Submitted:
08 August 2024
Posted:
09 August 2024
You are already at the latest version
Abstract
Keywords:
Introduction
Methods
arsR Plasmid Construction
Culturing Conditions
Protein Extraction
Shotgun Proteomics
Curation of Metalloproteome from Identified Proteins
Statistical Analysis of the Metalloproteome
Total Metal Analysis of Soluble Fractions
Results
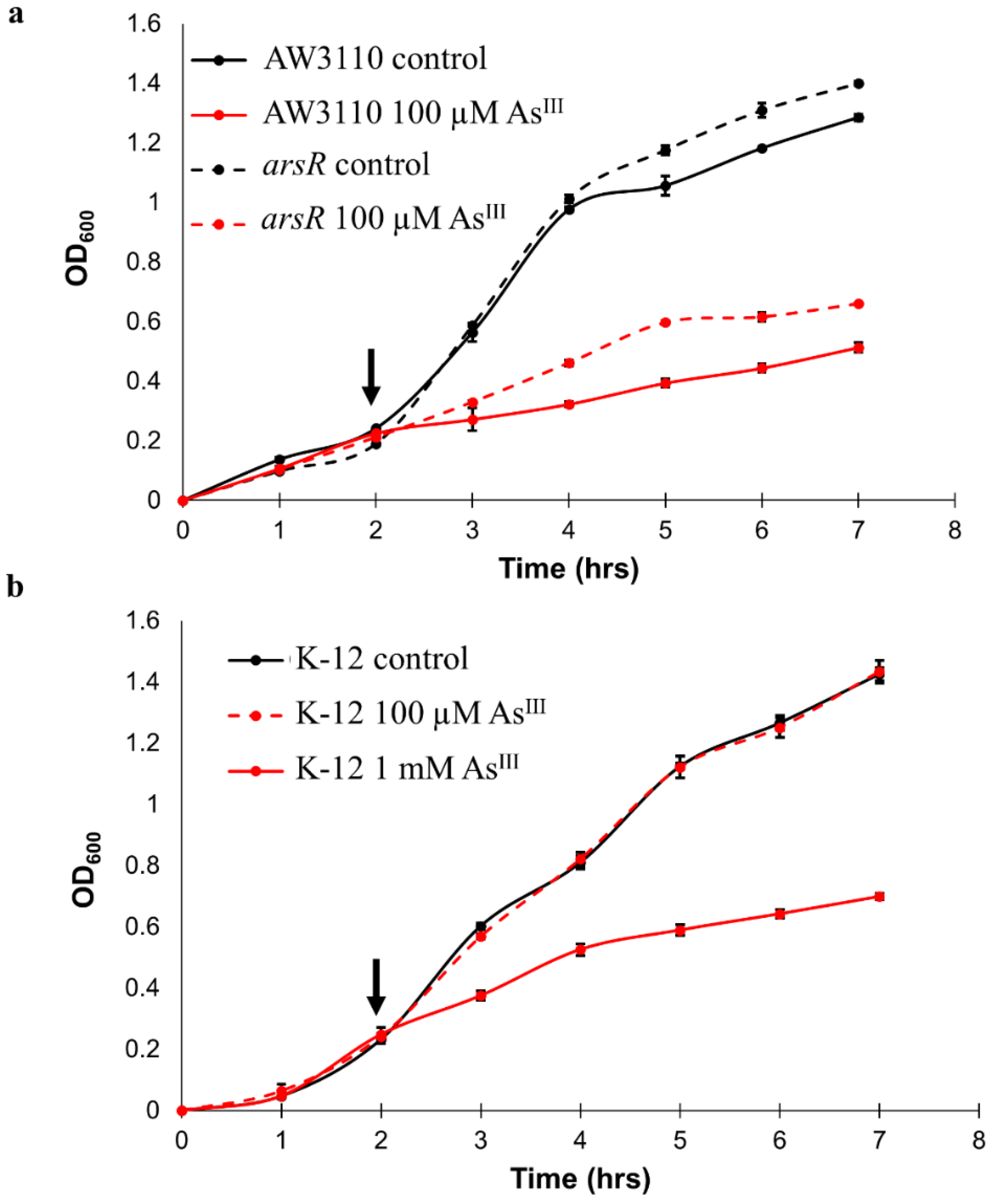
arsR Confers Arsenic Resistance
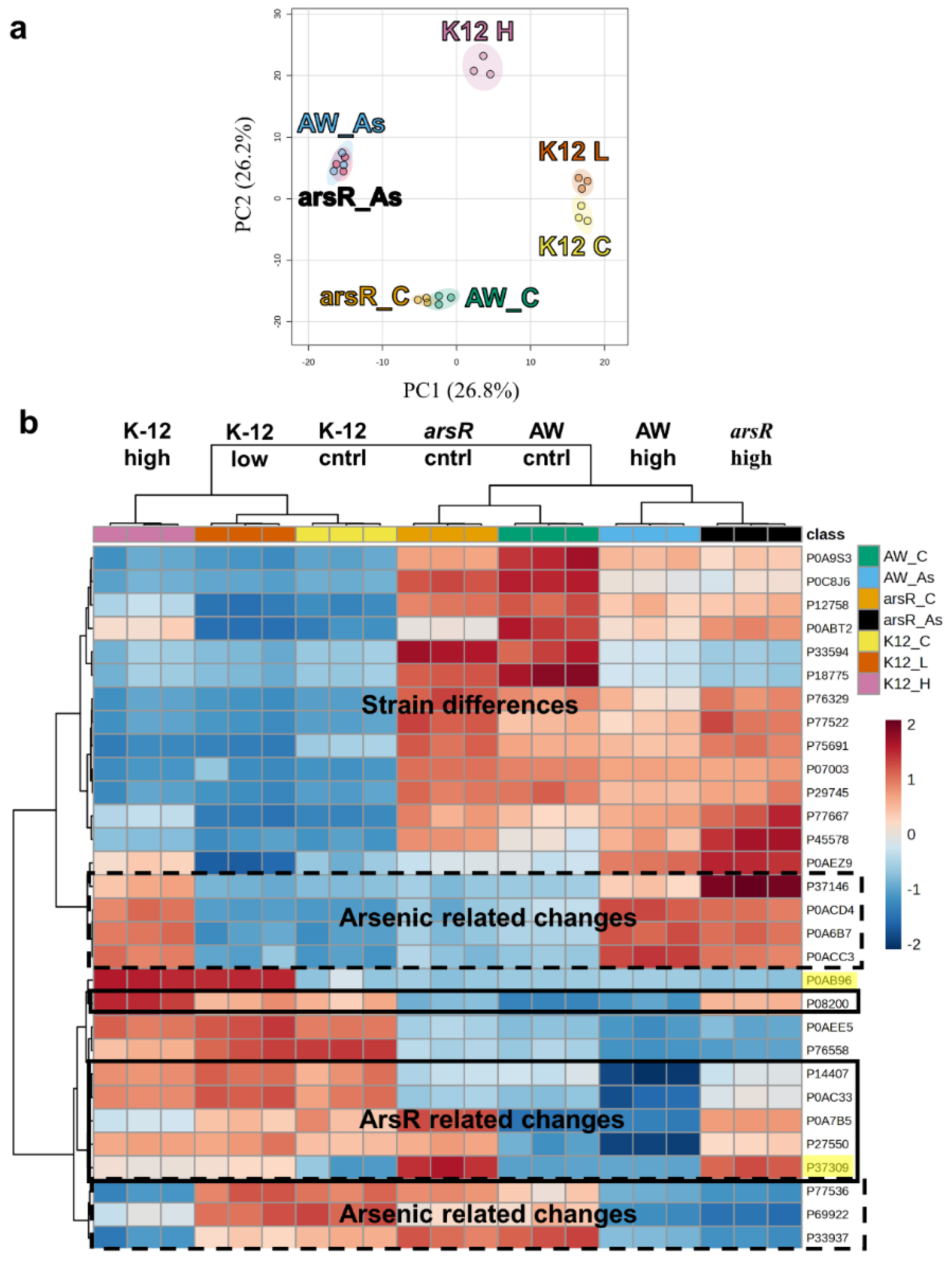
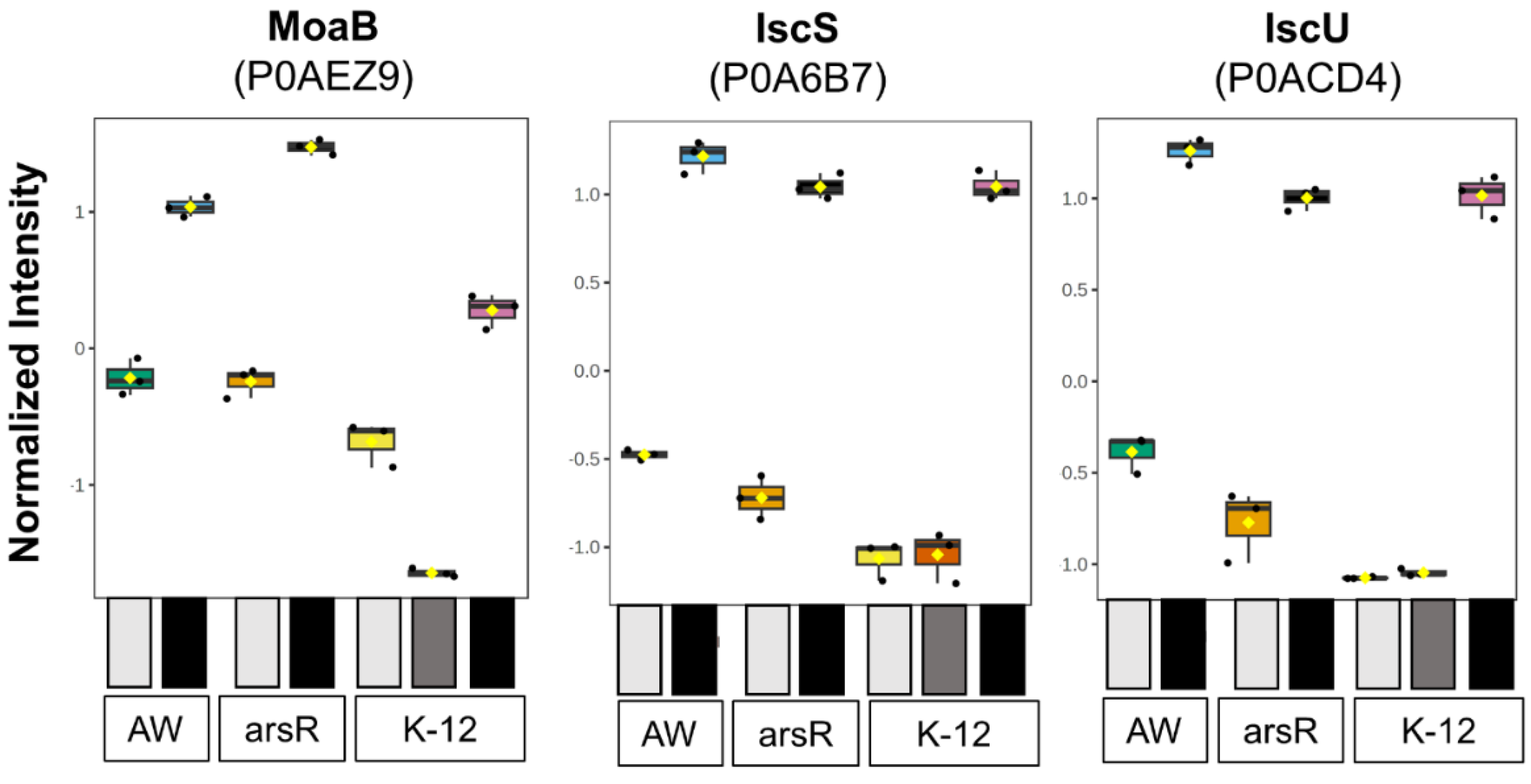
Changes to the Metalloproteome during AsIII stress
