |
79.35 |
62.79 |
89.17 |
84.13 |
79.55 |
78.06 |
| 10 |
16 |
10 |
8 |
80.43 |
65.72 |
93.77 |
76.32 |
77.90 |
65.58 |
92.06 |
83.76 |
82.95 |
79.83 |
| 10 |
16 |
12 |
4 |
81.49 |
64.66 |
93.04 |
73.68 |
74.28 |
58.60 |
94.58 |
84.13 |
79.17 |
78.18 |
| 10 |
16 |
12 |
6 |
83.63 |
63.96 |
92.67 |
69.74 |
77.90 |
63.26 |
92.78 |
82.66 |
82.58 |
78.80 |
| 10 |
16 |
12 |
8 |
81.14 |
62.90 |
93.77 |
73.68 |
81.88 |
66.51 |
92.42 |
82.66 |
83.71 |
79.85 |
| 10 |
16 |
14 |
4 |
81.49 |
60.78 |
94.14 |
76.32 |
79.35 |
65.12 |
92.06 |
84.87 |
82.20 |
79.59 |
| 10 |
16 |
14 |
6 |
80.43 |
66.08 |
95.24 |
75.44 |
77.17 |
68.84 |
91.34 |
83.39 |
78.41 |
79.59 |
| 10 |
16 |
14 |
8 |
82.21 |
64.66 |
93.04 |
77.63 |
77.54 |
66.98 |
89.53 |
84.13 |
79.17 |
79.43 |
| 10 |
32 |
10 |
4 |
83.27 |
64.31 |
95.24 |
74.56 |
74.64 |
62.33 |
89.17 |
81.92 |
81.44 |
78.54 |
| 10 |
32 |
10 |
6 |
85.05 |
69.61 |
94.14 |
79.82 |
77.90 |
62.79 |
89.53 |
80.81 |
84.09 |
80.42 |
| 10 |
32 |
10 |
8 |
85.05 |
68.90 |
95.60 |
76.32 |
76.81 |
66.05 |
90.61 |
83.76 |
84.85 |
80.88 |
| 10 |
32 |
12 |
4 |
81.85 |
58.66 |
95.97 |
69.30 |
79.71 |
63.72 |
90.25 |
83.76 |
79.17 |
78.04 |
| 10 |
32 |
12 |
6 |
82.92 |
65.02 |
93.77 |
71.93 |
79.71 |
61.40 |
89.89 |
85.24 |
84.47 |
79.37 |
| 10 |
32 |
12 |
8 |
81.85 |
69.96 |
93.41 |
74.12 |
80.43 |
66.51 |
89.17 |
84.87 |
86.36 |
80.74 |
| 10 |
32 |
14 |
4 |
84.70 |
62.90 |
94.14 |
70.18 |
77.90 |
60.93 |
89.89 |
83.03 |
85.23 |
78.76 |
| 10 |
32 |
14 |
6 |
83.99 |
66.78 |
95.24 |
71.93 |
83.33 |
61.86 |
92.06 |
83.03 |
84.47 |
80.30 |
| 10 |
32 |
14 |
8 |
82.92 |
62.54 |
94.87 |
74.12 |
81.52 |
68.37 |
90.25 |
86.72 |
85.98 |
80.81 |
| 10 |
48 |
10 |
4 |
83.63 |
61.13 |
94.14 |
68.42 |
78.62 |
58.60 |
90.97 |
83.39 |
71.97 |
76.77 |
| 10 |
48 |
10 |
6 |
84.70 |
62.54 |
93.77 |
73.25 |
76.45 |
66.98 |
89.89 |
83.39 |
85.98 |
79.66 |
| 10 |
48 |
10 |
8 |
84.70 |
65.72 |
93.77 |
73.25 |
76.45 |
62.79 |
82.67 |
85.24 |
83.71 |
78.70 |
| 10 |
48 |
12 |
4 |
82.21 |
65.02 |
94.87 |
69.74 |
76.09 |
59.53 |
77.26 |
85.24 |
82.20 |
76.91 |
| 10 |
48 |
12 |
6 |
83.63 |
72.79 |
93.41 |
69.30 |
78.99 |
64.19 |
90.61 |
84.13 |
85.23 |
80.25 |
| 10 |
48 |
12 |
8 |
86.12 |
71.02 |
94.87 |
80.26 |
76.81 |
69.30 |
87.00 |
81.18 |
85.23 |
81.31 |
| 10 |
48 |
14 |
4 |
81.85 |
59.72 |
94.87 |
72.37 |
77.54 |
56.74 |
90.25 |
84.50 |
81.44 |
77.70 |
| 10 |
48 |
14 |
6 |
82.21 |
68.90 |
96.70 |
77.19 |
78.99 |
60.93 |
91.34 |
87.82 |
79.55 |
80.40 |
| 10 |
48 |
14 |
8 |
86.12 |
69.96 |
95.60 |
75.88 |
80.43 |
60.93 |
88.09 |
83.76 |
82.95 |
80.42 |
Appendix A.2. Experiment EEG-TCNet with Different Parameters of Chebyshev Type 2 Filter
Table A2 shows the accuracy of 9 subjects when training EEG-TCNet with different parameters
N,
in the bandpass filter
Table A2.
Accuracy of EEG-TCNet with different bandpass filter parameters
Table A2.
Accuracy of EEG-TCNet with different bandpass filter parameters
| N |
|
S1 |
S2 |
S3 |
S4 |
S5 |
S6 |
S7 |
S8 |
S9 |
Average Accuracy (%) |
| 4 |
20 |
76.87 |
51.24 |
92.31 |
65.35 |
37.68 |
50.70 |
75.09 |
78.97 |
79.17 |
67.49 |
| 4 |
30 |
80.43 |
52.65 |
91.58 |
58.33 |
35.87 |
45.58 |
71.84 |
83.39 |
79.17 |
66.54 |
| 4 |
40 |
77.94 |
48.41 |
89.38 |
52.63 |
34.78 |
46.98 |
73.29 |
83.39 |
82.20 |
65.44 |
| 4 |
50 |
76.87 |
47.35 |
91.94 |
56.58 |
33.33 |
47.44 |
68.59 |
78.60 |
79.17 |
64.43 |
| 4 |
60 |
77.94 |
38.52 |
91.21 |
51.75 |
35.87 |
44.65 |
61.37 |
85.24 |
84.09 |
63.40 |
| 4 |
70 |
80.78 |
33.57 |
91.58 |
50.00 |
35.87 |
46.51 |
63.54 |
81.92 |
78.03 |
62.42 |
| 4 |
80 |
81.49 |
41.34 |
92.31 |
46.49 |
35.51 |
45.58 |
54.87 |
77.12 |
76.14 |
61.21 |
| 4 |
90 |
79.72 |
34.98 |
93.77 |
45.61 |
34.06 |
43.72 |
51.62 |
