Submitted:
08 August 2024
Posted:
09 August 2024
You are already at the latest version
Abstract

Keywords:
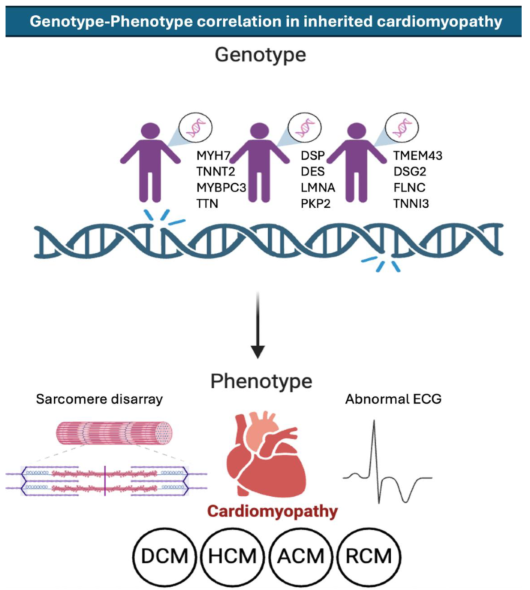
Introduction
Inherited Cardiomyopathies
Hypertrophic Cardiomyopathy (HCM)
Dilated Cardiomyopathy (DCM)
Arrhythmogenic Right Ventricular Cardiomyopathy (ARVC)
Restrictive Cardiomyopathy (RCM)
Multi-Omics Approaches in Cardiomyopathies
Genomics
- ○
- Genomic studies have identified numerous genetic mutations associated with inherited cardiomyopathies. Whole-exome sequencing (WES) and whole-genome sequencing (WGS) have been particularly useful in identifying rare and novel variants [44]. These technologies have also facilitated the study of genetic modifiers that influence disease severity and penetrance [45].
Transcriptomics
- ○
- Transcriptomic analyses, such as RNA sequencing, provide insights into gene expression changes associated with cardiomyopathies [46,47]. These studies have identified differentially expressed genes and pathways that contribute to disease pathogenesis [48]. For example, transcriptomic studies in HCM have revealed upregulation of hypertrophic signaling pathways and downregulation of energy metabolism genes [49].
Proteomics
- ○
- Proteomic approaches, including mass spectrometry, have been used to study protein expression and post-translational modifications in cardiomyopathies [50,51]. These studies have identified altered protein networks and signaling pathways that contribute to disease phenotypes [52]. For instance, proteomic analyses in DCM have revealed dysregulation of cytoskeletal and mitochondrial proteins [53].
Metabolomics
- ○
- Metabolomic studies provide insights into metabolic alterations associated with cardiomyopathies [50,54]. These studies have identified changes in metabolites and metabolic pathways that contribute to disease progression [55]. For example, metabolomic analyses in ARVC have revealed alterations in lipid metabolism and energy production [56].