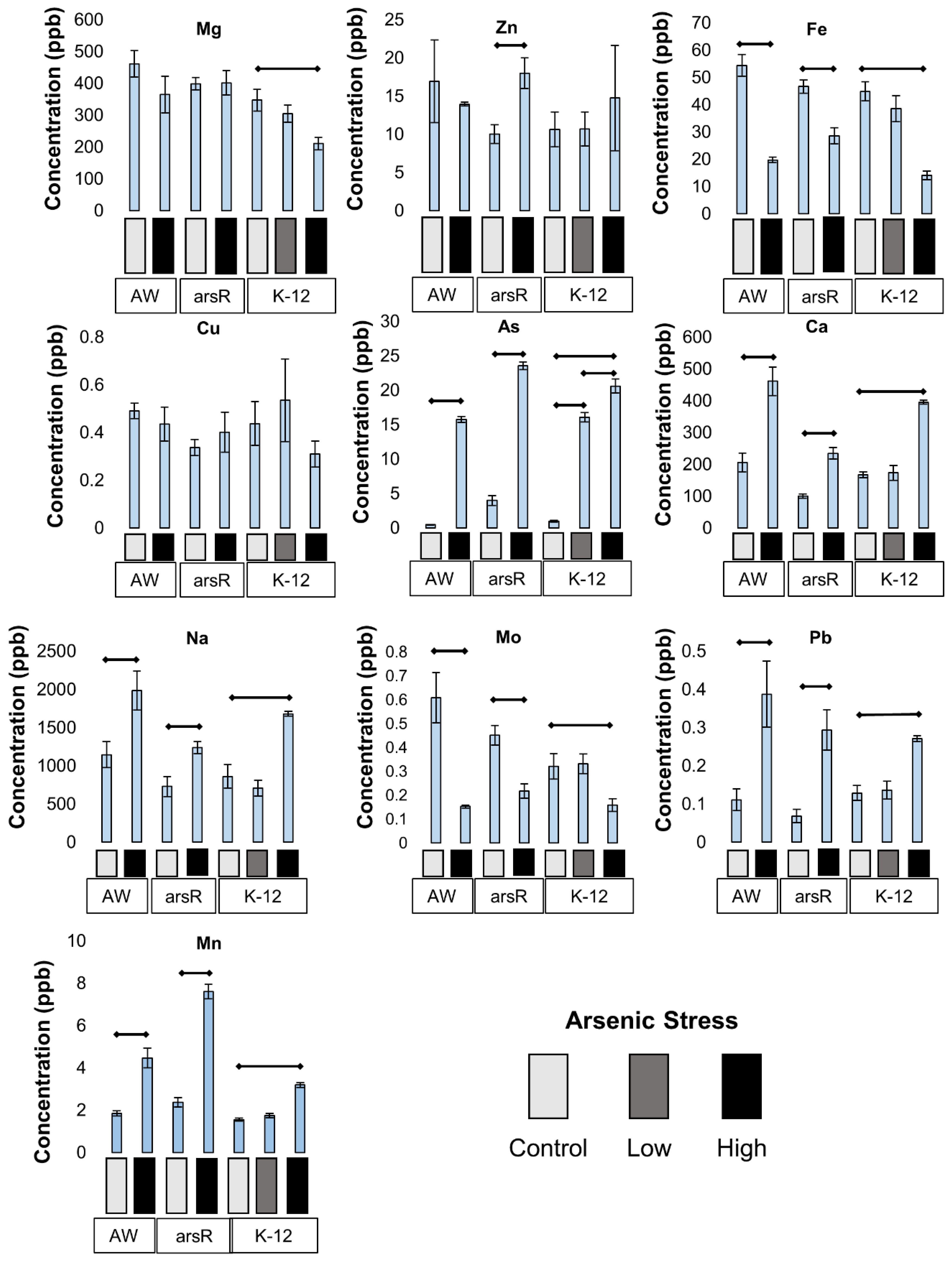
Changes to Intracellular Metals
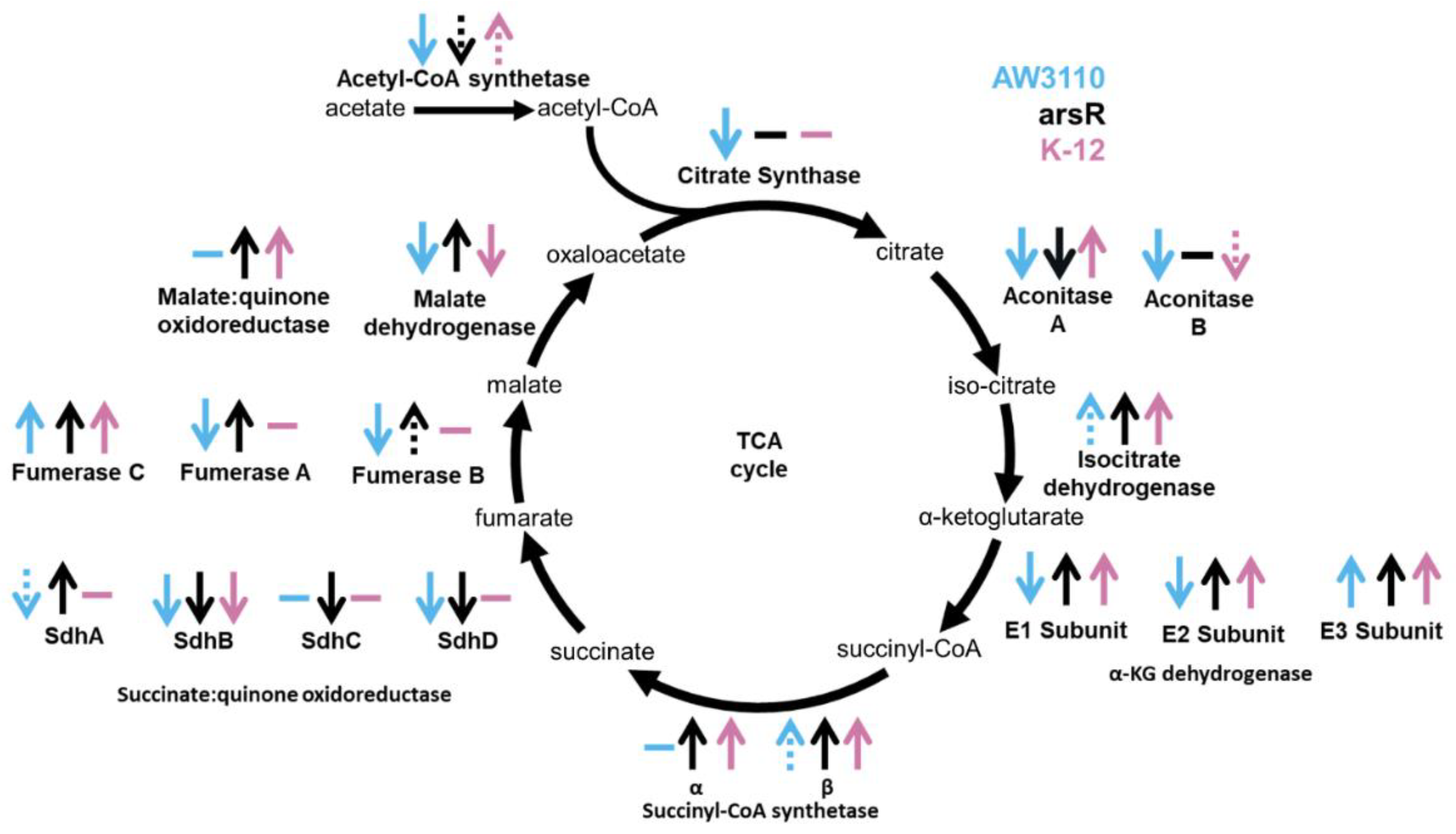
Discussion
Conclusions
Supplementary Materials
Acknowledgments
References
- Ng, J. C.; Wang, J.; Shraim, A. A Global Health Problem Caused by Arsenic from Natural Sources. Chemosphere 2003, 52, 1353–1359. [Google Scholar] [CrossRef] [PubMed]
- Kuivenhoven, M.; Mason, K. Arsenic Toxicity. StatPearls 2023. [Google Scholar]
- Styblo, M.; Del Razo, L. M.; Vega, L.; Germolec, D. R.; LeCluyse, E. L.; Hamilton, G. A.; Reed, W.; Wang, C.; Cullen, W. R.; Thomas, D. J. Comparative Toxicity of Trivalent and Pentavalent Inorganic and Methylated Arsenicals in Rat and Human Cells. Arch Toxicol 2000, 74, 289–299. [Google Scholar] [CrossRef] [PubMed]
- WHO. Guidelines for Drinking-Water Quality, 4th Edition, Incorporating the 1st Addendum. 2017.