59.78 |
66.29 |
56.62 |
| 4 |
100 |
77.94 |
36.04 |
91.21 |
43.42 |
37.32 |
41.86 |
48.74 |
53.87 |
48.11 |
53.17 |
| 5 |
20 |
78.65 |
54.06 |
92.31 |
66.23 |
46.74 |
43.26 |
74.73 |
84.13 |
79.55 |
68.85 |
| 5 |
30 |
81.85 |
51.24 |
89.01 |
66.67 |
35.87 |
45.12 |
74.73 |
83.39 |
81.44 |
67.70 |
| 5 |
40 |
78.65 |
50.53 |
91.94 |
58.33 |
32.61 |
48.37 |
77.98 |
83.39 |
80.30 |
66.90 |
| 5 |
50 |
76.51 |
48.76 |
91.58 |
54.39 |
35.51 |
44.19 |
71.12 |
82.29 |
82.20 |
65.17 |
| 5 |
60 |
73.67 |
49.12 |
91.21 |
57.46 |
37.68 |
49.77 |
69.31 |
82.66 |
80.68 |
65.73 |
| 5 |
70 |
76.16 |
49.82 |
89.74 |
56.14 |
35.87 |
43.26 |
63.90 |
81.55 |
81.82 |
64.25 |
| 5 |
80 |
82.21 |
34.63 |
91.94 |
54.82 |
36.96 |
51.16 |
64.98 |
80.07 |
82.58 |
64.37 |
| 5 |
90 |
72.95 |
36.04 |
91.58 |
47.37 |
34.06 |
50.23 |
58.84 |
81.55 |
80.68 |
61.48 |
| 5 |
100 |
77.58 |
35.34 |
94.51 |
42.98 |
36.59 |
45.12 |
51.99 |
74.17 |
78.03 |
59.59 |
| 6 |
20 |
81.49 |
56.18 |
90.48 |
59.21 |
35.51 |
40.00 |
84.84 |
79.34 |
82.58 |
67.74 |
| 6 |
30 |
78.65 |
52.30 |
89.01 |
56.58 |
34.42 |
48.84 |
79.78 |
84.50 |
76.89 |
66.77 |
| 6 |
40 |
78.65 |
49.82 |
89.74 |
64.47 |
37.32 |
45.12 |
73.65 |
79.70 |
83.33 |
66.87 |
| 6 |
50 |
80.43 |
47.00 |
92.67 |
60.96 |
36.59 |
46.05 |
81.95 |
79.34 |
80.30 |
67.25 |
| 6 |
60 |
76.51 |
50.88 |
89.74 |
49.56 |
34.78 |
44.65 |
72.20 |
80.44 |
74.62 |
63.71 |
| 6 |
70 |
78.29 |
53.36 |
92.31 |
57.02 |
37.68 |
47.91 |
67.87 |
81.92 |
83.33 |
66.63 |
| 6 |
80 |
78.65 |
36.75 |
91.58 |
52.19 |
31.52 |
49.77 |
68.23 |
83.39 |
81.06 |
63.68 |
| 6 |
90 |
76.16 |
40.64 |
90.48 |
57.89 |
33.70 |
54.88 |
66.06 |
80.81 |
84.47 |
65.01 |
| 6 |
100 |
79.36 |
37.46 |
92.67 |
52.63 |
34.78 |
49.77 |
66.43 |
82.29 |
82.95 |
64.26 |
| 7 |
20 |
77.94 |
54.42 |
89.74 |
69.74 |
36.23 |
46.05 |
75.09 |
81.92 |
73.86 |
67.22 |
| 7 |
30 |
81.85 |
56.18 |
91.21 |
64.91 |
44.57 |
45.12 |
78.70 |
81.55 |
81.44 |
69.50 |
| 7 |
40 |
79.00 |
51.94 |
90.48 |
72.37 |
56.88 |
51.16 |
82.67 |
82.29 |
78.79 |
71.73 |
| 7 |
50 |
73.67 |
51.59 |
88.64 |
51.32 |
32.25 |
43.26 |
74.01 |
83.03 |
78.03 |
63.98 |
| 7 |
60 |
80.07 |
48.41 |
90.84 |
58.77 |
34.78 |
49.30 |
76.17 |
81.92 |
78.03 |
66.48 |
| 7 |
70 |
79.00 |
50.18 |
90.48 |
49.56 |
38.41 |
47.91 |
70.40 |
83.03 |
81.06 |
65.56 |
| 7 |
80 |
74.73 |
49.12 |
90.48 |
54.39 |
32.97 |
40.00 |
72.92 |
82.29 |
80.68 |
64.18 |
| 7 |
90 |
80.43 |
35.34 |
91.21 |
49.56 |
38.77 |
54.88 |
66.06 |
79.70 |
81.44 |
64.15 |
| 7 |
100 |
79.72 |
35.34 |
90.48 |
53.95 |
35.51 |
48.84 |
67.87 |
83.76 |
83.33 |
64.31 |
| 8 |
20 |
75.09 |
50.18 |
87.55 |
55.26 |
36.59 |
46.98 |
76.17 |
83.39 |
75.00 |
65.13 |
| 8 |
30 |
80.07 |
55.12 |
92.31 |
63.60 |
38.77 |
44.19 |
81.95 |
80.81 |
81.82 |
68.74 |
| 8 |
40 |
76.16 |
52.30 |
91.58 |
63.60 |
38.41 |
52.09 |
83.39 |
81.55 |
79.55 |
68.73 |
| 8 |
50 |
75.80 |
50.88 |
90.48 |
59.65 |
38.77 |
47.91 |
80.51 |
80.81 |
84.09 |
67.65 |
| 8 |
60 |
81.49 |
48.06 |
93.04 |
54.82 |
37.32 |
48.84 |
80.51 |
80.44 |
81.82 |
67.37 |
| 8 |
70 |
79.00 |
54.06 |
89.74 |
51.32 |
29.35 |
40.00 |
77.62 |
81.55 |
81.44 |
64.90 |
| 8 |
80 |
79.00 |
47.00 |
90.11 |
48.68 |
35.14 |
43.72 |
71.48 |
80.44 |
84.85 |
64.49 |
| 8 |
90 |
78.29 |
45.94 |
90.11 |
45.18 |
38.77 |
52.09 |
75.81 |
81.92 |
80.68 |
65.42 |
| 8 |
100 |
76.87 |
46.64 |
91.58 |
49.56 |
37.32 |
46.51 |
67.15 |
80.81 |
80.30 |
64.08 |
Appendix B. EEG-Conformer Training Results
Appendix B.1. Experiment EEG-Conformer with Different Hyperparameters
Table A3 reveals the accuracy of 9 subjects that experiment with different hyperparameters
in EEG-Conformer architecture
Table A3.
Accuracy of EEG-Conformer with different hyperparameters
Table A3.