Disease Modeling in Cardiomyopathies
Induced Pluripotent Stem Cells (iPSCs)
- ○
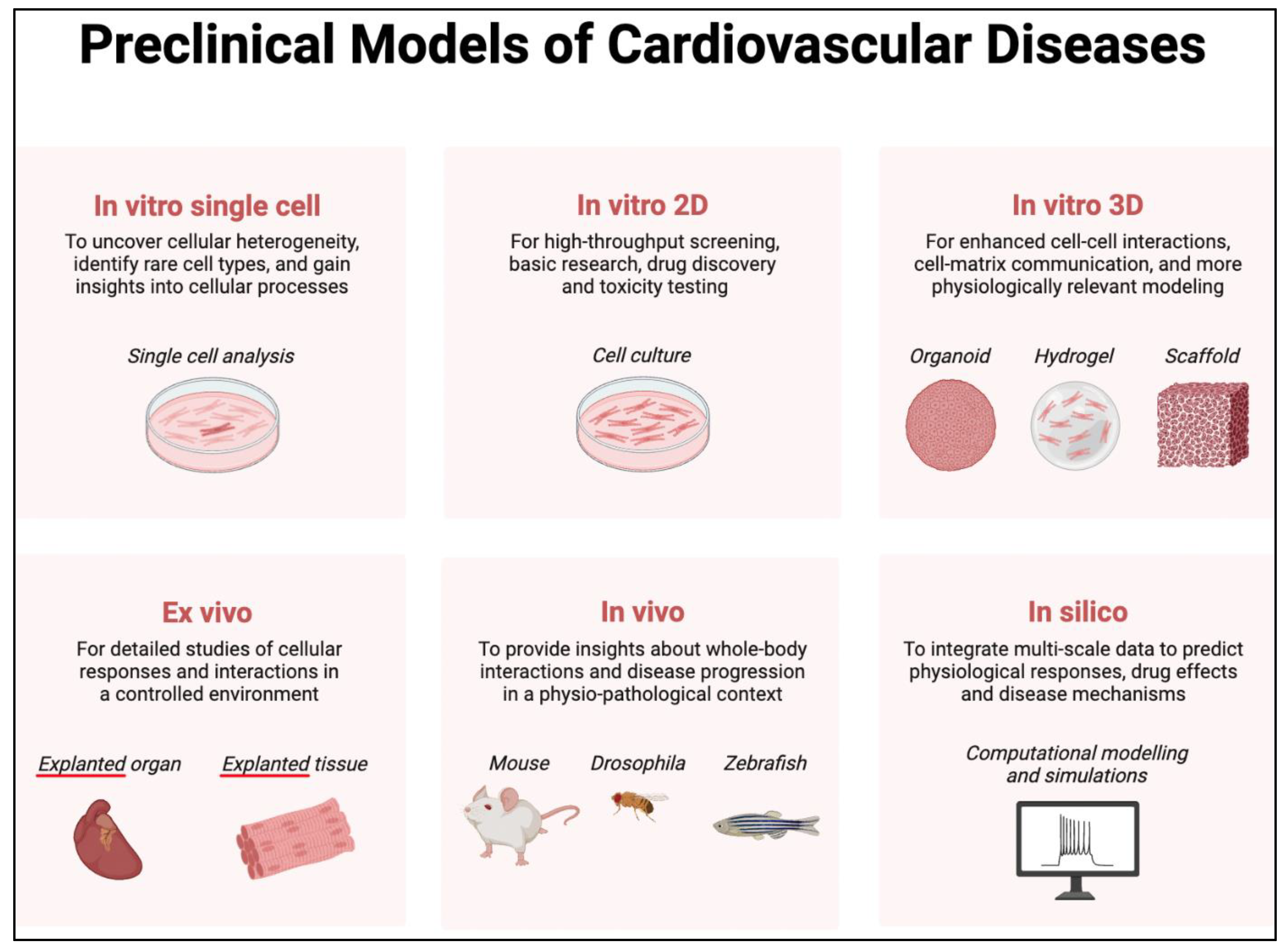
- iPSCs are derived from patient-specific somatic cells and can be differentiated into cardiomyocytes [58]. While monolayer iPSC cultures exhibit considerable scalability, they yield a model that is inherently simplified and less intricate in comparison to the complexity observed in both 3D in vitro models and in vivo systems (Figure 1). Despite this, iPSC-derived cardiomyocytes can still recapitulate many aspects of the patient's disease phenotype, allowing for the study of disease mechanisms and drug testing in a patient-specific context. For example, iPSC models of HCM have been used to study the effects of MYBPC3 mutations on sarcomere function and calcium handling [59]. Indeed, the technology of using iPSC-derived cardiomyocytes has been used to model numerous inherited cardiomyopathies, whether by producing patient-specific cells, editing the genome of healthy cells, or even overexpressing mutated ion channels.
Animal Models
- ○
- Animal models, including transgenic mice and zebrafish, have been widely used to study cardiomyopathies [60]. These models allow for the investigation of gene function and the study of disease progression in a whole-organism context [61]. For instance, transgenic mouse models of DCM have been used to study the effects of TTN mutations on cardiac function and remodeling [62].