- De Francisco, P.; Martín-González, A.; Rodriguez-Martín, D.; Díaz, S. Interactions with Arsenic: Mechanisms of Toxicity and Cellular Resistance in Eukaryotic Microorganisms. Int J Environ Res Public Health 2021, 18, 12226. [Google Scholar] [CrossRef] [PubMed]
- Rosen, B. P. Biochemistry of Arsenic Detoxification. FEBS Lett 2002, 529, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Páez-Espino, D.; Tamames, J.; De Lorenzo, V.; Cánovas, D. Microbial Responses to Environmental Arsenic. BioMetals 2009, 22, 117–130. [Google Scholar] [CrossRef]
- Garbinski, L. D.; Rosen, B. P.; Chen, J. Pathways of Arsenic Uptake and Efflux. Environ Int 2019, 126, 585–597. [Google Scholar] [CrossRef] [PubMed]
- Fekih, I. Ben; Zhang, C.; Li, Y. P.; Zhao, Y.; Alwathnani, H. A.; Saquib, Q.; Rensing, C.; Cervantes, C. Distribution of Arsenic Resistance Genes in Prokaryotes. Front Microbiol 2018, 9, 407801. [Google Scholar] [CrossRef]
- Saltikov, C. W.; Olson, B. H. Homology of Escherichia Coli R773 ArsA, ArsB, and ArsC Genes in Arsenic-Resistant Bacteria Isolated from Raw Sewage and Arsenic-Enriched Creek Waters. Appl Environ Microbiol 2002, 68, 280. [Google Scholar] [CrossRef]
- Carlin, A.; Shi, W.; Dey, S.; Rosen, B. P. The Ars Operon of Escherichia Coli Confers Arsenical and Antimonial Resistance. J Bacteriol 1995, 177, 981. [Google Scholar] [CrossRef]
- Rawle, R. A.; Kang, Y. S.; Bothner, B.; Wang, G.; McDermott, T. R. Transcriptomics Analysis Defines Global Cellular Response of Agrobacterium Tumefaciens 5A to Arsenite Exposure Regulated through the Histidine Kinases PhoR and AioS. Environ Microbiol 2019, 21, 2659–2676. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, D. R.; Botero, L. M.; Franck, W. L.; Hassett, D. J.; McDermott, T. R. Complex Regulation of Arsenite Oxidation in Agrobacterium Tumefaciens. J Bacteriol 2006, 188, 1081–1088. [Google Scholar] [CrossRef]
- Rawle, R.; Saley, T. C.; Kang, Y. S.; Wang, Q.; Walk, S.; Bothner, B.; McDermott, T. R. Introducing the ArsR-Regulated Arsenic Stimulon. Front Microbiol 2021, 12, 162. [Google Scholar] [CrossRef] [PubMed]
- Larson, J.; Tokmina-Lukaszewska, M.; Fausset, H.; Spurzem, S.; Cox, S.; Cooper, G.; Copié, V.; Bothner, B. Arsenic Exposure Causes Global Changes in the Metalloproteome of Escherichia Coli. Microorganisms 2023, 11, 382. [Google Scholar] [CrossRef] [PubMed]
- Nesvizhskii, A. I.; Keller, A.; Kolker, E.; Aebersold, R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal Chem 2003, 75, 4646–4658. [Google Scholar] [CrossRef] [PubMed]
- Huang, D. W.; Sherman, B. T.; Lempicki, R. A. Bioinformatics Enrichment Tools: Paths toward the Comprehensive Functional Analysis of Large Gene Lists. Nucleic Acids Res 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Huang, D. W.; Sherman, B. T.; Lempicki, R. A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat Protoc 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC-HRMS Spectra Processing, Multi-Omics Integration and Covariate Adjustment of Global Metabolomics Data. [CrossRef]
- Keseler, I. M.; Gama-Castro, S.; Mackie, A.; Billington, R.; Bonavides-Martínez, C.; Caspi, R.; Kothari, A.; Krummenacker, M.; Midford, P. E.; Muñiz-Rascado, L.; Ong, W. K.; Paley, S.; Santos-Zavaleta, A.; Subhraveti, P.; Tierrafría, V. H.; Wolfe, A. J.; Collado-Vides, J.; Paulsen, I. T.; Karp, P. D. The EcoCyc Database in 2021. Front Microbiol 2021, 12, 711077. [Google Scholar] [CrossRef] [PubMed]