Accuracy of EEG-Conformer with different hyperparameters
| k |
N |
S1 |
S2 |
S3 |
S4 |
S5 |
S6 |
S7 |
S8 |
S9 |
Average Accuracy (%) |
| 20 |
6 |
88.26 |
58.66 |
94.87 |
73.25 |
76.45 |
62.79 |
87.00 |
87.08 |
79.55 |
78.66 |
| 30 |
6 |
86.48 |
57.95 |
95.24 |
73.68 |
75.36 |
59.07 |
87.00 |
86.35 |
81.06 |
78.02 |
| 40 |
6 |
84.03 |
53.82 |
95.49 |
78.13 |
79.51 |
64.58 |
89.58 |
87.15 |
83.33 |
79.51 |
| 50 |
6 |
86.12 |
54.42 |
93.77 |
74.12 |
75.36 |
58.60 |
89.53 |
87.82 |
81.06 |
77.87 |
| 60 |
6 |
84.34 |
57.60 |
93.77 |
72.37 |
78.62 |
60.47 |
85.20 |
87.82 |
79.55 |
77.75 |
| 20 |
7 |
86.83 |
61.13 |
94.87 |
73.25 |
76.81 |
59.07 |
85.56 |
88.19 |
79.17 |
78.32 |
| 30 |
7 |
86.48 |
59.01 |
93.77 |
74.12 |
76.45 |
63.26 |
90.97 |
87.82 |
81.82 |
79.30 |
| 40 |
7 |
85.41 |
55.12 |
94.87 |
71.93 |
76.45 |
58.14 |
90.61 |
89.30 |
82.58 |
78.27 |
| 50 |
7 |
84.70 |
52.65 |
94.87 |
73.25 |
76.09 |
58.14 |
88.45 |
88.19 |
79.55 |
77.32 |
| 60 |
7 |
85.05 |
51.94 |
94.87 |
74.56 |
77.54 |
60.00 |
89.53 |
88.19 |
82.20 |
78.21 |
| 20 |
8 |
85.41 |
61.48 |
93.41 |
71.49 |
76.81 |
61.40 |
84.48 |
88.56 |
82.95 |
78.44 |
| 30 |
8 |
86.83 |
53.71 |
94.87 |
71.49 |
75.72 |
60.00 |
90.25 |
89.30 |
81.44 |
78.18 |
| 40 |
8 |
85.77 |
56.18 |
94.14 |
71.93 |
78.26 |
60.00 |
91.34 |
87.45 |
81.44 |
78.50 |
| 50 |
8 |
87.54 |
54.06 |
93.41 |
71.49 |
78.99 |
63.72 |
84.84 |
88.19 |
79.92 |
78.02 |
| 60 |
8 |
84.34 |
53.36 |
93.41 |
73.68 |
78.26 |
62.33 |
90.25 |
88.19 |
80.30 |
78.24 |
| 20 |
9 |
87.54 |
60.42 |
94.14 |
77.63 |
76.45 |
65.58 |
81.95 |
89.30 |
80.68 |
79.30 |
| 30 |
9 |
86.83 |
55.48 |
93.77 |
76.32 |
78.26 |
61.40 |
90.25 |
88.19 |
81.06 |
79.06 |
| 40 |
9 |
85.77 |
54.42 |
92.67 |
74.56 |
77.90 |
59.07 |
92.06 |
87.82 |
82.95 |
78.58 |
| 50 |
9 |
86.12 |
54.77 |
94.51 |
74.56 |
75.36 |
60.47 |
90.25 |
86.72 |
83.33 |
78.45 |
| 60 |
9 |
87.19 |
53.36 |
93.41 |
71.93 |
76.45 |
59.53 |
90.61 |
87.82 |
82.20 |
78.06 |
| 20 |
10 |
87.54 |
60.42 |
91.94 |
75.88 |
77.90 |
64.19 |
88.09 |
88.56 |
82.95 |
79.72 |
| 30 |
10 |
87.54 |
54.06 |
94.14 |
73.25 |
77.54 |
60.93 |
92.42 |
89.30 |
84.09 |
79.25 |
| 40 |
10 |
85.77 |
50.88 |
94.51 |
78.51 |
79.35 |
59.07 |
91.70 |
88.19 |
79.92 |
78.65 |
| 50 |
10 |
87.19 |
56.18 |
94.14 |
72.37 |
77.90 |
59.07 |
85.56 |
88.56 |
81.82 |
78.09 |
| 60 |
10 |
86.48 |
52.30 |
94.51 |
75.44 |
77.54 |
60.47 |
88.81 |
89.30 |
83.71 |
78.73 |
Appendix B.2. Experiment EEG-Conformer with Different Parameters of Chebyshev Type 2 Filter
Table A4 below illustrates the accuracy of 9 subjects trained with different parameters
in the bandpass filter
Table A4.
Accuracy of EEG-Conformer with different bandpass filter parameters
Table A4.
Accuracy of EEG-Conformer with different bandpass filter parameters
| N |
|
S1 |
S2 |
S3 |
S4 |
S5 |
S6 |
S7 |
S8 |
S9 |
Average Accuracy (%) |
| 4 |
20 |
90.04 |
63.96 |
92.67 |
81.58 |
79.71 |
66.98 |
94.95 |
88.56 |
88.26 |
82.97 |
| 4 |
30 |
90.75 |
60.07 |
92.31 |
81.58 |
65.94 |
65.58 |
93.86 |
88.56 |
87.12 |
80.64 |
| 4 |
40 |
91.10 |
56.89 |
92.67 |
81.58 |
57.97 |
65.58 |
92.78 |
88.56 |
87.50 |
79.40 |
| 4 |
50 |
89.68 |
59.36 |
93.04 |
78.95 |
55.07 |
65.58 |
90.61 |
87.82 |
88.26 |
78.71 |
| 4 |
60 |
87.90 |
60.07 |
93.77 |
78.07 |
49.64 |
64.65 |
84.12 |
88.56 |
87.50 |
77.14 |
| 4 |
70 |
86.48 |
49.82 |
93.77 |
71.05 |
46.01 |
63.72 |
80.87 |
87.45 |
86.36 |
73.95 |
| 4 |
80 |
87.19 |
48.76 |
93.77 |
62.28 |
45.65 |
64.65 |
77.26 |
84.50 |
85.61 |
72.19 |
| 4 |
90 |
86.48 |
47.35 |
93.41 |
59.65 |
43.12 |
57.67 |
68.23 |
78.23 |
75.00 |
67.68 |
| 4 |
100 |
81.85 |
45.94 |
91.58 |
56.58 |
39.86 |
53.95 |
59.21 |
67.53 |
66.29 |
62.53 |
| 5 |
20 |
90.39 |
59.36 |
93.77 |
86.40 |
75.36 |
71.16 |
95.31 |
87.08 |
87.50 |
82.93 |
| 5 |
30 |
91.81 |
60.42 |
93.41 |
82.89 |
67.75 |
63.26 |
94.22 |
87.08 |
86.74 |
80.84 |
| 5 |
40 |
90.39 |
62.90 |
94.14 |
85.09 |
59.42 |
64.65 |
92.42 |
86.72 |
87.12 |
80.32 |
| 5 |
50 |
90.39 |
58.30 |
92.67 |
83.33 |
56.16 |
65.58 |
93.14 |
88.19 |
87.50 |
79.48 |
| 5 |
60 |
88.97 |
59.36 |
93.04 |
79.82 |
54.71 |
64.19 |
91.34 |
88.56 |
85.98 |
78.44 |
| 5 |
70 |
87.90 |
61.48 |
93.77 |
77.19 |
50.00 |
66.51 |
84.84 |
88.19 |
87.50 |
77.49 |
| 5 |
80 |
85.41 |
55.83 |
92.67 |
73.25 |
48.91 |
65.12 |
83.75 |
88.56 |
87.88 |
75.71 |
| 5 |
90 |
87.54 |
50.18 |
93.41 |
65.35 |
43.84 |
68.84 |
78.70 |
86.35 |
88.64 |
73.65 |
| 5 |
100 |
86.12 |
49.12 |
94.14 |
61.40 |
44.57 |
66.05 |
70.76 |
84.87 |
82.95 |
71.11 |
| 6 |
20 |
88.61 |
59.01 |
92.31 |
80.70 |
78.26 |
68.84 |
96.39 |
88.56 |
87.50 |
82.24 |
| 6 |
30 |
91.46 |
57.60 |
93.77 |
82.46 |
73.55 |
63.26 |
94.58 |
87.45 |
87.12 |
81.25 |
| 6 |
40 |
91.10 |
59.72 |
93.41 |
84.21 |
64.13 |
66.51 |
93.86 |
86.72 |
87.12 |
80.75 |
| 6 |
50 |
89.68 |
61.13 |
91.94 |
83.77 |
59.42 |
66.05 |
94.22 |
88.56 |
86.74 |
80.17 |
| 6 |
60 |
89.58 |
60.76 |
93.40 |
79.17 |
52.78 |
62.85 |
91.32 |