Future Impact of Cardiac Regenerative Medicine in Finding Innovative Treatment for Cardiomyopathies
Implications for Personalized Medicine
Tailored Therapeutic Strategies
- ○
- Personalized treatment plans based on genetic profiles have shown promise in improving outcomes for patients with cardiomyopathies [82,83]. For example, genotype-specific therapies, such as small-molecule inhibitors and gene therapies, are being developed for patients with specific mutations [84]. Additionally, personalized exercise and lifestyle recommendations can be made based on the patient's genetic risk factors [85].
Predictive Diagnostics
- ○
- Predictive diagnostics based on genotype-phenotype correlations can help identify individuals at high risk for developing cardiomyopathies and guide early intervention strategies [86,87]. For example, genetic testing can identify individuals with pathogenic mutations, allowing for close monitoring and preventive measures to reduce the risk of adverse outcomes [88].
Challenges and Future Directions
- ○
- Despite the promise of personalized medicine, there are several challenges to its implementation. These include the complexity of genotype-phenotype correlations, the need for large-scale data integration, and the ethical considerations of genetic testing [81,89,90]. Future research should focus on addressing these challenges and further refining personalized treatment and diagnostic approaches.
Gaps in Current Research
Unresolved Correlations
- ○
- Certain genotype-phenotype correlations remain unclear, with some mutations leading to highly variable clinical presentations. For example, mutations in the LMNA gene can cause a wide range of phenotypes, from mild dilatation to severe heart failure and arrhythmias [93]. Further research is needed to elucidate the factors that contribute to this variability.
Technological and Methodological Limitations
- ○
- Current research methodologies have limitations that hinder the full understanding of genotype-phenotype correlations. For instance, WES and WGS may miss certain types of genetic variants, such as structural variants and deep intronic mutations [94]. Additionally, the integration of multi-omics data requires advanced computational tools and techniques, which are still in development [95].
Recommendations for Future Research
- ○
- To address these gaps, future research should focus on improving genetic testing techniques, developing better data integration methods, and conducting large-scale studies to validate genotype-phenotype correlations. Collaborative efforts and the use of advanced technologies, such as machine learning, will be essential in advancing our understanding of inherited cardiomyopathies.
Future Directions
Advances in Multi-Omics
- ○
- Emerging technologies in multi-omics research, such as single-cell RNA sequencing and spatial transcriptomics, have the potential to provide deeper insights into the cellular and molecular mechanisms of cardiomyopathies [96]. These technologies can help identify novel therapeutic targets and biomarkers for personalized treatment [97].
Innovations in Disease Modeling
- ○
- New approaches to disease modeling, such as the use of organ-on-a-chip technology and 3D bioprinting, can enhance our ability to study cardiomyopathies in a more physiologically relevant context [98]. These models can be used to test the efficacy and safety of new therapies and to study the interactions between different cell types in the heart [99].
Translational Research and Clinical Applications
- ○
- Bridging the gap between research and clinical practice is essential for translating scientific discoveries into effective treatments for patients with cardiomyopathies [100]. Collaborative efforts between researchers, clinicians, and industry partners will be crucial in developing and implementing personalized therapeutic strategies.
Summary
Acknowledgments
Conflicts of Interest
References
- Lukas Laws, J.; et al. Arrhythmias as Presentation of Genetic Cardiomyopathy. Circ Res 2022, 130, 1698–1722. [Google Scholar] [CrossRef]
- Fan, X.; et al. Arrhythmogenic Cardiomyopathy: from Preclinical Models to Genotype–phenotype Correlation and Pathophysiology. Stem Cell Reviews and Reports 2023, 19, 2683–2708. [Google Scholar] [CrossRef]
- Wu, Y. and L. Xie, AI-driven multi-omics integration for multi-scale predictive modeling of causal genotype-environment-phenotype relationships. arXiv preprint arXiv:2407.06405, 2024.
- Bueno, C.O.P. and M.L.O. Jurado, Cardiomyopathies: A Historical Journey. 2024.