- Karp, P. D.; Ouzounis, C. A.; Moore-Kochlacs, C.; Goldovsky, L.; Kaipa, P.; Ahrén, D.; Tsoka, S.; Darzentas, N.; Kunin, V.; López-Bigas, N. Expansion of the BioCyc Collection of Pathway/Genome Databases to 160 Genomes. Nucleic Acids Res 2005, 33, 6083–6089. [Google Scholar] [CrossRef]
- Xu, Z.; Wang, P.; Wang, H.; HangYu, Z.; Au-Yeung, H. Y.; Hirayama, T.; Sun, H.; Yan, A. Zinc Excess Increases Cellular Demand for Iron and Decreases Tolerance to Copper in Escherichia Coli. Journal of Biological Chemistry 2019, 294, 16978–16991. [Google Scholar] [CrossRef]
- Riley, M.; Abe, T.; Arnaud, M. B.; Berlyn, M. K. B.; Blattner, F. R.; Chaudhuri, R. R.; Glasner, J. D.; Horiuchi, T.; Keseler, I. M.; Kosuge, T.; Mori, H.; Perna, N. T.; Plunkett, G.; Rudd, K. E.; Serres, M. H.; Thomas, G. H.; Thomson, N. R.; Wishart, D.; Wanner, B. L. Escherichia Coli K-12: A Cooperatively Developed Annotation Snapshot—2005. Nucleic Acids Res 2006, 34, 1–9. [Google Scholar] [CrossRef]
- Hayashi, K.; Morooka, N.; Yamamoto, Y.; Fujita, K.; Isono, K.; Choi, S.; Ohtsubo, E.; Baba, T.; Wanner, B. L.; Mori, H.; Horiuchi, T. Highly Accurate Genome Sequences of Escherichia Coli K-12 Strains MG1655 and W3110. Mol Syst Biol 2006, 2. [Google Scholar] [CrossRef] [PubMed]
- Rajagopalan, K. V. The Pterin Molybdenum Cofactor. Article in Journal of Biological Chemistry 1992. [Google Scholar] [CrossRef]
- Miralles-Robledillo, J. M.; Torregrosa-Crespo, J.; Martínez-Espinosa, R. M.; Pire, C. DMSO Reductase Family: Phylogenetics and Applications of Extremophiles. Int J Mol Sci 2019, 20. [Google Scholar] [CrossRef] [PubMed]
- Lénon, M.; Arias-Cartín, R.; Barras, F. The Fe–S Proteome of Escherichia Coli: Prediction, Function, and Fate. Metallomics 2022, 14, 22. [Google Scholar] [CrossRef]
- Yokoyama, K.; Leimkühler, S. The Role of FeS Clusters for Molybdenum Cofactor Biosynthesis and Molybdoenzymes in Bacteria. Biochim Biophys Acta 2015, 1853, 1335. [Google Scholar] [CrossRef]
- Outten, F. W.; Djaman, O.; Storz, G. A Suf Operon Requirement for Fe–S Cluster Assembly during Iron Starvation in Escherichia Coli. Mol Microbiol 2004, 52, 861–872. [Google Scholar] [CrossRef]
- Lee, -H; Yeo, W. -S.; Roe, J.-H.; Lee, J.-H.; Yeo, W.-S.; Roe, J.-H. Induction of the SufA Operon Encoding Fe-S Assembly Proteins by Superoxide Generators and Hydrogen Peroxide: Involvement of OxyR, IHF and an Unidentified Oxidant-Responsive Factor. Mol Microbiol 2004, 51, 1745–1755. [Google Scholar] [CrossRef] [PubMed]
- Boyd, E. S.; Thomas, K. M.; Dai, Y.; Boyd, J. M.; Outten, F. W. Interplay between Oxygen and Fe–S Cluster Biogenesis: Insights from the Suf Pathway. Biochemistry 2014, 53, 5834. [Google Scholar] [CrossRef] [PubMed]
- Flora, S. J. S. Arsenic-Induced Oxidative Stress and Its Reversibility. Free Radic Biol Med 2011, 51, 257–281. [Google Scholar] [CrossRef]
- Kitchin, K. T.; Ahmad, S. Oxidative Stress as a Possible Mode of Action for Arsenic Carcinogenesis. Toxicol Lett 2003, 137, (1–2). [Google Scholar] [CrossRef] [PubMed]
- Gupta, D. K.; Inouhe, M.; Rodríguez-Serrano, M.; Romero-Puertas, M. C.; Sandalio, L. M. Oxidative Stress and Arsenic Toxicity: Role of NADPH Oxidases. Chemosphere 2013, 90, 1987–1996. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Cai, J. F.; Chiu, J. F. Arsenic Induces Oxidative Stress and Activates Stress Gene Expressions in Cultured Lung Epithelial Cells. J Cell Biochem 2002, 87, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Li, X. F.; Cullen, W. R.; Weinfeld, M.; Le, X. C. Arsenic Binding to Proteins. Chem Rev 2013, 113, 7769. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Wang, H.; Wang, Z.; Li, X. F.; Le, X. C. Identification of Reactive Cysteines in a Protein Using Arsenic Labeling and Collision-Induced Dissociation Tandem Mass Spectrometry. J Proteome Res 2008, 7, 3080–3090. [Google Scholar] [CrossRef] [PubMed]