86.81 |
87.85 |
78.28 |
| 6 |
70 |
90.04 |
60.07 |
93.04 |
79.39 |
53.99 |
66.05 |
92.42 |
87.08 |
86.36 |
78.71 |
| 6 |
80 |
87.54 |
60.78 |
93.77 |
78.07 |
51.81 |
66.51 |
87.73 |
88.56 |
88.26 |
78.11 |
| 6 |
90 |
85.77 |
58.30 |
93.41 |
76.32 |
49.64 |
65.58 |
84.84 |
89.30 |
86.36 |
76.61 |
| 6 |
100 |
85.05 |
53.36 |
93.41 |
72.81 |
47.10 |
65.12 |
81.59 |
85.98 |
87.88 |
74.70 |
| 7 |
20 |
89.32 |
61.48 |
93.04 |
85.96 |
75.00 |
70.70 |
95.31 |
86.72 |
87.12 |
82.74 |
| 7 |
30 |
89.68 |
59.72 |
93.41 |
85.09 |
69.93 |
67.91 |
94.95 |
86.72 |
87.88 |
81.70 |
| 7 |
40 |
91.10 |
59.01 |
94.14 |
84.21 |
68.48 |
64.65 |
93.86 |
87.45 |
86.74 |
81.07 |
| 7 |
50 |
91.10 |
60.07 |
93.41 |
84.21 |
64.49 |
66.05 |
91.34 |
87.45 |
86.74 |
80.54 |
| 7 |
60 |
89.68 |
60.78 |
93.04 |
84.21 |
60.14 |
66.98 |
92.78 |
88.93 |
85.98 |
80.28 |
| 7 |
70 |
89.32 |
58.66 |
93.41 |
81.14 |
54.71 |
67.44 |
93.14 |
87.45 |
85.98 |
79.03 |
| 7 |
80 |
90.04 |
59.36 |
92.67 |
80.70 |
53.62 |
69.30 |
88.09 |
87.08 |
87.88 |
78.75 |
| 7 |
90 |
87.90 |
61.84 |
92.31 |
78.07 |
50.36 |
65.12 |
90.25 |
88.93 |
87.50 |
78.03 |
| 7 |
100 |
85.41 |
60.07 |
93.41 |
77.19 |
48.55 |
63.72 |
85.56 |
88.93 |
88.26 |
76.79 |
| 8 |
20 |
90.28 |
59.72 |
93.75 |
82.29 |
69.10 |
63.54 |
95.49 |
88.54 |
87.85 |
81.17 |
| 8 |
30 |
91.10 |
61.48 |
93.41 |
85.53 |
71.38 |
65.58 |
94.58 |
86.72 |
86.74 |
81.84 |
| 8 |
40 |
90.04 |
62.90 |
93.04 |
83.33 |
69.20 |
67.91 |
94.22 |
88.19 |
88.26 |
81.90 |
| 8 |
50 |
91.81 |
60.78 |
93.77 |
82.89 |
67.75 |
65.58 |
93.14 |
87.08 |
85.61 |
80.94 |
| 8 |
60 |
91.10 |
60.78 |
93.77 |
82.46 |
62.32 |
65.58 |
93.14 |
87.82 |
86.74 |
80.41 |
| 8 |
70 |
89.68 |
59.72 |
92.31 |
82.02 |
57.97 |
66.51 |
91.70 |
88.56 |
87.50 |
79.55 |
| 8 |
80 |
90.39 |
59.01 |
93.41 |
82.02 |
56.16 |
66.51 |
91.34 |
87.45 |
85.61 |
79.10 |
| 8 |
90 |
90.04 |
58.66 |
92.67 |
80.70 |
55.80 |
68.37 |
89.17 |
86.72 |
87.50 |
78.85 |
| 8 |
100 |
88.97 |
60.78 |
92.31 |
75.44 |
51.81 |
66.05 |
91.34 |
88.56 |
87.12 |
78.04 |
Appendix C. EEG-TCNTransformer Training Results
Appendix C.1. Experiment EEG-TCNTransformer with Different Hyperparameters
Table A5 displays the accuracy of 9 subjects that train the EEG-TCNTransformer with different number of TCNet blocks
T and number of filters
k in TCN block
Table A5.
Accuracy of EEG-TCNTransformer with different hyperparameters
Table A5.
Accuracy of EEG-TCNTransformer with different hyperparameters
| T |
k |
S1 |
S2 |
S3 |
S4 |
S5 |
S6 |
S7 |
S8 |
S9 |
Average Accuracy (%) |
| 1 |
10 |
85.41 |
60.42 |
91.21 |
79.82 |
56.52 |
60.00 |
85.20 |
84.87 |
79.17 |
75.85 |
| 1 |
20 |
82.56 |
66.43 |
92.67 |
78.07 |
63.41 |
59.07 |
86.28 |
85.61 |
76.89 |
76.78 |
| 1 |
30 |
85.05 |
66.08 |
91.94 |
84.21 |
74.64 |
66.98 |
88.81 |
84.13 |
83.33 |
80.57 |
| 1 |
40 |
84.34 |
69.26 |
93.41 |
81.14 |
76.09 |
59.07 |
90.25 |
85.24 |
84.85 |
80.40 |
| 1 |
50 |
85.05 |
62.19 |
93.41 |
80.26 |
76.81 |
57.67 |
89.17 |
84.50 |
84.85 |
79.32 |
| 1 |
60 |
87.19 |
66.08 |
93.41 |
80.70 |
78.26 |
64.65 |
89.53 |
85.61 |
87.50 |
81.44 |
| 1 |
70 |
83.99 |
66.43 |
95.24 |
77.63 |
77.54 |
65.12 |
89.17 |
86.72 |
88.26 |
81.12 |
| 1 |
80 |
86.48 |
63.96 |
95.60 |
79.39 |
76.45 |
66.98 |
90.25 |
84.50 |
87.88 |
81.28 |
| 1 |
90 |
85.41 |
63.60 |
95.24 |
80.26 |
77.54 |
59.53 |
91.34 |
88.19 |
87.50 |
80.96 |
| 1 |
100 |
87.54 |
61.84 |
94.87 |
81.58 |
77.90 |
58.14 |
90.97 |
86.72 |
84.85 |
80.49 |
| 2 |
10 |
85.77 |
65.02 |
90.48 |
70.18 |
68.12 |
58.60 |
89.53 |
84.87 |
76.14 |
76.52 |
| 2 |
20 |
85.05 |
71.38 |
92.31 |
80.70 |
66.30 |
64.65 |
87.73 |
86.35 |
76.89 |
79.04 |
| 2 |
30 |
83.63 |
67.49 |
93.04 |
84.21 |
81.52 |
65.12 |
89.53 |
86.35 |
78.03 |
80.99 |
| 2 |
40 |
83.27 |
66.43 |
94.14 |
82.46 |
84.06 |
67.44 |
88.81 |
84.50 |
83.33 |
81.60 |
| 2 |
50 |
84.34 |
68.90 |
93.41 |
82.89 |
81.16 |
65.12 |
89.17 |
85.98 |
86.74 |
81.97 |
| 2 |
60 |
83.27 |
67.14 |
93.41 |
86.40 |
80.43 |
66.05 |
92.78 |
86.72 |
87.88 |
82.68 |
| 2 |
70 |
86.48 |
65.37 |
94.51 |
84.21 |
80.80 |
68.37 |
89.53 |
85.98 |
87.88 |
82.57 |
| 2 |
80 |
86.48 |
66.43 |
94.87 |
84.21 |
81.52 |
62.33 |
89.89 |
85.98 |
87.88 |
82.18 |
| 2 |
90 |
83.63 |
65.02 |
94.51 |
83.33 |
81.16 |
64.19 |
90.25 |
86.72 |
87.50 |
81.81 |
| 2 |
100 |
85.77 |
64.66 |
94.51 |
83.77 |
80.80 |
67.44 |
87.73 |
87.82 |
86.74 |
82.14 |
| 3 |
10 |
80.07 |
72.79 |
90.48 |
71.05 |
67.39 |
58.14 |
89.17 |
81.92 |
74.62 |
76.18 |
| 3 |
20 |
84.70 |
68.55 |
93.77 |
79.82 |
72.10 |
63.72 |
90.97 |
83.03 |
73.11 |
78.86 |
| 3 |
30 |
87.19 |
65.72 |
94.87 |
83.77 |
80.80 |
69.30 |
91.70 |
85.24 |
79.92 |
82.06 |
| 3 |
40 |
88.61 |
66.08 |
94.51 |
87.28 |
79.35 |
68.37 |
91.34 |
84.87 |
84.85 |
82.81 |
| 3 |
50 |
86.12 |
67.49 |
93.77 |
83.33 |
83.70 |
66.98 |
89.89 |
87.45 |
85.98 |
82.75 |
| 3 |
60 |
83.99 |
66.43 |
93.41 |
80.70 |
81.52 |
65.58 |
89.53 |
86.35 |
87.12 |
81.63 |
| 3 |
70 |
87.90 |
66.78 |