- Members, W.C.; et al. 2024 AHA/ACC/AMSSM/HRS/PACES/SCMR Guideline for the Management of Hypertrophic Cardiomyopathy: A Report of the American Heart Association/American College of Cardiology Joint Committee on Clinical Practice Guidelines. Journal of the American College of Cardiology 2024, 83, 2324–2405. [Google Scholar]
- Lopes, L.R. ; C.Y. Ho, and P.M. Elliott, Genetics of hypertrophic cardiomyopathy: established and emerging implications for clinical practice. European Heart Journal, 2024, ehae421.
- Franke, M.; et al. A MYH7 variant in a five-generation-family with hypertrophic cardiomyopathy. Frontiers in Genetics 2024, 15, 1306333. [Google Scholar] [CrossRef]
- Ananthamohan, K. ; J.E. Stelzer, and S. Sadayappan, Hypertrophic cardiomyopathy in MYBPC3 carriers in aging. The journal of cardiovascular aging 2024, 4(1).
- Ireland, C.G. and C.Y. Ho. Genetic Testing in Hypertrophic Cardiomyopathy. The American Journal of Cardiology 2024, 212, S4–S13. [Google Scholar] [CrossRef] [PubMed]
- Jansen, M.; et al. Penetrance and prognosis of MYH7 variant-associated cardiomyopathies: results from a Dutch multicenter cohort study. Heart Failure 2024, 12, 134–147. [Google Scholar]
- Hutt, E. and M.Y. Desai. Medical Treatment Strategies for Hypertrophic Cardiomyopathy. The American Journal of Cardiology 2024, 212, S33–S41. [Google Scholar] [CrossRef]
- Eda, Y.; et al. Non-dilated left ventricular cardiomyopathy vs. dilated cardiomyopathy: clinical background and outcomes. ESC Heart Failure. 2024. [Google Scholar]
- Eldemire, R.; L. Mestroni, and M.R. Taylor. Genetics of Dilated Cardiomyopathy. Annual review of medicine 2024, 75, 417–426. [Google Scholar] [CrossRef] [PubMed]
- Voinescu, O.R.; et al. Genetic Characterization of Dilated Cardiomyopathy in Romanian Adult Patients. International Journal of Molecular Sciences 2024, 25, 2562. [Google Scholar] [CrossRef] [PubMed]
- Voinescu, O.R.; et al. Genotype-Phenotype Insights of Inherited Cardiomyopathies—A Review. Medicina 2024, 60, 543. [Google Scholar] [CrossRef]
- Irene, B.; et al. Dilated cardiomyopathy due to a novel combination of TTN and BAG3 genetic variants: from acute heart failure to subclinical phenotypes. Cardiovascular Pathology 2024, 107675. [Google Scholar]
- León, P.; et al. TTN novel splice variant in familial dilated cardiomyopathy and splice variants review: a case report. Frontiers in Cardiovascular Medicine 2024, 11, 1387063. [Google Scholar] [CrossRef]
- Mariani, M.V.; et al. Inherited arrhythmias in the pediatric population: an updated overview. Medicina 2024, 60, 94. [Google Scholar] [CrossRef] [PubMed]
- Blich, M.; et al. The role of early cardiac resynchronization therapy implantation in dilated cardiomyopathy patients with narrow QRS carrying lamin A/C mutation. American Journal of Cardiovascular Disease 2024, 14, 47. [Google Scholar] [CrossRef]
- Arnautu, D.-A.; et al. Risk Assessment and Personalized Treatment Options in Inherited Dilated Cardiomyopathies: A Narrative Review. Biomedicines 2024, 12, 1643. [Google Scholar] [CrossRef]
- Malinow, I.; et al. Pediatric dilated cardiomyopathy: a review of current clinical approaches and pathogenesis. Frontiers in Pediatrics 2024, 12, 1404942. [Google Scholar] [CrossRef] [PubMed]
- Al-Aidarous, S.; et al. Management of arrhythmogenic right ventricular cardiomyopathy. Heart 2024, 110, 156–162. [Google Scholar] [CrossRef] [PubMed]