- Cline, D. J.; Thorpe, C.; Schneider, J. P. Effects of As(III) Binding on α-Helical Structure. J Am Chem Soc 2003, 125, 2923–2929. [Google Scholar] [CrossRef] [PubMed]
- Esquilin-Lebron, K.; Dubrac, S.; Barras, F.; Boyd, J. M. Bacterial Approaches for Assembling Iron-Sulfur Proteins. mBio 2021, 12. [Google Scholar] [CrossRef]
- McHugh, J. P.; Rodríguez-Quiñones, F.; Abdul-Tehrani, H.; Svistunenko, D. A.; Poole, R. K.; Cooper, C. E.; Andrews, S. C. Global Iron-Dependent Gene Regulation in Escherichia Coli: A NEW MECHANISM FOR IRON HOMEOSTASIS. Journal of Biological Chemistry 2003, 278, 29478–29486. [Google Scholar] [CrossRef]
- Semsey, S.; Andersson, A. M. C.; Krishna, S.; Jensen, M. H.; Massé, E.; Sneppen, K. Genetic Regulation of Fluxes: Iron Homeostasis of Escherichia Coli. Nucleic Acids Res 2006, 34, 4960. [Google Scholar] [CrossRef]
- Lee, K.-C.; Yeo, W.-S.; Roe, J.-H. Oxidant-Responsive Induction of the Suf Operon, Encoding a Fe-S Assembly System, through Fur and IscR in Escherichia Coli. J Bacteriol 2008, 190, 8244–8247. [Google Scholar] [CrossRef]
- Lee, C.; Lee, S. M.; Mukhopadhyay, P.; Kim, S. J.; Lee, S. C.; Ahn, W. S.; Yu, M. H.; Storz, G.; Ryu, S. E. Redox Regulation of OxyR Requires Specific Disulfide Bond Formation Involving a Rapid Kinetic Reaction Path. Nat Struct Mol Biol 2004, 11, 1179–1185. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y. S.; Brame, K.; Jetter, J.; Bothner, B. B.; Wang, G.; Thiyagarajan, S.; McDermott, T. R. Regulatory Activities of Four ArsR Proteins in Agrobacterium Tumefaciens 5A. Appl Environ Microbiol 2016, 82, 3471–3480. [Google Scholar] [CrossRef] [PubMed]
- Ke, C.; Zhao, C.; Rensing, C.; Yang, S.; Zhang, Y. Characterization of Recombinant E. Coli Expressing ArsR from Rhodopseudomonas Palustris CGA009 That Displays Highly Selective Arsenic Adsorption. Appl Microbiol Biotechnol 2018, 102, 6247–6255. [Google Scholar] [CrossRef] [PubMed]
- Tokmina-Lukaszewska, M.; Shi, Z.; Tripet, B.; McDermott, T. R.; Copié, V.; Bothner, B.; Wang, G. Metabolic Response of Agrobacterium Tumefaciens 5A to Arsenite. Environ Microbiol 2017, 19, 710–721. [Google Scholar] [CrossRef] [PubMed]
- Jozefczuk, S.; Klie, S.; Catchpole, G.; Szymanski, J.; Cuadros-Inostroza, A.; Steinhauser, D.; Selbig, J.; Willmitzer, L. Metabolomic and Transcriptomic Stress Response of Escherichia Coli. Mol Syst Biol 2010, 6, 364. [Google Scholar] [CrossRef] [PubMed]
- MacLean, A.; Legendre, F.; Appanna, V. D. The Tricarboxylic Acid (TCA) Cycle: A Malleable Metabolic Network to Counter Cellular Stress. Crit Rev Biochem Mol Biol 2023, 58, 81–97. [Google Scholar] [CrossRef] [PubMed]
- Naujokas, M. F.; Anderson, B.; Ahsan, H.; Vasken Aposhian, H.; Graziano, J. H.; Thompson, C.; Suk, W. A. The Broad Scope of Health Effects from Chronic Arsenic Exposure: Update on a Worldwide Public Health Problem. Environ Health Perspect 2013, 121, 295. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, P.; Chatterjee, D.; Singh, K. K.; Giri, A. K. Systems Biology Approaches to Evaluate Arsenic Toxicity and Carcinogenicity: An Overview. Int J Hyg Environ Health 2013, 216, 574–586. [Google Scholar] [CrossRef]
- McDermott, T. R.; Stolz, J. F.; Oremland, R. S. Arsenic and the Gastrointestinal Tract Microbiome. Environ Microbiol Rep 2020, 12, 136–159. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