94.51 |
82.02 |
81.88 |
66.51 |
89.53 |
88.56 |
89.02 |
82.97 |
| 3 |
80 |
82.21 |
67.14 |
91.94 |
81.58 |
82.25 |
63.26 |
89.89 |
86.35 |
87.12 |
81.30 |
| 3 |
90 |
86.83 |
65.72 |
93.77 |
82.46 |
78.99 |
68.37 |
83.03 |
87.45 |
89.39 |
81.78 |
| 3 |
100 |
85.41 |
62.19 |
93.04 |
82.89 |
78.62 |
66.05 |
88.81 |
86.72 |
87.50 |
81.25 |
Appendix C.2. Experiment EEG-TCNTransformer with Different Parameters of Chebyshev Type 2 Filter
Table A6 shows the accuracy of 9 subjects trained EEG-TCNTransformer with different parameters
in the bandpass filter
Table A6.
Accuracy of EEG-TCNTransformer with different bandpass filter parameters
Table A6.
Accuracy of EEG-TCNTransformer with different bandpass filter parameters
| T |
k |
S1 |
S2 |
S3 |
S4 |
S5 |
S6 |
S7 |
S8 |
S9 |
Average Accuracy (%) |
| 1 |
10 |
79.72 |
53.71 |
90.48 |
76.32 |
68.84 |
54.42 |
81.59 |
78.97 |
86.74 |
74.53 |
| 2 |
10 |
83.63 |
63.25 |
93.41 |
79.39 |
76.45 |
61.86 |
91.34 |
85.98 |
87.12 |
80.27 |
| 3 |
10 |
79.72 |
54.42 |
87.91 |
82.02 |
74.28 |
55.35 |
85.56 |
80.07 |
83.71 |
75.89 |
| 4 |
10 |
81.85 |
66.43 |
93.41 |
71.93 |
78.62 |
63.26 |
90.25 |
83.39 |
86.36 |
79.50 |
| 5 |
10 |
77.94 |
59.01 |
92.31 |
78.51 |
77.17 |
61.86 |
87.73 |
83.76 |
85.61 |
78.21 |
| 6 |
10 |
80.07 |
63.25 |
93.77 |
75.00 |
77.17 |
63.26 |
87.36 |
82.29 |
87.12 |
78.81 |
| 7 |
10 |
78.29 |
62.19 |
91.58 |
77.19 |
77.90 |
63.26 |
89.89 |
83.39 |
84.85 |
78.73 |
| 8 |
10 |
78.29 |
61.13 |
93.77 |
80.26 |
75.00 |
64.19 |
89.17 |
83.39 |
87.50 |
79.19 |
| 9 |
10 |
80.07 |
61.84 |
91.21 |
79.39 |
79.71 |
63.72 |
89.89 |
83.76 |
86.74 |
79.59 |
| 10 |
10 |
79.00 |
62.90 |
93.41 |
80.70 |
71.74 |
62.33 |
89.17 |
83.39 |
85.61 |
78.69 |
| 1 |
20 |
78.65 |
56.89 |
90.11 |
66.23 |
66.30 |
59.07 |
74.01 |
77.86 |
87.88 |
73.00 |
| 2 |
20 |
79.00 |
56.54 |
93.41 |
78.51 |
73.55 |
64.65 |
89.53 |
81.92 |
86.74 |
78.21 |
| 3 |
20 |
78.65 |
52.30 |
89.38 |
75.44 |
57.25 |
52.09 |
80.14 |
79.70 |
86.36 |
72.37 |
| 4 |
20 |
77.58 |
54.77 |
89.01 |
73.25 |
67.03 |
53.02 |
87.36 |
80.44 |
86.36 |
74.31 |
| 5 |
20 |
80.07 |
54.42 |
85.71 |
77.19 |
68.12 |
50.70 |
83.03 |
79.34 |
84.85 |
73.71 |
| 6 |
20 |
81.14 |
54.42 |
89.38 |
70.61 |
69.57 |
53.02 |
86.64 |
80.44 |
86.74 |
74.66 |
| 7 |
20 |
80.07 |
53.71 |
86.45 |
75.00 |
63.77 |
51.16 |
83.39 |
81.55 |
85.23 |
73.37 |
| 8 |
20 |
80.43 |
53.00 |
88.64 |
75.88 |
69.93 |
51.16 |
84.12 |
80.44 |
86.74 |
74.48 |
| 9 |
20 |
79.00 |
53.00 |
87.55 |
74.12 |
65.22 |
53.95 |
84.48 |
81.18 |
86.74 |
73.92 |
| 10 |
20 |
80.07 |
53.71 |
89.01 |
75.44 |
67.03 |
48.37 |
82.67 |
81.18 |
86.36 |
73.76 |
| 1 |
30 |
78.29 |
54.42 |
90.48 |
51.75 |
63.04 |
56.28 |
74.01 |
78.97 |
88.26 |
70.61 |
| 2 |
30 |
76.16 |
54.42 |
90.84 |
76.32 |
72.46 |
68.37 |
85.92 |
79.70 |
87.12 |
76.81 |
| 3 |
30 |
78.65 |
54.42 |
90.84 |
76.32 |
51.09 |
53.02 |
75.09 |
79.34 |
86.74 |
71.72 |
| 4 |
30 |
79.72 |
53.71 |
89.74 |
78.51 |
55.43 |
53.02 |
85.56 |
79.34 |
86.74 |
73.53 |
| 5 |
30 |
77.58 |
50.53 |
90.84 |
75.88 |
52.90 |
51.16 |
81.59 |
79.34 |
86.36 |
71.80 |
| 6 |
30 |
79.72 |
50.88 |
90.48 |
76.32 |
51.45 |
46.98 |
84.12 |
79.70 |
85.61 |
71.69 |
| 7 |
30 |
79.00 |
50.88 |
87.18 |
77.63 |
56.52 |
46.05 |
79.78 |
79.70 |
85.61 |
71.37 |
| 8 |
30 |
79.72 |
51.24 |
87.18 |
72.81 |
52.54 |
46.98 |
86.28 |
80.07 |
85.61 |
71.38 |
| 9 |
30 |
79.72 |
50.88 |
86.08 |
79.39 |
56.16 |
47.44 |
83.39 |
80.07 |
85.98 |
72.12 |
| 10 |
30 |
79.72 |
49.12 |
87.18 |
71.05 |
53.26 |
49.77 |
83.75 |
80.07 |
86.36 |
71.14 |
References
- Zhang, X.; Zhang, T.; Jiang, Y.; Zhang, W.; Lu, Z.; Wang, Y.; Tao, Q. A novel brain-controlled prosthetic hand method integrating AR-SSVEP augmentation, asynchronous control, and machine vision assistance. Heliyon 2024, 10. [Google Scholar] [CrossRef] [PubMed]
- Chai, R.; Ling, S.H.; Hunter, G.P.; Tran, Y.; Nguyen, H.T. Brain-Computer Interface Classifier for Wheelchair Commands Using Neural Network with Fuzzy Particle Swarm Optimization. IEEE Journal of Biomedical and Health Informatics 2014, 18, 1614–1624. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, L.; Soriano-Segura, P.; Navarro, J.; Jones, O.; Ortiz, M.; Iáñez, E.; Azorín, J.M.; Contreras-Vidal, J.L. Brain–machine interface based on deep learning to control asynchronously a lower-limb robotic exoskeleton: a case-of-study. Journal of NeuroEngineering and Rehabilitation 2024, 21. [Google Scholar] [CrossRef] [PubMed]
- Armour, B.S.; Courtney-Long, E.A.; Fox, M.H.; Fredine, H.; Cahill, A. Prevalence and causes of paralysis - United States, 2013. American Journal of Public Health 2016, 106, 1855–1857. [Google Scholar] [CrossRef] [PubMed]
- Berger, H. Über das Elektrenkephalogramm des Menschen. Archiv für Psychiatrie und Nervenkrankheiten 1929, 87, 527–570. [Google Scholar] [CrossRef]
- Hicks, R.J. Principles and Practice of Positron Emission Tomography. The Journal of Nuclear Medicine 2004, 45, 1973–1974. [Google Scholar]