- La Vecchia, G.; et al. Redefining Genotype-phenotype Correlation Among Patients with Arrhythmogenic Cardiomyopathy: A Cardiovascular Magnetic Resonance Cohort Study. Journal of Cardiovascular Magnetic Resonance 2024, 26. [Google Scholar] [CrossRef]
- Phan, D.P.; et al. Detection of gene mutation in the prognosis of a patient with arrhythmogenic right ventricular cardiomyopathy: a case report. Journal of Medical Case Reports 2024, 18, 49. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.D.; et al. Desmoplakin cardiomyopathy, a fibrotic and inflammatory form of cardiomyopathy distinct from typical dilated or arrhythmogenic right ventricular cardiomyopathy. Circulation 2020, 141, 1872–1884. [Google Scholar] [CrossRef]
- Cianci, V.; et al. Arrhythmogenic Right Ventricular Cardiomyopathy Post-Mortem Assessment: A Systematic Review. International Journal of Molecular Sciences 2024, 25, 2467. [Google Scholar] [CrossRef] [PubMed]
- Hespe, S.; et al. The role of genetic testing in management and prognosis of individuals with inherited cardiomyopathies. Trends in Cardiovascular Medicine 2024. [CrossRef] [PubMed]
- Moisa, S.M.; et al. Arrhythmogenic Right Ventricular Cardiomyopathy in Children: A Systematic Review. Diagnostics 2024, 14, 175. [Google Scholar] [CrossRef] [PubMed]
- Cimiotti, D.; et al. Genetic Restrictive Cardiomyopathy: Causes and Consequences—An Integrative Approach. International Journal of Molecular Sciences 2021, 22, 558. [Google Scholar] [CrossRef] [PubMed]
- Rapezzi, C.; et al. Restrictive cardiomyopathy: definition and diagnosis. European heart journal 2022, 43, 4679–4693. [Google Scholar] [CrossRef]
- Yang, Z.; et al. Genotype-phenotype associations with restrictive cardiomyopathy induced by pathogenic genetic mutations. Reviews in Cardiovascular Medicine 2022, 23, 185. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, U.; et al. Homozygous TNNI3 mutations and severe early onset dilated cardiomyopathy: patient report and review of the literature. Genes 2023, 14, 748. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; et al. Challenging of ECMO application in pediatric restrictive cardiomyopathy: case report of a novel TNNI3 variant. Frontiers in Cardiovascular Medicine 2024, 11, 1365209. [Google Scholar] [CrossRef] [PubMed]
- Catrina, B.I.; et al. A Family with Myh7 Mutation and Different Forms of Cardiomyopathies. Biomedicines 2023, 11, 2065. [Google Scholar] [CrossRef]
- Yogasundaram, H.; et al. Cardiomyopathies and genetic testing in heart failure: role in defining phenotype-targeted approaches and management. Canadian Journal of Cardiology 2021, 37, 547–559. [Google Scholar] [CrossRef]
- Ueno, M.; et al. A case report: Twin sisters with restrictive cardiomyopathy associated with rare mutations in the cardiac troponin I gene. Journal of Cardiology Cases 2021, 23, 154–157. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.-L.; et al. Pediatric restrictive cardiomyopathy: a case report. Journal of International Medical Research 2023, 51, 03000605231188276. [Google Scholar] [CrossRef] [PubMed]
- Vepsäläinen, T.; et al. MYH7 genotype–phenotype correlation in a cohort of Finnish patients. Cardiogenetics 2022, 12, 122–132. [Google Scholar] [CrossRef]
- Kian, W.; et al. Cardiomyopathy Etiologies, Symptoms and Management, in Cardiomyopathy-Disease of the Heart Muscle. 2021, IntechOpen.