- Mellinger, J.; Schalk, G.; Braun, C.; Preissl, H.; Rosenstiel, W.; Birbaumer, N.; Kübler, A. An MEG-based brain-computer interface (BCI). NeuroImage 2007, 36, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Naseer, N.; Hong, K.S. fNIRS-based brain-computer interfaces: a review. Frontiers in Human Neuroscience 2015. Name - Pusan National University Copyright - © 2015. This work is licensed under http://creativecommons.org/licenses/by/4.0/ (the “License”). Notwithstanding the ProQuest Terms and Conditions, you may use this content in accordance with the terms of the License. Last updated - 2023-11-24 SubjectsTermNotLitGenreText - South Korea. [CrossRef]
- Sorger, B.; Reithler, J.; Dahmen, B.; Goebel, R. A Real-Time fMRI-Based Spelling Device Immediately Enabling Robust Motor-Independent Communication. Current Biology 2012, 22, 1333–1338. [Google Scholar] [CrossRef]
- Ramoser, H.; Muller-Gerking, J.; Pfurtscheller, G. Optimal spatial filtering of single trial EEG during imagined hand movement. IEEE Transactions on Rehabilitation Engineering 2000, 8, 441–446. [Google Scholar] [CrossRef]
- Ang, K.K.; Chin, Z.Y.; Zhang, H.; Guan, C. Filter Bank Common Spatial Pattern (FBCSP) in Brain-Computer Interface. 2008, pp. 2390–2397. [CrossRef]
- Tabar, Y.R.; Halici, U. A novel deep learning approach for classification of EEG motor imagery signals. Journal of Neural Engineering 2017, 14. [Google Scholar] [CrossRef] [PubMed]
- Ieracitano, C.; Mammone, N.; Hussain, A.; Morabito, F.C. A novel multi-modal machine learning based approach for automatic classification of EEG recordings in dementia. Neural Networks 2020, 123, 176–190. [Google Scholar] [CrossRef] [PubMed]
- Kousarrizi, M.R.N.; Ghanbari, A.A.; Teshnehlab, M.; Shorehdeli, M.A.; Gharaviri, A. Feature Extraction and Classification of EEG Signals Using Wavelet Transform, SVM and Artificial Neural Networks for Brain Computer Interfaces. 2009, pp. 352–355. [CrossRef]
- Chen, C.Y.; Wu, C.W.; Lin, C.T.; Chen, S.A. A novel classification method for motor imagery based on Brain-Computer Interface. 2014, pp. 4099–4102. [CrossRef]
- Lawhern, V.J.; Solon, A.J.; Waytowich, N.R.; Gordon, S.M.; Hung, C.P.; Lance, B.J. EEGNet: A compact convolutional neural network for EEG-based brain-computer interfaces. Journal of Neural Engineering 2018, 15. [Google Scholar] [CrossRef] [PubMed]
- Schirrmeister, R.T.; Springenberg, J.T.; Fiederer, L.D.J.; Glasstetter, M.; Eggensperger, K.; Tangermann, M.; Hutter, F.; Burgard, W.; Ball, T. Deep learning with convolutional neural networks for EEG decoding and visualization. Human Brain Mapping 2017, 38, 5391–5420. [Google Scholar] [CrossRef] [PubMed]
- Ingolfsson, T.M.; Hersche, M.; Wang, X.; Kobayashi, N.; Cavigelli, L.; Benini, L. EEG-TCNet: An Accurate Temporal Convolutional Network for Embedded Motor-Imagery Brain-Machine Interfaces. Institute of Electrical and Electronics Engineers Inc., 10 2020, Vol. 2020-October, pp. 2958–2965. [CrossRef]
- Bai, S.; Kolter, J.Z.; Koltun, V. An Empirical Evaluation of Generic Convolutional and Recurrent Networks for Sequence Modeling 2018.
- Altaheri, H.; Muhammad, G.; Alsulaiman, M.; Amin, S.U.; Altuwaijri, G.A.; Abdul, W.; Bencherif, M.A.; Faisal, M. Deep learning techniques for classification of electroencephalogram (EEG) motor imagery (MI) signals: a review. Neural Computing and Applications 2023, 35, 14681–14722. [Google Scholar] [CrossRef]
- Pfeffer, M.A.; Ling, S.S.H.; Wong, J.K.W. Exploring the frontier: Transformer-based models in EEG signal analysis for brain-computer interfaces, 2024. [CrossRef]
- Liu, H.; Liu, Y.; Wang, Y.; Liu, B.; Bao, X. EEG classification algorithm of motor imagery based on CNN-Transformer fusion network. Institute of Electrical and Electronics Engineers Inc., 2022, pp. 1302–1309. [CrossRef]
- Ma, Y.; Song, Y.; Gao, F. A novel hybrid CNN-Transformer model for EEG Motor Imagery classification. Institute of Electrical and Electronics Engineers Inc., 2022, Vol. 2022-July. [CrossRef]
- Song, Y.; Zheng, Q.; Liu, B.; Gao, X. EEG Conformer: Convolutional Transformer for EEG Decoding and Visualization. IEEE Transactions on Neural Systems and Rehabilitation Engineering 2023, 31, 710–719. [Google Scholar] [CrossRef] [PubMed]
- Brunner, C.; Leeb, R.; Müller-Putz, G. BCI Competition 2008–Graz data set A 2024. [CrossRef]
- Wriessnegger, S.C.; Brunner, C.; Müller-Putz, G.R. Frequency specific cortical dynamics during motor imagery are influenced by prior physical activity. Frontiers in Psychology 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Ouadfeul, S.A.; Aliouane, L. , Wave-Number Filtering; Springer International Publishing, 2020; pp. 1–6. [CrossRef]
- Lotte, F. Signal processing approaches to minimize or suppress calibration time in oscillatory activity-based brain-computer interfaces. Proceedings of the IEEE 2015, 103, 871–890. [Google Scholar] [CrossRef]
Figure 1.