- Akhtar, M.M.; et al. Clinical phenotypes and prognosis of dilated cardiomyopathy caused by truncating variants in the TTN gene. Circulation: Heart Failure 2020, 13, e006832. [Google Scholar] [CrossRef] [PubMed]
- Spadotto, A.; et al. The challenges of diagnosis and treatment of arrhythmogenic cardiomyopathy: are we there yet? Reviews in Cardiovascular Medicine 2022, 23, 283. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.-S.; B.A. Maron, and J. Loscalzo. Multiomics network medicine approaches to precision medicine and therapeutics in cardiovascular diseases. Arteriosclerosis, thrombosis, and vascular biology 2023, 43, 493–503. [Google Scholar] [CrossRef] [PubMed]
- Doran, S.; et al. Multi-omics approaches for revealing the complexity of cardiovascular disease. Briefings in bioinformatics 2021, 22, bbab061. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-J.; et al. Clinical application of whole exome sequencing to identify rare but remediable neurologic disorders. Journal of Clinical Medicine 2020, 9, 3724. [Google Scholar] [CrossRef] [PubMed]
- Brlek, P.; et al. implementing whole genome sequencing (WGS) in clinical practice: advantages, challenges, and future perspectives. Cells 2024, 13, 504. [Google Scholar] [CrossRef] [PubMed]
- Wehrens, M.; et al. Single-cell transcriptomics provides insights into hypertrophic cardiomyopathy. Cell reports 2022, 39(6).
- Simonson, B.; et al. Single-nucleus RNA sequencing in ischemic cardiomyopathy reveals common transcriptional profile underlying end-stage heart failure. Cell reports 2023, 42(2).
- Li, C.-x., et al. Whole-transcriptome RNA sequencing reveals significant differentially expressed mRNAs, miRNAs, and lncRNAs and related regulating biological pathways in the peripheral blood of COVID-19 patients. Mediators of inflammation 2021, 2021, 6635925. [Google Scholar]
- Chen, S.; et al. Transcriptome analysis of human hypertrophic cardiomyopathy reveals inhibited cardiac development pathways in children. Iscience 2024, 27(1).
- Previs, M.J.; et al. Defects in the proteome and metabolome in human hypertrophic cardiomyopathy. Circulation: Heart Failure 2022, 15, e009521. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; et al. RNF207 exacerbates pathological cardiac hypertrophy via post-translational modification of TAB1. Cardiovascular Research 2023, 119, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Shimada, Y.J.; et al. Comprehensive proteomics profiling reveals circulating biomarkers of hypertrophic cardiomyopathy. Circulation: Heart Failure 2021, 14, e007849. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; et al. Integrative proteomic analysis reveals the cytoskeleton regulation and mitophagy difference between ischemic cardiomyopathy and dilated cardiomyopathy. Molecular & Cellular Proteomics 2023, 22. [Google Scholar]
- Gonzalez-Covarrubias, V.; E. Martínez-Martínez, and L. del Bosque-Plata. The potential of metabolomics in biomedical applications. Metabolites 2022, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Lopaschuk, G.D.; et al. Cardiac energy metabolism in heart failure. Circulation research 2021, 128, 1487–1513. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; et al. Multiomics Analysis Reveals Extensive Remodeling of the Extracellular Matrix and Cellular Metabolism Due to Plakophilin-2 Knockdown in Guinea Pigs. bioRxiv, 2024, 2024.03. 11.584401.
- Kawaguchi, N. and T. Nakanishi. Animal Disease Models and Patient-iPS-Cell-Derived In Vitro Disease Models for Cardiovascular Biology—How Close to Disease? Biology 2023, 12, 468. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; et al. Engineering considerations of iPSC-based personalized medicine. Biomaterials Research 2023, 27, 67. [Google Scholar] [CrossRef] [PubMed]
- Dababneh, S.; et al. Advances in Hypertrophic Cardiomyopathy Disease Modeling using Human iPSC-derived Cardiomyocytes. Canadian Journal of Cardiology 2023.
- Blackwell, D.J. ; J. Schmeckpeper, and B.C. Knollmann, Animal models to study cardiac arrhythmias. Circulation research 2022, 130, 1926–1964. [Google Scholar]
- Niu, Y.; et al. Using Zebrafish Animal Model to Study the Genetic Underpinning and Mechanism of Arrhythmogenic Cardiomyopathy. International journal of molecular sciences 2023, 24, 4106. [Google Scholar] [CrossRef] [PubMed]
- Inoue, S.; et al. Association Between Clonal Hematopoiesis and Left Ventricular Reverse Remodeling in Nonischemic Dilated Cardiomyopathy. JACC: Basic to Translational Science 2024,.