Time scheme of data record. [
25]
Figure 1.
Time scheme of data record. [
25]
Figure 2.
EEG signals visualised in EEG-LAB.
Figure 2.
EEG signals visualised in EEG-LAB.
Figure 3.
Transfer function of the Chebyshev filter (Type II). [
27]
Figure 3.
Transfer function of the Chebyshev filter (Type II). [
27]
Figure 4.
Segmentation and Reconstruction (R&S) augmentation in time domain. [
28]
Figure 4.
Segmentation and Reconstruction (R&S) augmentation in time domain. [
28]
Figure 5.
Architecture elements in a TCN. (a): Stacking of two residual blocks with dilated causal convolution with dilation factors d=1,2 and kernel size k=3. (b) Detailed layers in TCN residual block [
19]
Figure 5.
Architecture elements in a TCN. (a): Stacking of two residual blocks with dilated causal convolution with dilation factors d=1,2 and kernel size k=3. (b) Detailed layers in TCN residual block [
19]
Figure 6.
Architecture of the EEG-TCNet [
18]
Figure 6.
Architecture of the EEG-TCNet [
18]
Figure 7.
Architecture of the EEG-Conformer [
24]
Figure 7.
Architecture of the EEG-Conformer [
24]
Figure 8.
Architecture of the EEG-TCNTransformer
Figure 8.
Architecture of the EEG-TCNTransformer
Figure 9.
The column chart shows the accuracy between the EEG-TCNet baseline model, fixed model and variable model
Figure 9.
The column chart shows the accuracy between the EEG-TCNet baseline model, fixed model and variable model
Figure 10.
The Column chart shows the accuracy of the baseline EEG-TCNet, EEG-TCNet with fixed bandpass filter and EEG-TCNet with variable bandpass filter
Figure 10.
The Column chart shows the accuracy of the baseline EEG-TCNet, EEG-TCNet with fixed bandpass filter and EEG-TCNet with variable bandpass filter
Figure 11.
The column chart shows the accuracy between the EEG-Conformer baseline model, fixed model and variable.
Figure 11.
The column chart shows the accuracy between the EEG-Conformer baseline model, fixed model and variable.
Figure 12.
The Column chart shows accuracy of the baseline EEG- Conformert, EEG-Conformer with fixed bandpass filter and EEG- Conformert with variable bandpass filter.
Figure 12.
The Column chart shows accuracy of the baseline EEG- Conformert, EEG-Conformer with fixed bandpass filter and EEG- Conformert with variable bandpass filter.
Table 1.
EEG-TCNet architecture [
18].
Table 1.
EEG-TCNet architecture [
18].
| Layer |
Type |
#Filter |
Kernel |
Output |
|
Input |
|
|
(1,C,T) |
| |
Conv2D |
|
(1,) |
(,C,T) |
| |
BatchNorm |
|
|
|
|
DepthwiseConv2D |
|
(C,1) |
(,1,T) |
| |
BatchNorm |
|
|
|
| |
EluAct |
|
|
|
| |
AveragePool2D |
|
|
(,1,T//8) |
| |
Dropout |
|
|
|
|
SeparableConv2D |
|
(1,16) |
(,1,T//8) |
| |
BatchNorm |
|
|
|
| |
EluAct |
|
|
|
| |
AveragePool2D |
|
|
(,1,T//64) |
| |
Dropout |
|
|
|
|
TCN |
|
|
|
|
Dense |
|
|
4 |
| |
SoftMaxAct |
|
|
|
Table 2.
Experiment EEG-TCNet with different hyperparameters.
Table 2.
Experiment EEG-TCNet with different hyperparameters.
| EEGNet module |
|
6, 8, 10 |
| |
|
16, 32, 48 |
| TCN module |
|
10, 12, 14 |
| |
|
4, 6, 8 |
Table 3.
Comparison of different numbers of filter and kernel sizes in EEGNet module and TCN module in average accuracy.
Table 3.
Comparison of different numbers of filter and kernel sizes in EEGNet module and TCN module in average accuracy.
|
|
|
|
Average accuracy (%) |
| 6 |
32 |
12 |
4 |
75.38 |
| 8 |
|
|
|
77.28 |
| 10 |
|
|
|
78.04 |
| 8 |
16 |
12 |
4 |
76.66 |
| |
32 |
|
|
77.28 |
| |
48 |
|
|
77.05 |
| 8 |
32 |
10 |
4 |
77.99 |
| |
|
12 |
|
77.28 |
| |
|
14 |
|
78.58 |
| 8 |
32 |
12 |
4 |
77.28 |
| |
|
|
6 |
79.96 |
| |
|
|
8 |
78.59 |
Table 4.
Variable hyperparameters of EEG-TCNet.
Table 4.
Variable hyperparameters of EEG-TCNet.
| |
S1 |
S2 |
S3 |
S4 |
S5 |
S6 |
S7 |
S8 |
S9 |
|
6 |
10 |
6 |
6 |
8 |
10 |
10 |
10 |
8 |
|
48 |
48 |
32 |
32 |
32 |
48 |
16 |
48 |
32 |
|
12 |
12 |
14 |
14 |
10 |
12 |
12 |
14 |
14 |
|
6 |
6 |
6 |
6 |
8 |
8 |
4 |
6 |
4 |
Table 5.
Accuracy of the EEG-TCNet baseline model, fixed model and variable models (%).
Table 5.
Accuracy of the EEG-TCNet baseline model, fixed model and variable models (%).
| |
Baseline |
Fixed model |
Variable model |
| S1 |
84.34 |
86.12 |
88.26 |
| S2 |
62.90 |
71.02 |
72.79 |
| S3 |
93.77 |
94.87 |
97.07 |
| S4 |
71.05 |
80.26 |
80.26 |
| S5 |
77.17 |
76.81 |
84.06 |
| S6 |
57.21 |
69.30 |
69.30 |
| S7 |
88.45 |
87.00 |
94.58 |
| S8 |
83.76 |
81.18 |
87.82 |
| S9 |
76.89 |
85.23 |
87.12 |
| Average |
77.28 |
81.31 |
84.59 |
Table 6.
Experiment EEG-TCNet with different parameters of Chebyshev type 2 filter.
Table 6.
Experiment EEG-TCNet with different parameters of Chebyshev type 2 filter.
| N |
4, 5, 6, 7, 8 |
|
20, 30, 40, 50, 60. 70, 80, 90, 100 |
Table 7.
Variable bandpass filter of EEG-TCNet.
Table 7.
Variable bandpass filter of EEG-TCNet.
| |
S1 |
S2 |
S3 |
S4 |
S5 |
S6 |
S7 |
S8 |
S9 |
| N |
5 |
6 |
5 |
7 |
7 |
6 |
6 |
4 |
8 |
|
80 |
20 |
100 |
40 |
40 |
90 |
20 |
60 |
80 |
Table 8.
Accuracy of the baseline EEG-TCNet, EEG-TCNet with fixed bandpass filter and with variable bandpass filter. (%)
Table 8.