- Kasai-Brunswick, T.H.; A.B. Carvalho, and A.C.C. de Carvalho. Stem cell therapies in cardiac diseases: current status and future possibilities. World Journal of Stem Cells 2021, 13, 1231. [Google Scholar] [CrossRef] [PubMed]
- Neef, K.; et al. Co-transplantation of mesenchymal stromal cells and induced pluripotent stem cell-derived cardiomyocytes improves cardiac function after myocardial damage. Frontiers in Cardiovascular Medicine 2022, 8, 794690. [Google Scholar] [CrossRef]
- Fujita, J.; et al. Clinical Application of iPSC-Derived Cardiomyocytes in Patients with Advanced Heart Failure, in Advanced Technologies in Cardiovascular Bioengineering. 2022, Springer. p. 361-374.
- Yoshida, S.; et al. Syngeneic mesenchymal stem cells reduce immune rejection after induced pluripotent stem cell-derived allogeneic cardiomyocyte transplantation. Scientific reports 2020, 10, 4593. [Google Scholar] [CrossRef] [PubMed]
- Vo, Q.D.; et al. Induced pluripotent stem cell-derived cardiomyocytes therapy for ischemic heart disease in animal model: A meta-analysis. International journal of molecular sciences 2024, 25, 987. [Google Scholar] [CrossRef] [PubMed]
- Müller, L.; et al. Immunomodulatory properties of mesenchymal stromal cells: an update. Frontiers in cell and developmental biology 2021, 9, 637725. [Google Scholar] [CrossRef] [PubMed]
- Yuce, K. ; The Application of Mesenchymal Stem Cells in Different Cardiovascular Disorders: Ways of Administration, and the Effectors. Stem Cell Reviews and Reports, 2024, 1-21.
- da Silva, J.S.; et al. Mesenchymal stem cell therapy in diabetic cardiomyopathy. Cells 2022, 11, 240. [Google Scholar] [CrossRef] [PubMed]
- Kou, M.; et al. Mesenchymal stem cell-derived extracellular vesicles for immunomodulation and regeneration: a next generation therapeutic tool? Cell Death & Disease 2022, 13, 580. [Google Scholar]
- de Jong, B.; et al. Recent advances in extracellular vesicles as drug delivery systems and their potential in precision medicine. Pharmaceutics 2020, 12, 1006. [Google Scholar] [CrossRef]
- Simeone, P.; et al. Extracellular vesicles as signaling mediators and disease biomarkers across biological barriers. International journal of molecular sciences 2020, 21, 2514. [Google Scholar] [CrossRef] [PubMed]
- Roacho-Pérez, J.A.; et al. Artificial scaffolds in cardiac tissue engineering. Life 2022, 12, 1117. [Google Scholar] [CrossRef] [PubMed]
- Jafari, A.; et al. Latest advances in 3D bioprinting of cardiac tissues. Advanced materials technologies 2022, 7, 2101636. [Google Scholar] [CrossRef]
- Wang, Z.; et al. 3D bioprinting in cardiac tissue engineering. Theranostics 2021, 11, 7948. [Google Scholar] [CrossRef] [PubMed]
- Silver, S.E.; R.W. Barrs, and Y. Mei. Transplantation of human pluripotent stem cell-derived cardiomyocytes for cardiac regenerative therapy. Frontiers in Cardiovascular Medicine 2021, 8, 707890. [Google Scholar] [CrossRef] [PubMed]
- Margiana, R.; et al. Clinical application of mesenchymal stem cell in regenerative medicine: a narrative review. Stem Cell Research & Therapy 2022, 13, 366. [Google Scholar]
- Häneke, T. and M. Sahara. Progress in bioengineering strategies for heart regenerative medicine. International Journal of Molecular Sciences 2022, 23, 3482. [Google Scholar]
- Mazzola, M. and E. Di Pasquale. Toward cardiac regeneration: combination of pluripotent stem cell-based therapies and bioengineering strategies. Frontiers in Bioengineering and Biotechnology 2020, 8, 455. [Google Scholar]
- Hassan, M.; et al. Innovations in genomics and big data analytics for personalized medicine and health care: A review. International journal of molecular Sciences 2022, 23, 4645. [Google Scholar] [CrossRef] [PubMed]