Accuracy of the baseline EEG-TCNet, EEG-TCNet with fixed bandpass filter and with variable bandpass filter. (%)
| |
Baseline |
Fixed bandpass filter |
Variable bandpass filter |
| S1 |
84.34 |
79.00 |
82.21 |
| S2 |
62.90 |
51.94 |
56.18 |
| S3 |
93.77 |
90.48 |
94.51 |
| S4 |
71.05 |
72.37 |
72.37 |
| S5 |
77.17 |
56.88 |
56.88 |
| S6 |
57.21 |
51.16 |
54.88 |
| S7 |
88.45 |
82.67 |
84.84 |
| S8 |
83.76 |
82.29 |
85.24 |
| S9 |
76.89 |
78.79 |
84.85 |
| Average |
77.28 |
71.73 |
74.66 |
Table 9.
The architecture of the convolution module in EEG-Conformer. [
24]
Table 9.
The architecture of the convolution module in EEG-Conformer. [
24]
| Layer |
In |
Out |
Kernel |
Stride |
| Temporal Conv |
1 |
k |
(1,25) |
(1,1) |
| Spatial Conv |
k |
k |
(ch,1) |
(1,1) |
| Avg Pooling |
k |
k |
(1,75) |
(1,15) |
| Rearrange |
(k,1,m) → (m,k) |
Table 10.
Experiment EEG-Conformer with different hyperparameters
Table 10.
Experiment EEG-Conformer with different hyperparameters
| k |
20, 30, 40, 50, 60 |
| N |
6, 7, 8, 9, 10 |
Table 11.
Comparison of different numbers of filters k and numbers of self-attention block N in EEG-Conformer in average accuracy.
Table 11.
Comparison of different numbers of filters k and numbers of self-attention block N in EEG-Conformer in average accuracy.
| k |
N |
Average accuracy (%) |
| 20 |
6 |
78.66 |
| 30 |
|
78.02 |
| 40 |
|
79.51 |
| 50 |
|
77.86 |
| 60 |
|
77.74 |
| 40 |
6 |
79.51 |
| |
7 |
78.27 |
| |
8 |
78.50 |
| |
9 |
78.58 |
| |
10 |
78.65 |
Table 12.
Variable hyperparameters of EEG-Conformer
Table 12.
Variable hyperparameters of EEG-Conformer
| |
S1 |
S2 |
S3 |
S4 |
S5 |
S6 |
S7 |
S8 |
S9 |
| k |
20 |
20 |
40 |
40 |
40 |
20 |
30 |
40 |
30 |
| N |
6 |
8 |
6 |
10 |
6 |
9 |
10 |
7 |
10 |
Table 13.
Accuracy of the EEG-Conformer baseline model, fixed model and variable models
Table 13.
Accuracy of the EEG-Conformer baseline model, fixed model and variable models
| |
Baseline |
Fixed model |
Variable model |
| S1 |
84.03 |
87.54 |
88.26 |
| S2 |
53.82 |
60.42 |
61.48 |
| S3 |
95.49 |
91.94 |
95.49 |
| S4 |
78.13 |
75.88 |
78.51 |
| S5 |
79.51 |
77.90 |
79.51 |
| S6 |
64.58 |
64.19 |
65.58 |
| S7 |
89.58 |
88.09 |
92.42 |
| S8 |
87.15 |
88.56 |
89.30 |
| S9 |
83.33 |
82.95 |
84.09 |
| Average |
79.51 |
79.72 |
81.63 |
Table 14.
Experiment EEG-Conformer with different parameters of Chebyshev type 2 filter
Table 14.
Experiment EEG-Conformer with different parameters of Chebyshev type 2 filter
| N |
4, 5, 6, 7, 8 |
|
20, 30, 40, 50, 60. 70, 80, 90, 100 |
Table 15.
Variable bandpass filter of EEG-Conformer
Table 15.
Variable bandpass filter of EEG-Conformer
| |
S1 |
S2 |
S3 |
S4 |
S5 |
S6 |
S7 |
S8 |
S9 |
| N |
5 |
4 |
5 |
5 |
4 |
5 |
6 |
6 |
5 |
|
30 |
20 |
40 |
20 |
20 |
20 |
20 |
90 |
90 |
Table 16.
Accuracy of the baseline EEG-Conformer, EEG-Conformer with fixed bandpass filter and EEG-Conformer with variable bandpass filter (%)
Table 16.
Accuracy of the baseline EEG-Conformer, EEG-Conformer with fixed bandpass filter and EEG-Conformer with variable bandpass filter (%)
| |
Baseline |
Fixed bandpass filter |
Variable bandpass filter |
| S1 |
84.03 |
90.04 |
91.81 |
| S2 |
53.82 |
63.96 |
63.96 |
| S3 |
95.49 |
92.67 |
94.14 |
| S4 |
78.13 |
81.58 |
86.40 |
| S5 |
79.51 |
79.71 |
79.71 |
| S6 |
64.58 |
66.98 |
71.16 |
| S7 |
89.58 |
94.95 |
96.39 |
| S8 |
87.15 |
88.56 |
89.30 |
| S9 |
83.33 |
88.26 |
88.64 |
| Average |
79.51 |
82.97 |
84.61 |
Table 17.
The average accuracy of different numbers of filters k and number of TCN block T
Table 17.
The average accuracy of different numbers of filters k and number of TCN block T
| |
|
T |
| k |
|
1 |
2 |
3 |
| |
10 |
75.85 |
76.52 |
76.16 |
| |
20 |
76.78 |
79.04 |
78.86 |
| |
30 |
80.57 |
80.99 |
82.06 |
| |
40 |
80.40 |
81.60 |
82.81 |
| |
50 |
79.32 |
81.96 |
82.75 |
| |
60 |
81.44 |
82.68 |
81.63 |
| |
70 |
81.12 |
82.57 |
82.97 |
| |
80 |
81.28 |
82.18 |
81.30 |
| |
90 |
80.96 |
81.81 |
81.78 |
| |
100 |
80.49 |
82.13 |
81.25 |
Table 18.
Comparisons EEG-TCNet baseline, EEG-Conformer baseline and EEG-TCNTransformer
Table 18.
Comparisons EEG-TCNet baseline, EEG-Conformer baseline and EEG-TCNTransformer
| |
EEG-TCNet |
EEG-Conformer |
EEG-TCNTransformer |
| S1 |
84.34 |
84.03 |
87.90 |
| S2 |
62.90 |
53.82 |
66.78 |
| S3 |
93.77 |
95.49 |
94.51 |
| S4 |
71.05 |
78.13 |
82.02 |
| S5 |
77.17 |
79.51 |
81.88 |
| S6 |
57.21 |
64.58 |
66.51 |
| S7 |
88.45 |
89.58 |
89.53 |
| S8 |
83.76 |
87.15 |
88.56 |
| S9 |
76.89 |
83.33 |
89.02 |
| Average |
77.28 |
79.51 |
82.97 |
Table 19.
The average accuracy of EEG-TCNTransformer with different bandpass filter parameters
Table 19.
The average accuracy of EEG-TCNTransformer with different bandpass filter parameters
| |
|
|
| |
|
10 |
20 |
30 |
| N |
1 |
74.53 |
73.00 |
70.61 |
| |
2 |
80.27 |
78.21 |
76.81 |
| |
3 |
75.89 |
72.37 |
71.72 |
| |
4 |
79.50 |
74.31 |
73.53 |
| |
5 |
78.21 |
73.71 |
71.80 |
| |
6 |
78.81 |
74.66 |
71.69 |
| |
7 |
78.73 |
73.37 |
71.37 |
| |
8 |
79.19 |
74.48 |
71.38 |
| |
9 |
79.59 |
73.92 |
72.12 |
| |
10 |
78.69 |
73.76 |
71.14 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).