- Crotti, L.; et al. From gene-discovery to gene-tailored clinical management: 25 years of research in channelopathies and cardiomyopathies. Europace 2023, 25, euad180. [Google Scholar] [CrossRef] [PubMed]
- Elliott, P.M. ; Personalized medicine for dilated cardiomyopathy. European Heart Journal 2021, 42, 175–177. [Google Scholar] [CrossRef] [PubMed]
- Chaput, D. and G. Andelfinger, Small molecule inhibition for RASopathy-associated hypertrophic cardiomyopathy: Clinical application of a basic concept. Canadian Journal of Cardiology. 2024. [Google Scholar]
- Farrokhi, M.; et al. Role of precision medicine and personalized medicine in the treatment of diseases. Kindle 2023, 3, 1–164. [Google Scholar]
- Paldino, A.; et al. Prognostic prediction of genotype vs phenotype in genetic cardiomyopathies. Journal of the American College of Cardiology 2022, 80, 1981–1994. [Google Scholar] [CrossRef]
- Njoroge, J.N.; et al. Emerging Genotype–Phenotype associations in dilated cardiomyopathy. Current Cardiology Reports 2022, 24, 1077–1084. [Google Scholar] [CrossRef] [PubMed]
- Ahluwalia, M. and C.Y. Ho. Cardiovascular genetics: the role of genetic testing in diagnosis and management of patients with hypertrophic cardiomyopathy. Heart 2021, 107, 183–189. [Google Scholar] [CrossRef]
- Magrinelli, F.; B. Balint, and K.P. Bhatia. Challenges in clinicogenetic correlations: one gene–many phenotypes. Movement Disorders Clinical Practice 2021, 8, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Balogun, O.D.; et al. The role of pharmacists in personalised medicine: a review of integrating pharmacogenomics into clinical practice. International Medical Science Research Journal 2024, 4, 19–36. [Google Scholar] [CrossRef]
- Kingdom, R. and C.F. Wright. Incomplete Penetrance and Variable Expressivity: From Clinical Studies to Population Cohorts. Front Genet 2022, 13, 920390. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Z.; et al. Precision Cardio-oncology: Update on Omics-Based Diagnostic Methods. Current Treatment Options in Oncology, 2024, 1-23.
- Tiwari, V.; et al. The structure and function of lamin A/C: Special focus on cardiomyopathy and therapeutic interventions. Life Sciences, 2024, 122489.
- Maggi, J.; et al. Limited Added Diagnostic Value of Whole Genome Sequencing in Genetic Testing of Inherited Retinal Diseases in a Swiss Patient Cohort. International Journal of Molecular Sciences 2024, 25, 6540. [Google Scholar] [CrossRef]
- Kaithal, P. ; S. Kanchan, and M. Kesheri, Recent Advances in Biological Omics Databases and Tools in Human Health. Microbial Omics in Environment and Health, 2024, 311-341.
- Palmer, J.A.; et al. Revisiting Cardiac Biology in the Era of Single Cell and Spatial Omics. Circulation Research 2024, 134, 1681–1702. [Google Scholar] [CrossRef] [PubMed]
- Kiessling, P. and C. Kuppe. Spatial multi-omics: novel tools to study the complexity of cardiovascular diseases. Genome Medicine 2024, 16, 14. [Google Scholar] [CrossRef]
- Mourad, O.; et al. Modeling heart diseases on a chip: advantages and future opportunities. Circulation Research 2023, 132, 483–497. [Google Scholar] [CrossRef] [PubMed]
- Liu, J. and Y. Wang. Advances in organ-on-a-chip for the treatment of cardiovascular diseases. MedComm–Biomaterials and Applications 2023, 2, e63. [Google Scholar]
- Mistrulli, R.; et al. Cardiomyopathy and Sudden Cardiac Death: Bridging Clinical Practice with Cutting-Edge Research. Biomedicines 2024, 12, 1602. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




